Abstract
Magnetic resonance imaging (MRI) is a sophisticated and versatile medical imaging modality. Traditionally, MR images are reconstructed from the raw measurements by a simple inverse 2D or 3D fast Fourier transform (FFT). However, there are a growing number of MRI applications where a simple inverse FFT is inadequate, e.g., due to non-Cartesian sampling patterns, non-Fourier physical effects, nonlinear magnetic fields, or deliberate under-sampling to reduce scan times. Such considerations have led to increasing interest in methods for model-based image reconstruction in MRI.
1. INTRODUCTION
The inverse fast Fourier transform (FFT) has served the MR community very well as the conventional image reconstruction method for k-space data with full Cartesian sampling. And for well sampled non-Cartesian data, the gridding method [1] with appropriate density compensation factors [2] is fast and effective. But when only under-sampled data is available, or when non-Fourier physical effects like field inhomogeneity are important, then gridding/FFT methods for image reconstruction are suboptimal, and iterative algorithms based on appropriate models can improve image quality, at the price of increased computation. This paper reviews the use of iterative algorithms for model-based MR image reconstruction. The references give pointers to some recent work but are by no means a comprehensive survey★.
2. MRI BACKGROUND
Any signal processing method aimed at forming images from measurement devices such as MRI scanners must consider the relevant physics. A survey in this magazine [3] and a book written from a signal processing perspective [4] have described MRI physics well. Here we review the physics in a somewhat unconventional way that facilitates describing some of the “non-Fourier” aspects of MRI.
2.1. MRI physics
Standard MRI scanners use a large static magnetic field
| (1) |
to induce a net magnetization M⃗ = Mxi⃗ + My j⃗+ Mz k⃗ at each point in space in the body being imaged, where i⃗, j⃗, k⃗ denote the unit vectors along the x, y and z axes respectively, and r⃗ = (x, y, z) denotes 3D spatial coordinates. Ideally the static field strength B0(r⃗) would be spatially uniform, i.e., a single constant B0. In practice it is never perfectly uniform, due to the unavoidable nonuniformities of all practical coil designs and due to the field strength variations that are induced by the nonuniform magnetic susceptibilities of different tissue types[5] [6]. The electron distributions in different molecules also influence the local magnetic environment experienced by an atom’s nucleus, called chemical shift. Some types of MRI scans are robust to such spatial variations of B0; others are sensitive to nonuniformities, necessitating correction methods[7].
At equilibrium (which is established within a few seconds for a stationary object), the magnetization M⃗ is aligned with the applied static field and its magnitude is proportional to the product of B0(r⃗) and the object-dependent local density of (predominately) hydrogen protons or “spins.” This proton density alone is of only modest interest in MRI; in practice one applies time-varying magnetic fields B⃗ (r⃗, t) that induce time-varying changes in the magnetization
| (2) |
These changes depend on time constants (tissue-dependent relaxation parameters) and other factors, and the goal in MRI is to form images of aspects of this magnetization. By manipulating the applied field B⃗0(r⃗, t) appropriately, sometimes in conjunction with injected or inhaled contrast agents, one can examine a multitude of different tissue properties.
An MRI scan consists of one or more alternations between two stages: excitation and readout. During the excitation stage, the applied magnetic field B⃗ (r⃗, t) is designed to tip the magnetization vectors M⃗ within some slice or slab away from equilibrium, so that they have a component in the transverse plane, i.e., the (x, y) plane. It is convenient to represent this transverse component mathematically using a complex function defined as follows:
| (3) |
where . Note that the field components Mx and My are real physical quantities; the “transverse magnetization” M(r⃗, t) is complex solely by definition. The excitation process can be quite complicated to model and is beyond the scope of this paper. See [3] for an introduction to the role that signal processing plays in the design of excitation pulses, and [8] [9] [10] for some recent model-based RF pulse design methods.
During the readout stage, the applied field B⃗ (r⃗, t) is manipulated in ways that help elucidate the transverse magnetization M(r⃗, t). For image reconstruction, it is essential to model the effects of the applied field on the transverse magnetization. The precise relationship is governed by the Bloch equation [11] [3]. For most image reconstruction purposes, it suffices to consider just two aspects of the full relationship: precession and transverse relaxation. The most important equation in MRI is the Larmor relation: ω = γ|B⃗|, which states that the magnetization precesses (around the axis of the applied field) at a frequency ω that is proportional to the magnitude of the applied field. The constant of proportionality γ is called the gyromagnetic ratio and is about 42.6 MHz per Tesla for hydrogen protons. During a readout, only the longitudinal component of B⃗ is varied usually, i.e.,
| (4) |
so the magnetization precesses around k⃗, i.e., within the transverse plane. This property is why the complex representation (3) is convenient, because precession can be expressed using a complex phase in this form. In general the applied longitudinal field strength Bz(r⃗, t) varies both spatially and temporally, so the Larmor relationship describes the instantaneous frequency at a given spatial location:
| (5) |
Without loss of generality, let t = 0 be the time when the excitation pulse is completed, and consider some time point t > 0 during the readout. The precession of the transverse magnetization between time 0 and time t corresponds to a net phase that is the integral of the instantaneous frequency (5), i.e., ideally we would have
In practice, microscopic variations in the magnetic field cause the spins within a given voxel to become out of phase over time. So the transverse magnetization vector’s magnitude decreases approximately exponentially with a time constant . Accounting for this decay, an accurate model for the temporal evolution of the transverse magnetization during a readout is
| (6) |
where f(r⃗) ≜ M(r⃗,0) denotes the object’s transverse magnetization immediately after excitation. A typical goal in MRI is to form an image of f(r⃗). The properties of f(r⃗) depend not only on spin density, but also on the type of excitation used. Note that for simplicity of exposition, we focus here on the case where the object is static so that f(r⃗) is not a function of time t. Generalizations to dynamic imaging are very active research areas in MR image reconstruction[12].
The relaxation factor varies spatially, and often is on the order of 10 ms. This relatively rapid decay is a significant limitation in MRI. If were longer, then a signal excitation stage followed by a (lengthy) readout stage could be sufficient to form a high-resolution image of f(r⃗). In practice, the rapid decay limits how much spatial information can be recorded in a single readout stage, so such “single shot” imaging, such as echo-planar imaging (EPI) [13], provides only modest spatial resolution. Therefore, high-resolution imaging uses multiple alternations between excitation stages and readout stages, each with different variations of the applied field Bz(r⃗, t).
2.2. Data acquisition: the MR signal
By Faraday’s law the time-varying magnetization M(r⃗, t) will induce an electromotive force (emf) in a nearby coil. The emf will be proportional to the volume integral of the time derivative of the magnetization M(r⃗, t) multiplied by the coil response pattern c(r⃗). The resulting electrical potential v(t) across the receive coil is
| (7) |
where real(·) denotes the real part of a complex number. The coil response c(r⃗) generally decreases with distance from the coil. If uncorrected, this nonuniformity causes spatial variations in signal strength that can be a challenge for image processing methods like segmentation algorithms. Numerous correction methods have been developed[14] [15] [16] [17] [18].
