Abstract
YfcG is one of eight glutathione (GSH) transferase homologues encoded in the Escherichia coli genome. The protein exhibits low or no GSH transferase activity toward a panel of electrophilic substrates. In contrast, it has a very robust disulfide-bond reductase activity toward 2-hydroxyethyldisulfide on par with mammalian and bacterial glutaredoxins. The structure of YfcG at 2.3 Å-resolution from crystals grown in the presence of GSH reveals a molecule of glutathione disulfide in the active site. The crystallographic results and the lack of functional cysteine residues in the active site of YfcG suggests that the reductase activity is unique in that no sulfhydryl groups in the YfcG protein are covalently involved in the redox chemistry.
The genome of Escherichia coli harbors eight genes that encode members of the glutathione (GSH) transferase superfamily (1, 2). The biochemical activities and biological functions of most of these proteins remain to be elucidated. The YfcG protein has been reported by others to have low GSH transferase and peroxidase activities and to be involved in defense against oxidative stress (3). However, it is unclear if the tepid peroxidase activity can be directly associated with resistance to oxidative stress.
The yfcG gene is unusual in that it is expressed predominantly in late stationary phase (2) here a significant fraction of GSH is found as glutathionylspermidine (GspSH) (4). This coincidence suggests that YfcG might interact with GspSH and perhaps utilize GspSH as a substrate or a regulatory molecule.
In this rapid report, we describe the three-dimensional structure of YfcG in complex with glutathione disulfide, GSSG, and provide compelling biochemical evidence that this structure represents an oxidized version of an efficient disulfide-bond oxido reductase. Furthermore, the evidence suggests that the previously reported tepid GSH transferase and organic hydroperoxidase activities of YfcG are probably promiscuous activities that are a vestige of the evolution of a disulfide-bond reductase.
The yfcG gene was cloned from E. coli K12 genomic DNA and moved into an expression vector, and the protein was expressed and purified. The protein was crystallized in the presence of GSH and the structure determined at a resolution of 2.3 Å. The dissociation constants for GSH, GspSH, and elated ligands were determined by fluorescence titration. The enzymatic activity was evaluated with a panel of electrophilic substrates. The details of these experiments are provided in Supporting Information.
Given the coincidence that the yfcG gene is expressed in late stationary phase when GspSH is produced, we measured the dissociation constant for GSH and GspSH by titration, monitoring the intrinsic fluorescence of the protein. The results are summarized in Table 1. GspSH binds about 10 times more tightly than GSH. Even more interesting is the fact that GSSG binds 100 times more tightly than GSH and 10 times more tightly than the GSH analogue, GSO3−.
Table 1.
Dissociation Constants of Thiol and Disulfide Ligands and Substrates as Determined by Fluorescence Titration at 25 °C
| compound | Kd (µM) |
|---|---|
| GSH | 330 ± 10 |
| GSO3− | 23 ± 5 |
| GspSH | 29 ± 7 |
| GspSH-disulfide | 180 ± 40 |
| GSSG | 2.4 ± 0.5 |
Activity assays of YfcG with GSH and a panel of electrophilic substrates that includes 1-chloro-2,4-dinitrobenzene (CDNB), ethacrynic acid, trans-4-phenyl-3-butene-2-one, 1,2-epoxy-3-(4-nitrophenoxy) propane, and 1,4-dichloro-2-nitrobenzene indicated that only CDNB is a substrate, albeit a very poor one, with kcat=0.1 ± 0.01 s−1 and kcat/KMGSH=29 ± 5 M−1 s−1. The enzyme has no peroxidase activity toward H2O2 and no hydroperoxidase toward t-butyl-hydroperoxide, benzyl peroxide, or lauroyl peroxide. YfcG does have a low, previously observed but not quantified (3), hydroperoxidase activity toward cumene hydroperoxide. In our hands, kcat=0.27 ± 0.03 s−1 and kcat/KMGSH = 520 ± 200 M−1 s−1. The enzyme exhibited no detectable activity toward the above substrates when GspSH was the nucleophile.
YfcG crystallized in the presence of GSH, 20% PEG 3000, and 0.1 M NaOAc (pH 4.5) in the tetragonal space group P41212 with one molecule in the asymmetric unit. The initial phases were obtained by molecular replacement, and the structure was determined at a resolution of 2.3 Å. Details of the data collection, structure determination, and refinement are provided in Supporting Information.
