Abstract
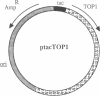
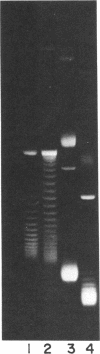
We show that, despite differences in primary structure, substrate preference, and mechanism of catalysis, yeast DNA topoisomerase I can functionally substitute for Escherichia coli DNA topoisomerase I. A family of plasmids expressing the yeast TOP1 gene or 5'-deletion mutations of it were used to complement the temperature-sensitive phenotype of an E. coli topA mutant. These plasmids were then isolated from the cells by a rapid lysis procedure and examined for their degrees of supercoiling. Functional complementation of a conditional-lethal mutation in topA, which encodes E. coli DNA topoisomerase I, correlates with the expression of a catalytically active yeast enzyme that reduces the degree of negative supercoiling of intracellular DNA. We also show that approximately 130 amino acids of the amino-terminal portion of the yeast enzyme can be deleted without affecting its activity in vitro; activity of the enzyme inside E. coli, however, is more sensitive to such deletions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baase W. A., Wang J. C. An omega protein from Drosophila melanogaster. Biochemistry. 1974 Oct 8;13(21):4299–4303. doi: 10.1021/bi00718a009. [DOI] [PubMed] [Google Scholar]
- Been M. D., Champoux J. J. Breakage of single-stranded DNA by eukaryotic type 1 topoisomerase occurs only at regions with the potential for base-pairing. J Mol Biol. 1984 Dec 15;180(3):515–531. doi: 10.1016/0022-2836(84)90025-1. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Dean F., Krasnow M. A., Otter R., Matzuk M. M., Spengler S. J., Cozzarelli N. R. Escherichia coli type-1 topoisomerases: identification, mechanism, and role in recombination. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):769–777. doi: 10.1101/sqb.1983.047.01.088. [DOI] [PubMed] [Google Scholar]
- Depew R. E., Liu L. F., Wang J. C. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J Biol Chem. 1978 Jan 25;253(2):511–518. [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Goto T., Wang J. C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7178–7182. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J Mol Biol. 1985 Oct 5;185(3):625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Mizusawa H., Kakefuda T. Unwinding of double-stranded DNA helix by dehydration. Proc Natl Acad Sci U S A. 1981 May;78(5):2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Raji A., Zabel D. J., Laufer C. S., Depew R. E. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J Bacteriol. 1985 Jun;162(3):1173–1179. doi: 10.1128/jb.162.3.1173-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivenugopal K. S., Lockshon D., Morris D. R. Escherichia coli DNA topoisomerase III: purification and characterization of a new type I enzyme. Biochemistry. 1984 Apr 24;23(9):1899–1906. doi: 10.1021/bi00304a002. [DOI] [PubMed] [Google Scholar]
- Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981 May;78(5):2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C., Bankier A. T., Barrell B. G., Sternglanz R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4374–4378. doi: 10.1073/pnas.82.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C., Voelkel K., DiNardo S., Sternglanz R. Identification of Saccharomyces cerevisiae mutants deficient in DNA topoisomerase I activity. J Biol Chem. 1984 Feb 10;259(3):1375–1377. [PubMed] [Google Scholar]
- Tse-Dinh Y. C., Wang J. C. Complete nucleotide sequence of the topA gene encoding Escherichia coli DNA topoisomerase I. J Mol Biol. 1986 Oct 5;191(3):321–331. doi: 10.1016/0022-2836(86)90129-4. [DOI] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Becherer K. Cloning of the gene topA encoding for DNA topoisomerase I and the physical mapping of the cysB-topA-trp region of Escherichia coli. Nucleic Acids Res. 1983 Mar 25;11(6):1773–1790. doi: 10.1093/nar/11.6.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Recent studies of DNA topoisomerases. Biochim Biophys Acta. 1987 Jun 6;909(1):1–9. doi: 10.1016/0167-4781(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Eliason J. L., States D. J. Dynamic filtering by two-dimensional 1H NMR with application to phage lambda repressor. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6019–6023. doi: 10.1073/pnas.81.19.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumstein L., Wang J. C. Probing the structural domains and function in vivo of Escherichia coli DNA topoisomerase I by mutagenesis. J Mol Biol. 1986 Oct 5;191(3):333–340. doi: 10.1016/0022-2836(86)90130-0. [DOI] [PubMed] [Google Scholar]