Because the time constant is on the order of milliseconds whereas the phase variations in (6) are many MHz, it is very reasonable to use a narrow-band approximation when evaluating the time derivative of M(r⃗, t) as needed in (7). The time derivative of a narrow-band signal is well approximated by a constant scaling factor: . We absorb this constant into the coil response pattern and rewrite (7) as
| (8) |
The receive coil’s signal is amplified and demodulated using some center frequency ω0. Ideally one would use ω0 = γB0 if the static magnetic field had uniform strength B0. Usually quadrature demodulation is used, yielding separate in-phase I(t) and quadrature Q(t) baseband signals. In the literature, the demodulated “MR signal” s(t) is defined (implicitly) as
| (9) |
where the lowpass operation selects the baseband component of the demodulated signal. This complex analog signal is just a mathematical definition; in practice, the I(t) and Q(t) signals are each sampled and digitized yielding two digital signals. (One can use two separate A/D converters, or a single A/D converter running at twice the normal rate to avoid I/Q imbalance.) Digitally, these two signals can be combined and stored as complex values, i.e., we record samples
where ΔT denotes the sampling rate (typically around 1 μs) and nd denotes the number of recorded samples, typically 64-512 for a given readout stage. Again, the physical quantities are real, but complex quantities are defined in terms of those physical quantities for convenience. (In some systems, digital demodulation is used, but the modeling remains identical.)
2.3. Signal model
To improve signal to noise ratio (SNR) and reduce acquisition times, the use of multiple receive coils has become increasingly popular in MRI. Although originally called phased array imaging [19], a term that resonates with other signal processing applications involving multiple receivers, today the use of multiple receive coils in MRI is usually called parallel imaging [20] [21] [22].
Let cl(r⃗) denote the sensitivity (response pattern) of the lth coil, for l = 1, …, L, where L denotes the number of coils. Let sl(t) denote the demodulated “MR signal” associated with the lth coil, defined as in (9). Substituting (6) into (9) and simplifying yields the following general forward model for the MR signal associated with the lth coil:
| (10) |
where the space- and time-varying phase is
| (11) |
In practice multiple such signals are recorded, one for each excitation/readout pair (“shots”). For simplicity of notation we consider “single shot” imaging; the extension to multiple shots is conceptually straightforward but notationally cumbersome. Note that the phase variations (11) are common to all receive coils; only the coil response patterns {cl(r⃗)} differ between coils.
2.4. Measurement model
The recorded measurements in a MR scan consist of noisy samples of the MR signal (10)
| (12) |
where yli denotes the ith sample of the lth coil’s signal at time ti and nd denotes the number of time samples. Usually the ti values are equally spaced, and often there are one or more time values where the signal is particularly strong due to alignment of the magnetization’s phases; these values are called echo times. The measurement errors εli are very well modeled by additive, complex, zero-mean, temporally white gaussian noise [23] [24] [25]. However, there can be coupling of the noise values between different coils for the same time points, i.e.,
| (13) |
where δ denotes the Kronecker impulse, and the L × L matrix Σ characterizes the noise covariance between coils [26] [22].
3. LINEAR RECONSTRUCTION PROBLEM
Using the measurement model (12) and the signal model (10), the “typical” image reconstruction problem in MRI is to estimate the object f(r⃗) from the measurement vector y = (y1, …, yL), where yl = (yl1, …, yl,nd). (All vectors are column vectors here.) We first consider model-based image reconstruction for this “basic” linear formulation. Because parallel imaging is of considerable interest, we continue to consider the general case of L receive coils. A standard single receive coil is a simple special case.
This is an ill-posed problem because the given measurements y are discrete whereas the object f(r⃗) is an unknown continuous-space function. To facilitate parametric estimation, we approximate the object f(r⃗) using a “finite series expansion” as follows:
| (14) |
where b(·) denotes the object basis function, r⃗j denotes the center of the jth translated basis function, and N is the number of parameters. Such approximations are classic in the tomographic image reconstruction literature [27] and are slowly taking root in the MR community. Minimum 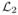 norm methods [28] can postpone the discretization (14) until the final step of displaying the image, but it is unclear if this approach provides image quality benefits that outweigh its computational requirements. For simplicity, hereafter we use rect basis functions b(r⃗) = rect(r⃗/Δ), i.e., square pixels of dimension Δ, so N is the number of pixels, or voxels in 3D scans. Many other possible basis function choices can be considered, all of which are imperfect because the true object never satisfies the parametric model (14) exactly. Nevertheless simple basis functions can provide useful approximations.
norm methods [28] can postpone the discretization (14) until the final step of displaying the image, but it is unclear if this approach provides image quality benefits that outweigh its computational requirements. For simplicity, hereafter we use rect basis functions b(r⃗) = rect(r⃗/Δ), i.e., square pixels of dimension Δ, so N is the number of pixels, or voxels in 3D scans. Many other possible basis function choices can be considered, all of which are imperfect because the true object never satisfies the parametric model (14) exactly. Nevertheless simple basis functions can provide useful approximations.
Substituting the basis expansion (14) into the signal model (10) and simplifying leads to the discrete forward model
| (15) |
where the elements {alij} of the system matrix Al associated with the lth coil are given by
| (16) |
In practice the basis functions are usually highly localized (e.g., voxels), so “center of voxel” approximations like the following are nearly always used, often implicitly:
| (17) |
For exceptions, see [29] [30].
Typically the decay due to is ignored, or it is assumed implicitly that the total readout time tnd− t1 is small relative to in which case one can make the approximation . Under this approximation, we can absorb the -weighting effect of into the unknown image f(r⃗).
Combining (12) and (15) in matrix-vector form yields:
where f = (f1, …, fN) is the vector of parameters (pixel values) that we wish to estimate from the data y. Stacking up all L measurement vectors as y = (y1, …, yL) and defining the (ndL) × N system matrix A = (A1, …, AL) yields the linear model
| (18) |
At first glance this linear model appears amenable to a variety of iterative solution methods. However, a significant challenge that arises is that in general the elements of A can be quite complicated in the form above, yet A is too large to store for typical problem sizes. Most iterative algorithms require matrix-vector multiplication by A and its transpose; there are fast algorithms for these operations (without storing A explicitly) in many special cases of interest [31] [30] [7].
Thus far we have allowed the phase function φ(r⃗j, ti) to be quite general, without the traditional focus on “Fourier encoding.” Recently there has been interest in investigating nonlinear magnetic field variations Bz(r⃗, t) in (4), and reconstruction algorithms have been proposed that use much of the generality in (16) [32] [33]. These are currently specialized research topics, so we now focus on the more common case of linear field gradients.
3.1. Fourier encoding
In typical MR scanners, the longitudinal component of the applied field Bz(r⃗, t) in (4) consists of three components:
| (19) |
The constant B0 denotes the advertised field strength of the main static field. The function ΔB0(r⃗) denotes the spatial deviations of the field strength from this nominal value. This function is often called a field map, and in general it is unknown, but it can be estimated by suitable types of acquisitions and data processing methods [34] [35]. The field gradients G⃗(t) = G⃗x(t)j⃗ + G⃗y(t)j⃗ + G⃗z(t)j⃗ consist of three user-controlled functions that are the historical key to providing spatial information in standard MR imaging[36]. Many different types of MR scans are possible by changing G⃗ (t).