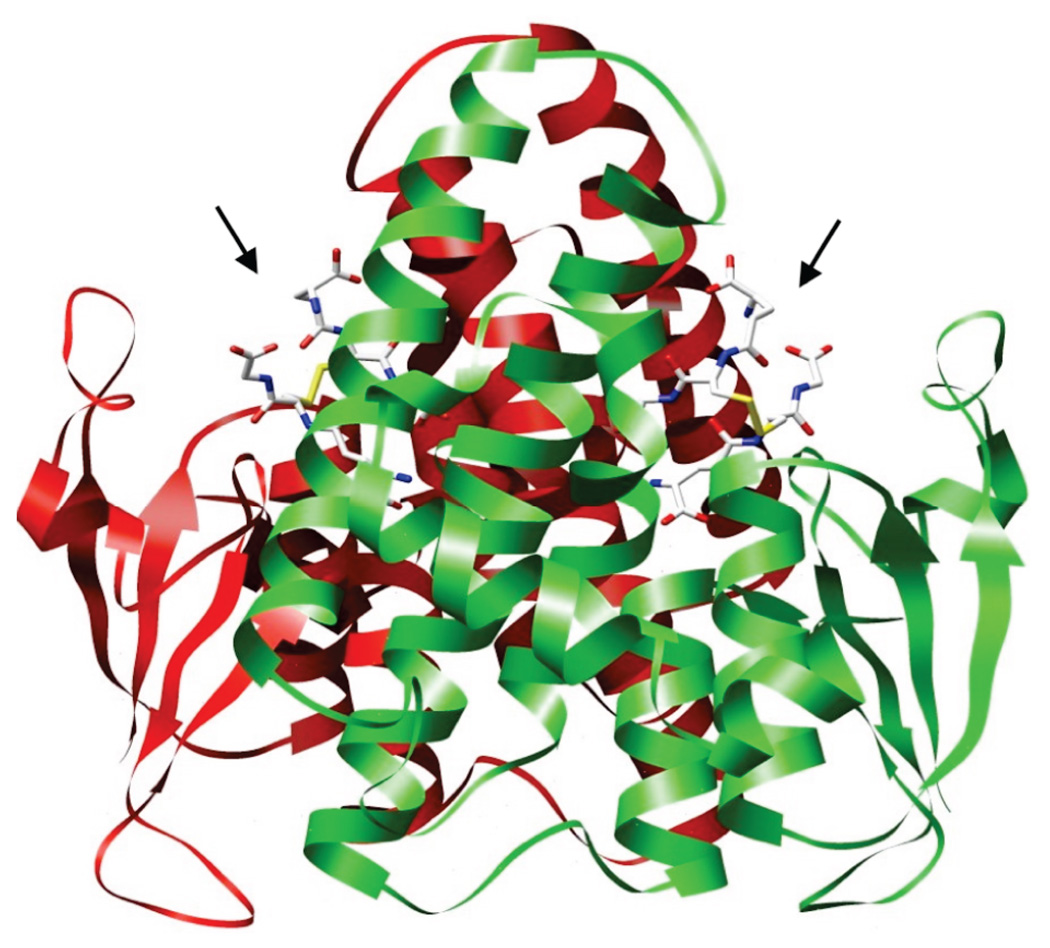
The structure (Figure 1) has the typical GSH transferase fold with an N-terminal thioredoxin-like domain and a C-terminal all α-helical domain (5). The most unusual aspect of the structure is the fact that a molecule of GSSG is found in the active site in spite of the fact that the crystals were grown in the presence of GSH. The electron density for GSSG in the structure is illustrated in Figure 2. One other structure of a GSH transferase with GSSG bound has been reported (PDB file 1YKC), but the accommodation of the bound disulfide in the active site of this mammalian protein is quite different from that described here.
FIGURE 1.
Ribbon diagram of the structure of the dimer of YfcG with GSSG bound in the active sites. The two subunits are shown in red and green, and the two molecules of GSSG (indicated by the arrows) are shown in stick representation. The β structure of the thioredoxin domains is apparent on the right and left sides.