Substituting (19) into (11) using ω0 ≜ γ B0 and simplifying yields
or equivalently
| (20) |
where Δω0(r⃗) ≜ γ ΔB0(r⃗) denotes the off-resonance frequency and the k-space trajectory is defined by
| (21) |
Usually the phase accrual e−i Δ ω0(r⃗)t due to off resonance is undesirable and can distort reconstructed images if ignored[37]. Therefore some image reconstruction methods, particularly in fMRI, account for its effects [38] [30]. In some cases the map Δω0(r⃗) is found from a separate “pre-scan;” in other cases it is estimated jointly with f [39] [40] [41]. In chemical shift imaging, e.g., to separate fat and water components, the term Δω0(r⃗) includes both useful information about the chemical shift effect as well as the undesirable variations due to field inhomogeneity [42] [43].
For the linear field gradients (19), substituting (20) into (17) yields simpler expressions for the system matrix:
| (22) |
where we define the “rate map” z(r⃗) by combining the relaxation and field maps:
| (23) |
When this rate map is assumed to be zero, i.e., if relaxation and off resonance are ignored, then alij is the product of a Fourier encoding matrix having elements e−i2πk⃗(ti) ·r⃗j with a diagonal sensitivity encoding matrix having elements cl(r⃗j).
If the k-space sample locations k⃗(ti) lie on an appropriate subset of a Cartesian grid, then FFT operations provide efficient multiplication by A and its transpose. If non-Cartesian k-space sampling is used, then a nonuniform FFT (NUFFT) is needed [44].
When z(r⃗) in (22) is nonzero, then the elements (22) no longer correspond to a standard Fourier transform. Approximations are needed to provide fast computation of matrix-vector products. In particular, often one can approximate the exponentials in (17) using an additively separable form:
for various choices for the basis functions bik and coefficients ckj [7]. With this type of approximation, we can rewrite matrix-vector multiplication as follows:
The inner sum is simply a FFT or NUFFT so this approach is relatively fast. Free software for this is available [45].
3.2. Reconstruction cost function
Having specified the linear model (18), we now turn to solution methods. Because the noise in MRI measurements is gaussian, a natural approach is to estimate f by minimizing a regularized least-squares cost function:
| (24) |
For a single coil, the noise variance in the k-space data is white (uncorrelated with uniform variance), so the usual Euclidian norm ||·|| is appropriate. For parallel MRI, noise is stationary across time samples (i), but the norm should include the inverse of the L × L covariance matrix Σ in (13) that describes the noise correlation between receive coils [26] [22].
If the k-space samples lie on an equally-spaced grid (Cartesian sampling) with appropriate sample spacings relative to the object field of view, and if the rate map z(r⃗) is zero (i.e., we ignore relaxation and field inhomogeneity), and if we consider just a single coil (L = 1) and treat the sensitivity pattern as uniform, i.e., c1(r⃗) = 1, then the system matrix Al is orthogonal. In this special case, no regularization is needed and and the solution is simply , which can be evaluated by an inverse FFT. This is the most common MR image reconstruction method. However, if any of these conditions do not hold, then typically the system matrix A is not well conditioned, and the unregularized LS solution can lead to undesirable noise amplification. To avoid this problem, some form of regularization is needed.
3.3. Regularization
An open problem in most image reconstruction problems, including MRI, is how to best choose the regularizer R(f). If this term is not included, then the image estimate f̂ will suffer from noise and artifacts for under-sampled and/or non-Cartesian data, because this inverse problem is ill-conditioned. The approach for iterative reconstruction that has been adopted in commercial PET scanners is to use an unregularized algorithm[46], initialize it with a uniform image, stop iterating just as the image gets unacceptably noisy, and then perhaps apply a bit of post-filtering to reduce the noise. One could adopt a similar approach for MR imaging[47]. However, introducing regularization can ensure that the iterative algorithm converges to a stable image, and can enforce prior information that improves image quality particularly for under-sampled data.
The simplest choice is Tikhonov regularization R(f) = ||f||2 or R(f) = ||f − f̄||2, where f̄ is some prior or reference image (possibly zero). The disadvantage of this choice is that it biases the estimate towards the reference image f̄. In particular, if the reference image is zero, then all pixel values in f̂ are diminished towards zero, possibly reducing contrast.
Another choice is a quadratic roughness penalty function, which in 1D would be written
| (25) |
This choice biases the reconstruction towards a smooth image where neighboring pixel values are similar. It is convenient for minimization [30], but it has the drawback of smoothing image edges, particularly if the regularization parameter β in (24) is too large. One can prove that using (25) guarantees that the cost function (24) has a unique minimizer.
More recently, total variation methods have been investigated for MR image reconstruction [48]. In 1D, these methods replace the squared differences between neighboring pixels above with absolute differences:
| (26) |
In 2D continuous space, the analogous functional is
The advantage of this type of regularization is that it biases the reconstructed image towards a piecewise smooth image, instead of a globally smooth image, thereby better preserving image edges. However it is harder to minimize and can lead to the appearance of “blocky” texture in images[49]. Numerous alternatives of the form
for various choices of the “potential function” ψ(·) have been proposed in the imaging literature. Many of these compromise between the quadratic case (25) and the absolute difference case (26), for example the hyperbola
| (27) |
is approximately quadratic near 0, which aids noise reduction, yet approximately linear away from 0, which helps preserve edges[50].
3.4. Algorithms
Iterative algorithms are needed to minimize (24). For differentiable regularizers such as (25), the conjugate gradient algorithm is a natural choice [26] [30]. For nondifferentiable regularizers like (26), more sophisticated algorithms are needed and this is an active research area[51, 52] [12].
4. RECONSTRUCTION CHALLENGES
Although a variety of useful problems can be solved in MRI using the formulation (24), there are numerous challenges that provide research opportunities.
4.1. Regularization parameter selection
A practical challenge with regularized methods is selection of the regularization parameter β in (24). For quadratic regularization, there is a well-developed theory for choosing β in terms of the desired spatial resolution properties of the reconstructed image [53] [54]. This theory extends readily to MR imaging with reasonably well sampled trajectories (and to parallel imaging with reasonable acceleration factors) for which the point spread function (PSF) of the reconstructed image is relatively close to a Kronecker impulse so that simple measures like full width at half maximum (FWHM) are reasonable resolution metrics[55]. For highly under-sampled trajectories the PSF can have “heavy tails” due to aliasing effects, and more investigation is needed to extend the above methods to MR applications.
For nonquadratic regularization such as the total variation method (26), the analysis in [53] [54] is inapplicable so one must resort to other methods for choosing β. Statisticians often use cross validation [56] [57] for choosing regularization parameters, with a goal of finding the parameter that minimizes the mean-squared error (MSE) between f̂ and the unknown f. However, MSE is the sum of variance and bias squared, and where bias is related to spatial resolution and artifacts, and it is unclear whether an equal weighting of noise variance and bias (squared) is optimal from an image quality perspective in medical imaging.
Another method for choosing β is the “L-curve” method[58] [59]. This method is expensive because it requires evaluating f̂ for several values of β, and it has some theoretical deficiencies [60].
In summary, choosing β for nonquadratic regularization remains a nontrivial issue in most ill-posed imaging problems including MRI, and remains an active research area [61] [62].
4.2. Partial k-space methods
If the object f(r⃗) were real, then its Fourier transform would be Hermitian symmetric so in principle only half of k-space would need to be sampled. In practice the magnetization (3) is complex due to a variety of physical effects. However, in many cases the phase of M(r⃗, t) can be assumed to be a smooth function. This property has led to a variety of partial k-space methods where one samples a bit more than half of k-space, then estimates the phase from the central portion of k-space (corresponding to low spatial frequencies), and then uses this estimated phase to reconstruct the entire image [63] [64] [65] [66]. Such methods are used routinely in many types of MR scans.
4.3. Under-sampled k-space data
The need for some type of regularization is essential when the k-space data is under sampled, i.e., when the number of measurements Lnd is less than the number of unknown voxels N. In MRI, the scan time is roughly proportional to the number of measurements, so collecting fewer samples can reduce scan time, which is particularly desirable in dynamic imaging.
In the broader field of tomographic image reconstruction, there is a long history of using prior information, such as assuming objects are piecewise smooth, to reconstruct images from an under-sampled set of projection views, e.g., [67] [68] [69] [70] [71]. Many of these methods involve cost functions of the form (24) with a suitable system matrix A for the application and appropriate regularizers R(f) that capture prior information about the object.
Recently it has become very popular to express prior information in terms of some type of sparsity of the object. Notions of sparsity have deep roots in statistical signal processing [72] [73] [74] [75]. Sparsity is especially apparent in MR angiography [76]. The moniker of compressed sensing or compressive sampling has become widespread for such techniques, and recently entire sessions at MR conferences are devoted to this topic [77] [12]. Some compressed sensing formulations ignore the noise in the data. In the presence of noise, a typical formulation is
where Ψ transforms the image f into a domain (such as wavelet coefficients) where one postulates that the signal is sparse.
Often this optimization problem is solved using a Lagrange multiplier approach:
which corresponds to a particular regularizer in (24). Rarely is the ℓ1 norm implemented exactly; in practice usually a continuously differentiable approximation is used, such as:
| (28) |
for some small value of δ > 0. This approximation is equivalent to the hyperbola (27) used frequently for edge-preserving image reconstruction. Non-convex methods that enforce sparsity even more strongly are also under investigation[78]. In the usual case where A corresponds to an under-sampled discrete Fourier transform (DFT), a variety of algorithms are available that have numerous potential applications in MR [12]. Challenges with this approach include choosing the sparsifying transform Ψ and regularization parameters β and δ appropriately. Furthermore, when δ is small, the regularizer (28) has very high curvature near 0, which can slow convergence. Choice of the k-space sampling pattern is also important. Nonuniform sampling has a long history in MRI, e.g., [79].
5. NONLINEAR RECONSTRUCTION PROBLEMS
The linear image reconstruction problem (24) is just one of many estimation problems of interest in MRI. Returning to the elements of the system matrix (22), there has been research on estimating essentially every component therein, as summarized below.
5.1. Field map estimation
For scans with long readout times, the effect of field inhomogeneity Δω0 in (22) is important. In practice the field map ω(r⃗) is not known a priori but rather it must be estimated from noisy MR scans. One can examine the phase differences between two scans having different echo times to determine Δω0. If these two scans have short readouts, then there are simple image-domain methods for estimating Δω0, which is known as B0 field mapping [80] [35]. Errors in the field map estimates may cause artifacts in reconstructed images that are based on models like (22).
In addition, object motion that occurs between the field map scans and subsequent scans of interest, e.g., in fMRI, will lead to an inconsistency between the actual scan data and the assumed model (22) used by the reconstruction algorithm. This possibility has motivated the development of dynamic field mapping methods that estimate the field map separately for each frame in a dynamic study, e.g., [38] [81] [82] [41]. For scans with long readout durations, the appearance of Δω0 in a complex exponential in (22) makes this a somewhat complicated nonlinear estimation problem.
5.2. Relaxation map estimation
In some MR applications, it is useful to estimate tissue relaxation parameters, particularly T2 or , on a pixel-by-pixel basis[83] [84] [85] [86]. One approach to measuring such relaxation parameters is to acquire a “baseline” scan of the object and then acquire one or more additional scans having different echo times. One then reconstructs images from each of those scans, and then performs linear regression on a voxel-by-voxel basis using the logarithm of the image voxel values. This approach can be adequate if the readout durations are sufficiently small. But for acquisitions with long readouts, the effect of time ti in the e− z(r⃗j) ti in (22) should be considered, i.e., we should account for relaxation during the signal readout[86]. This requires methods that estimate the relaxation map directly from the k-space data. These are more challenging nonlinear estimation problems because appears in an exponent in (22). Several methods for jointly estimating , Δω0, and f (r⃗) have been investigated [87] [88] [40] [89] [82].
5.3. Sensitivity map estimation
The coil sensitivity patterns cl(r⃗) in (22) also must be determined for parallel imaging based on sensitivity encoding. Normally this is done by acquiring well-sampled data both with local receive coils and with a reference body coil and dividing the two [22]. Acquiring the extra reference data can be inconvenient, so normalizing by the square root of the sum of squares of the local receive coils is also used[90]. A variety of other estimation methods have been proposed[91] [92] [93], including methods that jointly estimate the sensitivity maps {cl(r⃗)} and the image f(r⃗) [94] [95] [96]. Note that if f(r⃗) were known, then the problem of estimating cl(r⃗) would be a linear estimation problem because cl(r⃗) appears as a linear scaling in (22). But when both f(r⃗) and cl(r⃗) are to be estimated, the model is bilinear because f(r⃗) and cl(r⃗) appear as a product in (10). This complicates joint estimation.
5.4. Trajectory mapping
The k-space trajectory k⃗(ti), defined as an integral of the gradient waveforms in (21), should be calibrated carefully to ensure that the system model (22) is accurate. In practice, the field gradients induced by the gradient coils in the scanner are not exactly proportional to the waveforms applied to those coils due to eddy currents. Therefore the physical k-space trajectory realized in the system can depart somewhat from the desired k-space trajectory. These differences can degrade the reconstructed image, particularly for non-Cartesian trajectories with long readout durations. Therefore, a variety of techniques have been developed for mapping the actual k-space trajectory experimentally[97] [98] [99] [100] [101] [102] [103].
5.5. Within-voxel gradients
The model (23) treats the field inhomogeneity within each voxel as being a constant, ignoring within-voxel gradients of the off-resonance map. However, these gradients can be significant in functional magnetic resonance imaging (fMRI) based on the BOLD effect [104]. Accurate reconstruction of signals near air-tissue interfaces requires compensation for these within-voxel gradients, which complicates the reconstruction method [105] [106] [29] [107].