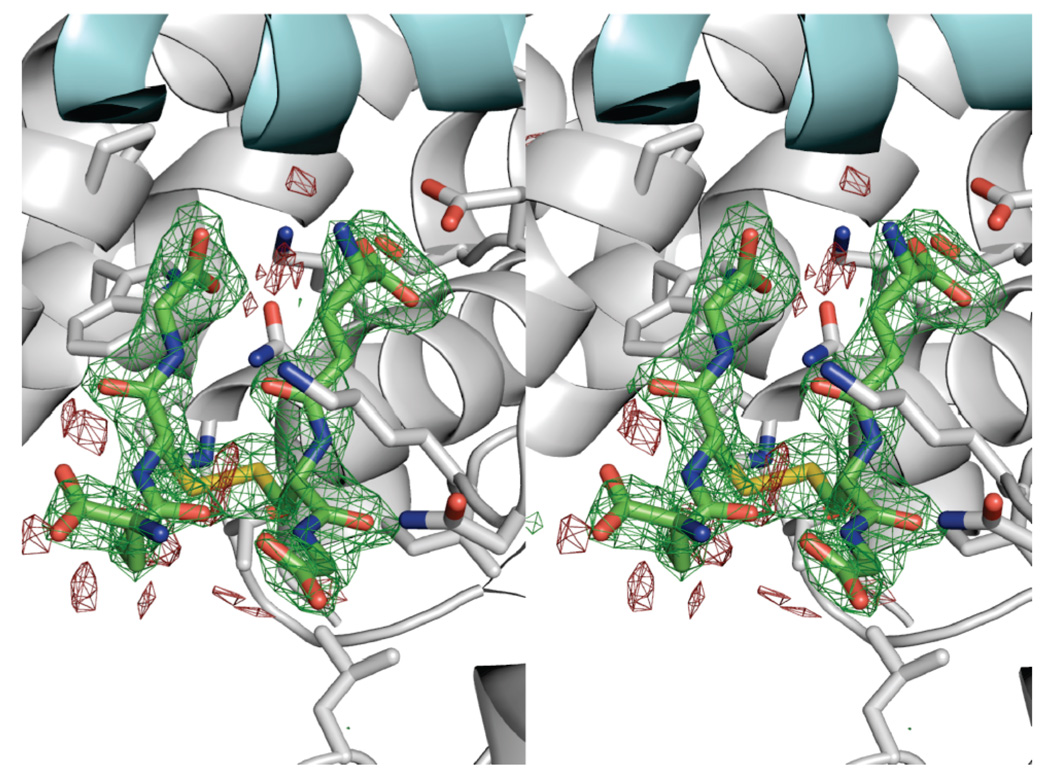
FIGURE 2.
Stereo view of the difference electron density for GSSG from an omit map contoured at 3σ. The blue helix is from the opposite subunit. The average B factor for the GSSG atoms in the final refined structure is 30.7 Å2.
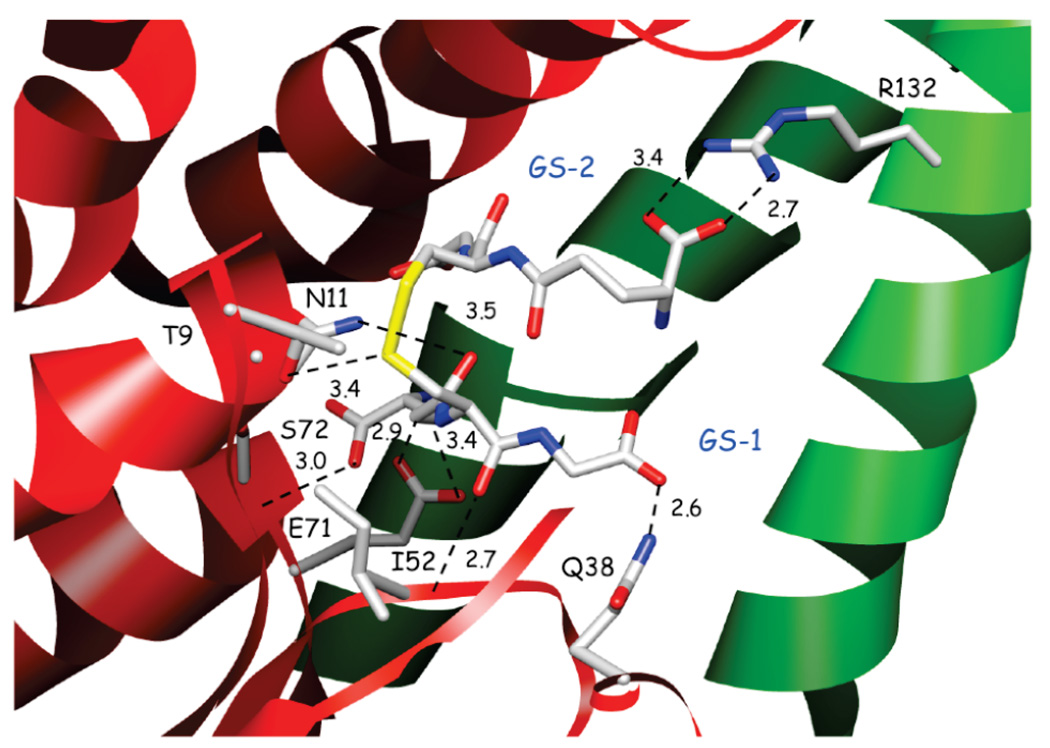
As illustrated in Figure 3, one-half of the GSSG molecule (GS-1) resides in the canonical GSH binding site with numerous hydrogen-bonding interactions to the primary subunit including the hydroxyl group of T9 at the end R-helix-1 and the sulfur of GS-1. In addition, there are interactions of GS-1 with the highly conserved ββα motif that recognizes the γ-glutamyl residue of GSH with side chains (E71 and S72) located in the turn between β4-strand and α-helix-3. The second half of the GSSG molecule (GS-2) has very few hydrogen-bonding interactions with YfcG, but reaches out across the molecule where the γ-glutamyl-α-carboxylate has an ionic interaction with the R132 side chain on α-helix-5 of the opposite subunit.