6. EXAMPLE
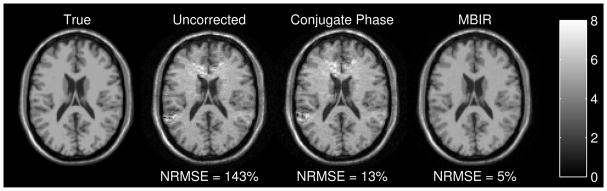
To illustrate the capabilities of model-based image reconstruction methods for MRI, we simulated k-space data for a 4-shot EPI sequence with matrix size 128 × 128 and 5 μsec sampling so the readout duration was 27.3 msec per shot. The field map ΔB0(r⃗) appears in Fig. 2 of [35] and is based on a brain slice above the sinuses and ear canals where susceptibility effects occur. Fig. 1 shows the true image used in the simulations and images from three different reconstruction methods. The “uncorrected” reconstruction simply uses an inverse 2D FFT, with no consideration of field inhomogeneity. The field inhomogeneity causes spatial distortion in the read-out (vertical) direction (that increases NRMSE dramatically), as well as significant intensity artifacts above the ears and sinuses where the susceptibility effects are largest. The classical conjugate phase reconstruction method, which corresponds to A′y in this single-coil case, reduces the spatial distortion but the intensity artifacts persist. Applying 15 iterations of a conjugate gradient algorithm with a monotonic line search [108] [7] to the cost function (24) with the edge-preserving hyperbola (27) yields the right-most image in Fig. 1. This model-based image reconstruction method yields the lowest RMS error, but it requires about 30 × more computation than the noniterative conjugate phase method [7] because each iteration requires multiplication by A and A′. The software that generated this figure is available online [45].
Fig. 1.
Comparison of model-based image reconstruction with conventional methods.
7. SUMMARY
Image reconstruction is not a single problem in MRI, but rather is a wide family of problems depending on what physical effects are included in the signal model. The most widely studied case, particularly in the signal processing community, is when nearly all physical effects are disregarded and the system model consists solely of sampled of the Fourier transform of the object. This basic model is amenable to familiar signal processing tools and is applicable to many MR scans. But there are also many interesting applications where other physical effects are relevant, and model-based methods that account for those effects are proving to be beneficial for improving image quality. Model-based methods themselves depend on estimates of a variety of model parameters, leading to interesting problems where those parameters are determined either by separate calibration scans or by jointly estimating the image and those parameters. Despite over 3 decades of MR research, there remain challenging and intriguing problems in MR image reconstruction.
The published version of this paper (with a short bibliography) is [109].
Acknowledgments
Supported in part by NIH grants CA87634 and NS058576. This is an extended version of a paper that appeared in July 2010 IEEE Signal Processing Magazine. The magazine limit was 30 citations whereas this version has many more.
References
- 1.Beatty PJ, Nishimura DG, Pauly JM. Rapid gridding reconstruction with a minimal oversampling ratio. IEEE Trans Med Imag. 2005 June;24(6):799–808. doi: 10.1109/TMI.2005.848376. [DOI] [PubMed] [Google Scholar]
- 2.Bydder M, Samsonov AA, Du J. Evaluation of optimal density weighting for regridding. Mag Res Im. 2007 June;25(5):695–702. doi: 10.1016/j.mri.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Wright GA. Magnetic resonance imaging. IEEE Sig Proc Mag. 1997 January;14(1):56–66. [Google Scholar]
- 4.Liang Z-P, Lauterber PC. Principles of magnetic resonance imaging. IEEE; New York: 2000. [Google Scholar]
- 5.Schenck JF. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med Phys. 1996 June;23(6):815–50. doi: 10.1118/1.597854. [DOI] [PubMed] [Google Scholar]
- 6.Li L. Magnetic susceptibility quantification for arbitrarily shaped objects in inhomogeneous fields. Mag Res Med. 2001 November;46(5):907–16. doi: 10.1002/mrm.1276. [DOI] [PubMed] [Google Scholar]
- 7.Fessler JA, Lee S, Olafsson VT, Shi HR, Noll DC. Toeplitz-based iterative image reconstruction for MRI with correction for magnetic field inhomogeneity. IEEE Trans Sig Proc. 2005 September;53(9):3393–402. [Google Scholar]
- 8.Yip C, Fessler JA, Noll DC. Iterative RF pulse design for multidimensional, small-tip-angle selective excitation. Mag Res Med. 2005 October;54(4):908–17. doi: 10.1002/mrm.20631. [DOI] [PubMed] [Google Scholar]
- 9.Zelinski AC, Wald LL, Setsompop K, Alagappan V, Gagoski BA, Goyal VK, Adalsteinsson E. Fast slice-selective radio-frequency excitation pulses for mitigating B+1 inhomogeneity in the human brain at 7 Tesla. Mag Res Med. 2008 June;59(6):1355–64. doi: 10.1002/mrm.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grissom WA, Xu D, Kerr AB, Fessler JA, Noll DC. Fast large-tip-angle multidimensional and parallel RF pulse design in MRI. IEEE Trans Med Imag. 2009 October;28(10):1548–59. doi: 10.1109/TMI.2009.2020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloch F. Nuclear induction. Phys Rev. 1946 October;70(7–8):460–74. [Google Scholar]
- 12.Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. IEEE Sig Proc Mag. 2008 March;25(2):72–82. [Google Scholar]
- 13.Mansfield P, Pykett IL. Biological and medical imaging by NMR. J Mag Res. 1978 February;29(2):355–73. doi: 10.1016/j.jmr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Tincher M, Meyer CR, Gupta R, Williams DM. Polynomial modeling and reduction of RF body coil spatial inhomogeneity in MRI. IEEE Trans Med Imag. 1993 June;12(2):361–5. doi: 10.1109/42.232267. [DOI] [PubMed] [Google Scholar]
- 15.Meyer CR, Bland PH, Pipe J. Retrospective correction of intensity inhomogeneities in MRI. IEEE Trans Med Imag. 1995 March;14(1):36–41. doi: 10.1109/42.370400. [DOI] [PubMed] [Google Scholar]