FIGURE 3.
Details of the GSSG binding site. Subunits 1 and 2 are colored in red and green, respectively. Potential hydrogen bonding interactions are shown as dashed lines with distances shown in Angstroms.
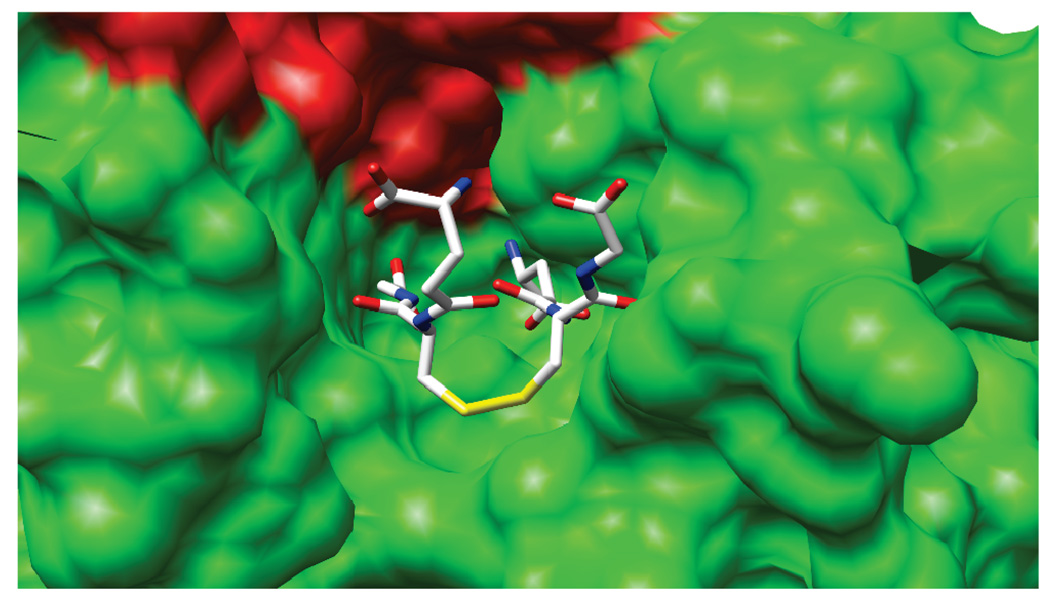
The GSSG molecule is bound in a crevice between the two subunits of the dimer as illustrated inFigure 4.The disulfide bond of GSSG is presented on the surface of the YfcG•GSSG complex, suggesting that it might be involved in disulfide bond chemistry with other molecules.
FIGURE 4.
Space-filling representation of the YfcG•GSSG complex illustrating the exposure of the disulfide bond of GSSG on the surface of the protein.
The fact that the disulfide bond of GSSG is on the surface of the protein suggested that YfcG might be involved in disulfide-bond oxido-reductase chemistry similar to that catalyzed by the thioredoxins and glutaredoxins (6–8). The disulfide bond reductase activity of the protein was measured by the coupled assay with GSH reductase (6) and 2-hydroxyethyldisulfide as the substrate. YfcG is a very efficient disulfide-bond reductase toward this model substrate (Table 2) and on par with the thioredoxins and glutaredoxins (7, 8).
Table 2.
Steady-State Kinetic Constants for the Reduction of 2-Hydroxyethyl Disufide (1.0 mM) by YfcG and Its C166A Mutant at 25 °C
| enzyme | kcat (s−1) | kcat/KMGSH (M−1s−1) | KMGSH (mM) |
|---|---|---|---|
| YfcG | 180 ± 10 | (1.1 ± 0.2) × 105 | 1.6 ± 0.3 |
| YfcG (C166A) | 180 ± 10 | (1.9 ± 0.2) × 105 | 1.0 ± 0.2 |
Most disulfide-bond oxido-reductases have either one or two protein cysteinyl residues that participate in redox chemistry (7–9). YfcG has but a single cysteine residue, C166, that is located 15.1 Å from the active site, suggesting that it does not participate in disulfide-bond reductase activity of the protein. This notion was confirmed by the catalytic characteristics of the C166A mutant protein (Table 2), which are very similar to those of the native enzyme.
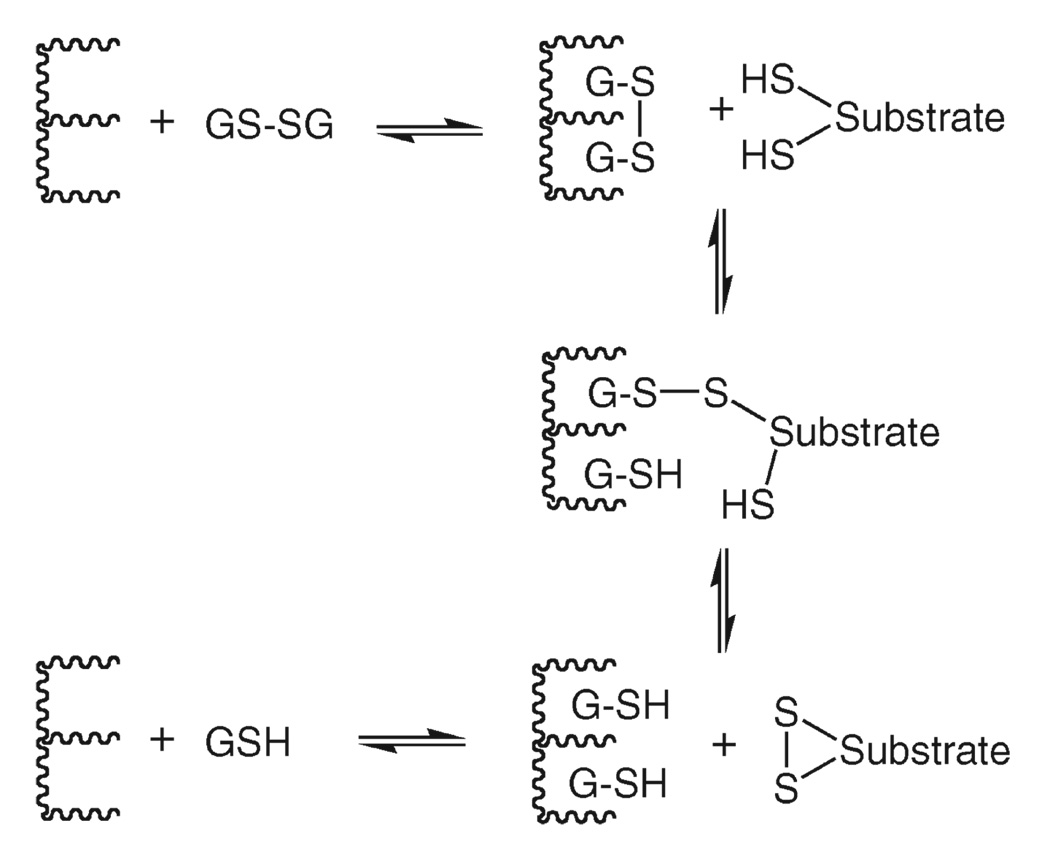
Although most disulfide-bond oxido-reductases utilize one or two cysteinyl residues that are part of the covalent structure of the protein, there is no reason, in principle, why a noncovalently bound thiol-disulfide couple, say, 2GSH/GSSG, cannot perform the same function, as illustrated in Scheme 1. Whether YfcG acts as an oxidant or reductant depends on the redox potential of the cellular compartment in which it resides and the redox potential of the protein-bound thiol–disulfide couple. The primary structure of the enzyme suggests that it resides in the cytoplasm of the cell where the GSH/GSSG ratio is typically 200 to 1. At this point, the actual substrates for YfcG are not known. The association/dissociation of the mixed disulfide intermediate (Scheme 1) from the enzyme in the mechanism cannot be excluded. It is possible that the biologically relevant substrates are glutathionylated proteins such as the mixed-disulfide intermediate illustrated in Scheme 1.