- 16.Brinkmann BH, Manduca A, Robb RA. Optimized homorphic unsharp masking for MR grayscale inhomogeneity correction. IEEE Trans Med Imag. 1998 April;17(2):161–71. doi: 10.1109/42.700729. [DOI] [PubMed] [Google Scholar]
- 17.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998 February;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 18.Lai S-H, Fang M. A new variational shape-from-orientation approach to correcting intensity inhomogeneities in magnetic resonance images. Med Im Anal. 1999 December;3(4):409–24. doi: 10.1016/s1361-8415(99)80033-4. [DOI] [PubMed] [Google Scholar]
- 19.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Mag Res Med. 1990 November;16(2):192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- 20.Ra JB, Rim CY. Fast imaging using subencoding data sets from multiple detectors. Mag Res Med. 1993 July;30(1):142–5. doi: 10.1002/mrm.1910300123. [DOI] [PubMed] [Google Scholar]
- 21.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): Fast imaging with radiofrequency coil arrays. Mag Res Med. 1997 October;38(4):591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- 22.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Mag Res Med. 1999 November;42(5):952–62. [PubMed] [Google Scholar]
- 23.McVeigh ER, Henkelman RM, Bronskill MJ. Noise and filtration in magnetic resonance imaging. Med Phys. 1985 September;12(5):586–91. doi: 10.1118/1.595679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edelstein WA, Glover GH, Hardy CJ, Redington RW. The intrinsic signal-to-noise ratio in NMR Imaging. Mag Res Med. 1986 August;3(4):606–18. doi: 10.1002/mrm.1910030413. [DOI] [PubMed] [Google Scholar]
- 25.Macovski A. Noise in MRI. Mag Res Med. 1996 September;36(3):494–7. doi: 10.1002/mrm.1910360327. [DOI] [PubMed] [Google Scholar]
- 26.Pruessmann KP, Weiger M, Börnert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Mag Res Med. 2001 October;46(4):638–51. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 27.Censor Y. Finite series expansion reconstruction methods. Proc IEEE. 1983 March;71(3):409–19. [Google Scholar]
- 28.Van de Walle R, Barrett HH, Myers KJ, Altbach MI, Desplanques B, Gmitro AF, Cornelis J, Lemahieu I. Reconstruction of MR images from data acquired on a general non-regular grid by pseudoinverse calculation. IEEE Trans Med Imag. 2000 December;19(12):1160–7. doi: 10.1109/42.897806. [DOI] [PubMed] [Google Scholar]
- 29.Sutton BP, Noll DC, Fessler JA. Compensating for within-voxel susceptibility gradients in BOLD fMRI. Proc Intl Soc Mag Res Med. 2004:349. [Google Scholar]
- 30.Sutton BP, Noll DC, Fessler JA. Fast, iterative image reconstruction for MRI in the presence of field inhomogeneities. IEEE Trans Med Imag. 2003 February;22(2):178–88. doi: 10.1109/tmi.2002.808360. [DOI] [PubMed] [Google Scholar]
- 31.Lee J-Y, Greengard L. The type 3 nonuniform FFT and its applications. J Comp Phys. 2005 June;206(1):1–5. [Google Scholar]
- 32.Hennig J, Welz AM, Schultz G, Korvink J, Liu Z, Speck O, Zaitsev M. Parallel imaging in non-bijective, curvilinear magnetic field gradients: a concept study. Mag Res Mat Bio Phys Med. 2008 March;21(1–2):5–14. doi: 10.1007/s10334-008-0105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockmann J, Ciris P, Constable RT. Efficient “O-space” parallel imaging with higher-order encoding gradients and no phase encoding. Proc Intl Soc Mag Res Med. 2009:761. doi: 10.1002/mrm.22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekihara K, Matsui S, Kohno H. A new method of measuring static field distribution using modified Fourier NMR imaging. J Phys E Sci Instrum. 1985 March;18(3):224–7. [Google Scholar]
- 35.Funai AK, Fessler JA, Yeo DTB, Olafsson VT, Noll DC. Regularized field map estimation in MRI. IEEE Trans Med Imag. 2008 October;27(10):1484–94. doi: 10.1109/TMI.2008.923956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauterbur PC. Image formation by induced local interations: examples employing nuclear magnetic resonance. Nature. 1973 March;242(5394):190–1. [PubMed] [Google Scholar]
- 37.Sekihara K, Kuroda M, Kohno H. Image restoration from non-uniform magnetic field influence for direct Fourier NMR imaging. Phys Med Biol. 1984 January;29(1):15–24. doi: 10.1088/0031-9155/29/1/002. [DOI] [PubMed] [Google Scholar]
- 38.Harshbarger TB, Twieg DB. Iterative reconstruction of single-shot spiral MRI with off-resonance. IEEE Trans Med Imag. 1999 March;18(3):196–205. doi: 10.1109/42.764889. [DOI] [PubMed] [Google Scholar]
- 39.Sutton BP, Fessler JA, Noll DC. Field-corrected imaging using joint estimation of image and field map. Proc Intl Soc Mag Res Med. 2002:737. [Google Scholar]
- 40.Twieg DB. Parsing local signal evolution directly from a single-shot MRI signal: A new approach for fMRI. Mag Res Med. 2003 November;50(5):1043–52. doi: 10.1002/mrm.10613. [DOI] [PubMed] [Google Scholar]
- 41.Sutton BP, Noll DC, Fessler JA. Dynamic field map estimation using a spiral-in/spiral-out acquisition. Mag Res Med. 2004 June;51(6):1194–204. doi: 10.1002/mrm.20079. [DOI] [PubMed] [Google Scholar]
- 42.Glover GH, Schneider E. Three-point Dixon technique for true water/fat decomposition with B0 inhomogeneity correction. Mag Res Med. 1991 April;18(2):371–83. doi: 10.1002/mrm.1910180211. [DOI] [PubMed] [Google Scholar]
- 43.Dixon W. Simple proton spectroscopic imaging. Radiology. 1984 October;153(1):189–94. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 44.Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans Sig Proc. 2003 February;51(2):560–74. [Google Scholar]
- 45.Fessler JA. Matlab tomography toolbox. 2004 Available from http://www.eecs.umich.edu/~fessler.
- 46.Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imag. 1994 December;13(4):601–9. doi: 10.1109/42.363108. [DOI] [PubMed] [Google Scholar]
- 47.Qu P, Zhong K, Zhang B, Wang J, Shen GX. Convergence behavior of iterative SENSE reconstruction with non-Cartesian trajectories. Mag Res Med. 2005 October;54(4):1040–5. doi: 10.1002/mrm.20648. [DOI] [PubMed] [Google Scholar]
- 48.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Mag Res Med. 2007 June;57(6):1086–98. doi: 10.1002/mrm.21236. [DOI] [PubMed] [Google Scholar]
- 49.Raj A, Singh G, Zabih R, Kressler B, Wang Y, Schuff N, Weiner M. Bayesian parallel imaging with edge-preserving priors. Mag Res Med. 2007 January;57(1):8–21. doi: 10.1002/mrm.21012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charbonnier P, Blanc-Féraud L, Aubert G, Barlaud M. Two deterministic half-quadratic regularization algorithms for computed imaging. Proc IEEE Intl Conf on Image Processing. 1994;2:168–71. doi: 10.1109/83.551699. [DOI] [PubMed] [Google Scholar]
- 51.Ramani S, Fessler JA. An accelerated iterative reweighted least squares algorithm for compressed sensing MRI. Proc IEEE Intl Symp Biomed Imag. 2010 To appear as 1166, oral WE-OS2.2. [Google Scholar]
- 52.Goldstein T, Osher S. The split Bregman method for L1-regularized problems. SIAM J Imaging Sci. 2009;2(2):43. [Google Scholar]
- 53.Qi J, Leahy RM. Resolution and noise properties of MAP reconstruction for fully 3D PET. IEEE Trans Med Imag. 2000 May;19(5):493–506. doi: 10.1109/42.870259. [DOI] [PubMed] [Google Scholar]
- 54.Fessler JA, Rogers WL. Spatial resolution properties of penalized-likelihood image reconstruction methods: Space-invariant tomographs. IEEE Trans Im Proc. 1996 September;5(9):1346–58. doi: 10.1109/83.535846. [DOI] [PubMed] [Google Scholar]
- 55.Olafsson V, Fessler JA, Noll DC. Spatial resolution analysis of iterative image reconstruction with separate regularization of real and imaginary parts. Proc IEEE Intl Symp Biomed Imag. 2006:5–8. [Google Scholar]
- 56.Stone M. Cross-validation: A review. Math Oper Stat Ser Stat. 1978;9(1):127–139. [Google Scholar]
- 57.Golub GH, Heath M, Wahba G. Generalized cross-validation as a method for choosing a good ridge parameter. Technometrics. 1979 May;21(2):215–23. [Google Scholar]
- 58.Hansen PC. Analysis of discrete ill-posed problems by means of the L-curve. SIAM Review. 1992 December;34(4):561–580. [Google Scholar]
- 59.Hansen PC, O’Leary DP. The use of the L-curve in the regularization of discrete ill-posed problems. SIAM J Sci Comp. 1993;14(6):1487–506. [Google Scholar]
- 60.Vogel CR. Non-convergence of the L-curve regularization parameter selection method. Inverse Prob. 1996 August;12(4):535–47. [Google Scholar]
- 61.Ramani S, Blu T, Unser M. Monte-Carlo SURE: A black-box optimization of regularization parameters for general denoising algorithms. IEEE Trans Im Proc. 2008 September;17(9):1540–54. doi: 10.1109/TIP.2008.2001404. [DOI] [PubMed] [Google Scholar]
- 62.Ahn S, Leahy RM. Analysis of resolution and noise properties of nonquadratically regularized image reconstruction methods for PET. IEEE Trans Med Imag. 2008 March;27(3):413–24. doi: 10.1109/TMI.2007.911549. [DOI] [PubMed] [Google Scholar]
- 63.McGibney G, Smith MR, Nichols ST, Crawley A. Quantitative evaluation of several partial Fourier reconstruction algorithms used in MRI. Mag Res Med. 1993 July;30(1):51–9. doi: 10.1002/mrm.1910300109. [DOI] [PubMed] [Google Scholar]
- 64.Fessler JA, Noll DC. Iterative image reconstruction in MRI with separate magnitude and phase regularization. Proc IEEE Intl Symp Biomed Imag. 2004:209–12. [Google Scholar]
- 65.Bydder M, Robson MD. Partial Fourier partially parallel imaging. Mag Res Med. 2005 June;53(6):1393–401. doi: 10.1002/mrm.20492. [DOI] [PubMed] [Google Scholar]
- 66.Noll DC, Nishimura DG, Macovski A. Homodyne detection in magnetic resonance imaging. IEEE Trans Med Imag. 1991 June;10(2):154–63. doi: 10.1109/42.79473. [DOI] [PubMed] [Google Scholar]
- 67.Hanson KM, Wecksung GW. Bayesian approach to limited-angle reconstruction in computed tomography. J Opt Soc Am. 1983 November;73(11):1501–9. [Google Scholar]
- 68.Mohammad-Djafari A, Demoment G. Maximum entropy image reconstruction in X-ray and diffraction tomography. IEEE Trans Med Imag. 1988 December;7(4):345–54. doi: 10.1109/42.14518. [DOI] [PubMed] [Google Scholar]
- 69.Bresler Y, Macovski A. Three-dimensional reconstruction from projections with incomplete and noisy data by object estimation. IEEE Trans Acoust Sp Sig Proc. 1987 August;35(8):1139–52. [Google Scholar]
- 70.Persson M, Bone D, Elmqvist H. Total variation norm for three-dimensional iterative reconstruction in limited view angle tomography. Phys Med Biol. 2001 March;46(3):853–66. doi: 10.1088/0031-9155/46/3/318. [DOI] [PubMed] [Google Scholar]
- 71.Delaney AH, Bresler Y. Globally convergent edge-preserving regularized reconstruction: an application to limited-angle tomography. IEEE Trans Im Proc. 1998 February;7(2):204–21. doi: 10.1109/83.660997. [DOI] [PubMed] [Google Scholar]
- 72.Natarajan BK. Sparse approximate solutions to linear systems. siamjc. 1995 April;24(2):227–34. [Google Scholar]
- 73.Gorodnitsky IF, Rao BD. Sparse signal reconstruction from limited data using FOCUSS: a re-weighted minimum norm algorithm. IEEE Trans Sig Proc. 1997 March;45(3):600–16. [Google Scholar]
- 74.Harikumar G, Couvreur C, Bresler Y. Fast optimal and suboptimal algorithms for sparse solutions to linear inverse problems. Proc IEEE Conf Acoust Speech Sig Proc. 1998;3:1877–80. [Google Scholar]
- 75.Harikumar G, Bresler Y. A new algorithm for computing sparse solutions to linear inverse problems. Proc IEEE Conf Acoust Speech Sig Proc. 1996;3:1331–4. [Google Scholar]
- 76.Candès E, Romberg J, Velikina J, Ron A, Mistretta C. Image reconstruction from highly undersampled data using total variation minimization. MRA Workshop; Oct. 8; London Ontario. 2004. p. 147. [Google Scholar]
- 77.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Mag Res Med. 2007 December;58(6):1182–95. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 78.Trzasko J, Manduca A. Highly undersampled magnetic resonance image reconstruction via homotopic l0-minimization. IEEE Trans Med Imag. 2009 January;28(1):106–21. doi: 10.1109/TMI.2008.927346. [DOI] [PubMed] [Google Scholar]
- 79.Cao Y, Levin DN. MR imaging with spatially variable resolution. J Mag Res Im. 1992 November;2(6):701–9. doi: 10.1002/jmri.1880020615. [DOI] [PubMed] [Google Scholar]