Scheme 1.
The redox potential of GSH in aqueous solution is estimated to be approximately −250 mV (8, 10–12). The observation that YfcG binds GSSG ≥100-fold more tightly than GSH (Table 1) and prefers to bind GSSG in the presence of at least a 10-fold excess of GSH (crystal structure) indicates that the redox potential of GSH when bound to the surface of YfcG is significantly perturbed. If the differential binding energy of GSSG and GSH is fully expressed in the redox potential of bound GSH, then the redox potential of the YfcG•2GSH complex could be as much as −50 mV more reducing (ca. −300 mV) than GSH in aqueous solution. The crystal structure of YfcG also provides a very rough estimate of the internal equilibrium constant for [YfcG3•2GSH ⇌ YfcG•GSSG] (Kint ≫ 1) in the presence of at least a 10-fold excess of external GSH under the crystallization conditions. The remarkable physical–chemical properties of YfcG are consistent with it being a disulfide bond reductant in the cytosol of E. coli. The actual physiological substrates that are reduced by the enzyme have not been identified.
Finally, the data presented in Table 1 indicate that YfcG preferentially binds GspSH about 10-fold more tightly than GSH. Crystals of YfcG grown in the presence of GspSH (and in the absence GSH) are of comparable quality to those grown in the presence of GSH alone. The structure refines, at a resolution of 2.3 Å, with respectable R and Rfree values of 0.205 and 0.233, respectively. However, the resulting protein structure is less well ordered, particularly in the region of the GSSG binding site. No electron density is observed in the peptide-binding site or for residues 32–50 or 110–133 of the protein. The reason for this disorder is not known. It is clear that there is room for a single molecule of GspSH in the active site of YfcG. However, it is equally clear that there is not enough room to tightly accommodate an entire molecule of GspSH-disulfide as suggested by its elevated Kd (Table 1). YfcG and GspSH are coincidentally elevated in late stationary phase (2, 4). Consequently, the potential role of YfcG in the biochemistry of GspSH is under active investigation.
Supplementary Material
Footnotes
This work was supported by NIH Grants R01 GM030910, P30 ES000267, and T32 GM008320.
The atomic coordinates and structure factors for YfcG have been deposited with the Protein Data Bank under file name 3GX0.
SUPPORTING INFORMATION AVAILABLE: Details of all the experimental procedures including the expression and purification of protein, enzyme assays, protein crystallization, X-ray diffraction data collection and refinement, and supporting references. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rife CL, Parsons JF, Xiao G, Gilliland GL, Armstrong RN. Proteins. 2003;53:777–782. doi: 10.1002/prot.10452. [DOI] [PubMed] [Google Scholar]
- 2.Stourman NV, Schaab MR, Wadington MC, Atkinson HJ, Babbitt PC, Armstrong RN. In: Proceedings of the 3rd International Beilstein Workshop on Experimental Standard Conditions of Enzyme Characterizations. Hicks MG, Kettner C, editors. 2008. pp. 1–12. [Google Scholar]
- 3.Kanai T, Takahashi K, Inoue H. J. Biochem. 2006;140:703–711. doi: 10.1093/jb/mvj199. [DOI] [PubMed] [Google Scholar]
- 4.Smith K, Borges A, Ariyanayagam MR, Fairlamb AH. Biochem. J. 1995;312:465–469. doi: 10.1042/bj3120465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong RN. Chem. Res. Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 6.Vlamis-Gardikas A, Åslund F, Spyrout G, Bergman T, Holmgren A. J. Biol. Chem. 1997;272:11236–11243. doi: 10.1074/jbc.272.17.11236. [DOI] [PubMed] [Google Scholar]
- 7.Berndt C, Lillig CH, Holmgren A. Biochim. Biophys. Acta. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Åslund F, Berndt KD, Holmgren A. J. Biol. Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 9.Faulkner MJ, Veeravalli K, Gon S, Georgiou G, Beckwith J. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6735–6740. doi: 10.1073/pnas.0801986105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert HF. Adv. Enzymol. Relat. Areas Mol. Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 11.Lees WJ, Whitesides GM. J. Org. Chem. 1993;58:642–647. [Google Scholar]
- 12.Millis KK, Weaver KH, Rabenstein DL. J. Org. Chem. 1993;58:4144–4146. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.