- 80.Fessler JA, Yeo D, Noll DC. Regularized field map estimation in MRI. Proc IEEE Intl Symp Biomed Imag. 2006:706–9. doi: 10.1109/TMI.2008.923956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schomberg H. Off-resonance correction of MR images. IEEE Trans Med Imag. 1999 June;18(6):481–95. doi: 10.1109/42.781014. [DOI] [PubMed] [Google Scholar]
- 82.Olafsson VT, Noll DC, Fessler JA. Fast joint reconstruction of dynamic and field maps in functional MRI. IEEE Trans Med Imag. 2008 September;27(9):1177–88. doi: 10.1109/TMI.2008.917247. [DOI] [PubMed] [Google Scholar]
- 83.McKenzie CA, Chen Z, Drost DJ, Prato FS. Fast acquisition of quantitative T2 maps. Mag Res Med. 1999 January;41(1):208–12. doi: 10.1002/(sici)1522-2594(199901)41:1<208::aid-mrm30>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 84.Ogawa S, Menon RS, Tank DW, Kim SG, Merkle H, Ellermann JM, Ugurbil K. Functional brain mapping by blood oxygenation level-dependent contrast magnetic resonance imaging. A comparison of signal characteristics with a biophysical model. Biophys J. 1993 March;64(3):803–12. doi: 10.1016/S0006-3495(93)81441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song HK, Dougherty L. K-space weighted image contrast (KWIC) for contrast manipulation in projection reconstruction MRI. Mag Res Med. 2000 December;44(6):825–32. doi: 10.1002/1522-2594(200012)44:6<825::aid-mrm2>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 86.Altbach MI, Bilgin A, Li Z, Clarkson EW, Trouard TP, Gmitro AF. Processing of radial fast spin-echo data for obtaining T2 estimates from a single k-space data set. Mag Res Med. 2005 September;54(3):549–59. doi: 10.1002/mrm.20611. [DOI] [PubMed] [Google Scholar]
- 87.Lee S, Fessler JA, Noll D. A simultaneous estimation of field inhomogeneity and R2* maps using extended rosette trajectory. Proc Intl Soc Mag Res Med. 2002:2327. [Google Scholar]
- 88.Sutton BP, Peltier SJ, Fessler JA, Noll DC. Simultaneous estimation of I0, , and field map using a multiecho spiral acquisition. Proc Intl Soc Mag Res Med. 2002:1323. [Google Scholar]
- 89.Olafsson V, Fessler JA, Noll DC. Dynamic update of R2* and field map in fMRI. Proc Intl Soc Mag Res Med. 2004:45. [Google Scholar]
- 90.Bydder M, Larkman DJ, Hajnal JV. Combination of signals from array coils using image-based estimation of coil sensitivity profiles. Mag Res Med. 2002 March;47(3):539–48. doi: 10.1002/mrm.10092. [DOI] [PubMed] [Google Scholar]
- 91.McKenzie CA, Yeh EN, Ohliger MA, Price MD, Sodickson DK. Self-calibrating parallel imaging with automatic coil sensitivity extraction. Mag Res Med. 2002 March;47(3):529–38. doi: 10.1002/mrm.10087. [DOI] [PubMed] [Google Scholar]
- 92.Lin FH, Chen YJ, Belliveau JW, Wald LL. A wavelet-based approximation of surface coil sensitivity profiles for correction of image intensity inhomogeneity and parallel imaging reconstruction. Hum Brain Map. 2003 March;19(2):96–111. doi: 10.1002/hbm.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cao M, Stenger VA, Eddy W. Estimation of images and sensitivities for multi-coil MRI. Proc Intl Soc Mag Res Med. 2005:2447. [Google Scholar]
- 94.Uecker M, Hohage T, Block KT, Frahm J. Image reconstruction by regularized nonlinear inversion - Joint estimation of coil sensitivities and image content. Mag Res Med. 2008 September;60(3):674–82. doi: 10.1002/mrm.21691. [DOI] [PubMed] [Google Scholar]
- 95.Ying L, Sheng J, Liu B. Joint estimation of image and coil sensitivities in parallel MRI. Proc IEEE Intl Symp Biomed Imag. 2006:17–20. [Google Scholar]
- 96.Ying L, Sheng J. Joint image reconstruction and sensitivity estimation in SENSE (JSENSE) Mag Res Med. 2007 June;57(6):1196–1202. doi: 10.1002/mrm.21245. [DOI] [PubMed] [Google Scholar]
- 97.Takahashi A, Peters T. Compensation of multidimensional selective excitation pulses using measured k-space trajectories. Mag Res Med. 1995 September;34(3):446–56. doi: 10.1002/mrm.1910340323. [DOI] [PubMed] [Google Scholar]
- 98.Mason GF, Harshbarger T, Twieg DB, et al. A method to measure arbitrary k-space trajectories for rapid MR imaging. Mag Res Med. 1997 September;38(3):492–6. doi: 10.1002/mrm.1910380318. [DOI] [PubMed] [Google Scholar]
- 99.Papadakis NG, Wilkinson AA, Carpenter TA, Hall LD. A general method for measurement of the time integral of variant magnetic field gradients: Application to 2D spiral imaging. Mag Res Im. 1997;15(5):567–78. doi: 10.1016/s0730-725x(97)00014-3. [DOI] [PubMed] [Google Scholar]
- 100.Alley MT, Glover GH, Pelc NJ. Gradient characterization using a Fourier-transform technique. Mag Res Med. 1998 April;39(4):581–7. doi: 10.1002/mrm.1910390411. [DOI] [PubMed] [Google Scholar]
- 101.Duyn JH, Yang Y, Frank JA, van der Veen JW. Simple correction method for k-space trajectory deviations in MRI. J Mag Res. 1998 May;132(1):150–3. doi: 10.1006/jmre.1998.1396. [DOI] [PubMed] [Google Scholar]
- 102.Grieve SM, Blamire AM, Styles P. Elimination of Nyquist ghosting caused by read-out to phase-encode gradient cross-terms in EPI. Mag Res Med. 2002 February;47(2):337–43. doi: 10.1002/mrm.10055. [DOI] [PubMed] [Google Scholar]
- 103.Goodyear DJ, Shea M, Beyea SD, Shah NJ, Balcom BJ. Single point measurements of magnetic field gradient waveform. J Mag Res. 2003 July;163(1):1–7. doi: 10.1016/s1090-7807(03)00152-6. [DOI] [PubMed] [Google Scholar]
- 104.Noll DC, Fessler JA, Sutton BP. Conjugate phase MRI reconstruction with spatially variant sample density correction. IEEE Trans Med Imag. 2005 March;24(3):325–36. doi: 10.1109/tmi.2004.842452. [DOI] [PubMed] [Google Scholar]
- 105.Fernández-Seara MA, Wehrli FW. Postprocessing technique to correct for background gradients in image-based measurements. Mag Res Med. 2000 September;44(3):358–36. doi: 10.1002/1522-2594(200009)44:3<358::aid-mrm3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 106.Sutton BP. PhD thesis. Univ. of Michigan; Ann Arbor, MI, 48109-2122, Ann Arbor, MI: 2003. Physics-based reconstruction of magnetic resonance images. [Google Scholar]
- 107.Fessler JA, Noll DC. Model-based MR image reconstruction with compensation for through-plane field inhomogeneity. Proc IEEE Intl Symp Biomed Imag. 2007:920–3. Invited paper. [Google Scholar]
- 108.Fessler JA, Booth SD. Conjugate-gradient preconditioning methods for shift-variant PET image reconstruction. IEEE Trans Im Proc. 1999 May;8(5):688–99. doi: 10.1109/83.760336. [DOI] [PubMed] [Google Scholar]
- 109.Fessler JA. Model-based image reconstruction for MRI. IEEE Sig Proc Mag. 2010 July;27(4):81–9. doi: 10.1109/MSP.2010.936726. invited submission to special issue on medical imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]