Abstract
Molecular docking is a widely-used computational tool for the study of molecular recognition, which aims to predict the binding mode and binding affinity of a complex formed by two or more constituent molecules with known structures. An important type of molecular docking is protein-ligand docking because of its therapeutic applications in modern structure-based drug design. Here, we review the recent advances of protein flexibility, ligand sampling, and scoring functions—the three important aspects in protein-ligand docking. Challenges and possible future directions are discussed in the Conclusion.
Keywords: protein flexibility, ligand sampling, scoring functions, molecular docking, protein-ligand interactions
1. Introduction
Molecular recognitions including enzyme-substrate, drug-protein, drug-nucleic acid, protein-nucleic acid, and protein-protein interactions play important roles in many biological processes such as signal transduction, cell regulation, and other macromolecular assemblies. Therefore, determination of the binding mode and affinity between the constituent molecules in molecular recognition is crucial to understanding the interaction mechanisms and to designing therapeutic interventions. Due to the difficulties and economic cost of the experimental methods for determining the structures of complexes, computational methods such as molecular docking are desired for predicting putative binding modes and affinities. In molecular docking, based on the protein structures, thousands of possible poses of association are tried and evaluated; the pose with the lowest energy score is predicted as the “best match”, i.e., the binding mode. Since Kuntz and colleagues’ pioneering work [1], significant progress has been made in docking research to improve the computational speed and accuracy. Among them, protein-ligand docking is a particularly vibrant research area because of its importance to structure-based drug design [2–8] and will be the subject of the present review.
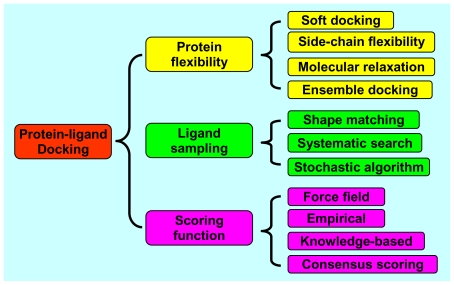
A protein-ligand docking program consists of two essential components, sampling and scoring. Sampling refers to the generation of putative ligand binding orientations/conformations near a binding site of a protein and can be further divided into two aspects, ligand sampling and protein flexibility. Scoring is the prediction of the binding tightness for individual ligand orientations/conformations with a physical or empirical energy function. The top orientation/conformation, namely the one with the lowest energy score, is predicted as the binding mode. Here, we will review the recent advances in protein-ligand docking on three important aspects: protein flexibility, ligand sampling, and scoring function, as illustrated in Figure 1. Challenges and future directions will also be discussed.
Figure 1.
Classification of the methods for protein-ligand docking.
2. Protein Flexibility
Ligand binding commonly induces protein conformational changes (referred to as “induced fit”), which range from local rearrangements of side-chains to large domain motions. Due to the large size and many degrees of freedom of proteins, their flexibility may be the most challenging issue in molecular docking. Current methods to account for protein flexibility can be grouped into four categories: soft docking, side-chain flexibility, molecular relaxation, and protein ensemble docking [9–14].
2.1. Soft Docking
Soft docking is the simplest method which considers protein flexibility implicitly. It works by allowing for a small degree of overlap between the ligand and the protein through softening the interatomic van der Waals interactions in docking calculations [15,16]. The advantages of soft docking are its computational efficiency and easiness for implementation. However soft docking can account for only small conformational changes.
2.2. Side-Chain Flexibility
Many of the early attempts to incorporate certain protein conformational changes into molecular docking focused on side-chain flexibility, in which backbones are kept fixed and side-chain conformations are sampled. One of the earliest studies is the ligand docking algorithm developed by Leach, in which discrete side-chain flexibility is included by using a rotamer library [17]. Since then, researchers have proposed many improved techniques to incorporate continuous or discrete side-chain flexibility in ligand docking [18–24].
2.3. Molecular Relaxation
The third type of methods account for protein flexibility by firstly using rigid-body docking to place the ligand into the binding site and then relaxing the protein backbone and side-chain atoms nearby. Specifically, the initial rigid-body docking allows for atomic clashes between the protein and the placed ligand orientations/conformations in order to consider the protein conformational changes. Then, the formed complexes are relaxed or minimized by Monte Carlo (MC), Molecular Dynamic simulations, or other methods [25,26]. The advantage of the molecular relaxation method is the inclusion of certain backbone flexibility in addition to the side-chain conformational changes. However, compared to the side-chain flexibility methods, the relaxation method is more demanding for the scoring function because it involves not only the side-chain movement but also the more challenging backbone sampling, thereby inaccuracies in the scoring function may lead to artifacts (e.g., improper backbone torsions) in the relaxed protein conformations. Moreover, the relaxation method is time-consuming.
2.4. Docking of Multiple Protein Structures
The most widely-used type of methods for incorporating protein flexibility utilize an ensemble of protein structures to represent different possible conformational changes [9–14]. One of the earliest studies was done by Knegtel et al. [27], in which an averaged energy grid was constructed by combining the energy grids generated from individual experimentally-determined protein structures using a weighting scheme, followed by standard ligand docking. Osterberg et al. extended the method to AutoDock [28] with a larger ensemble consisting of 21 different conformations of the HIV-1 protease [29]. The averaging nature of the method may miss the geometric accuracy of the protein. Claussen et al. developed a docking program FlexE to dock ligands into an ensemble of protein structures [30], in which the similar segments of the protein structures in the ensemble are aligned and merged while the dissimilar segments are used to combinatorially create new possible protein conformations for docking. In Wei et al.’s algorithm [31], a protein was decomposed into a rigid part and several flexible parts according to the crystal structures of the protein in the ensemble. For a given ligand placement, only the best-fit local conformer was kept for each flexible part in the protein, assuming the flexible regions move independently. The selected local conformers were joined with the rigid part to form the best-fit protein conformation. Compared to FlexE, this algorithm is significantly faster and scales linearly rather than exponentially with the protein flexibility. However, the algorithm may rely on the quality of ligand orientational/conformational sampling, namely whether proper initial ligand placement is included. Huang and Zou developed a fast ensemble docking algorithm by treating the protein conformational ensemble as an additional dimension to the traditional six degrees of freedom (three translational plus three rotational) for ligand energy optimization [32,33]. The algorithm is almost as fast as single docking whereas keeping the accuracy of sequential docking. The ensemble docking algorithm is not used for generating new protein structures, but instead for selecting the induced-fit structure from a given protein ensemble. Following a similar procedure, Abagyan and colleagues expanded Huang and Zou’s algorithm to create ICM’s ensemble docking algorithm, referred to as four-dimensional (4D) docking [34]. In addition to experimental structures such as NMR structures or crystal structures bound of the protein with different ligands, ensembles of protein conformations generated by molecular dynamic simulations, Monte Carlo simulations, or structure prediction have also been used to account for protein flexibility [35–43].
3. Ligand Sampling
Ligand sampling is the most basic element in protein-ligand docking. Given a protein target, the sampling algorithm generates putative ligand orientations/conformations (i.e., poses) around the chosen binding site of the protein. The binding site can be the experimentally determined active site, a dimer interface or other site of interest. Ligand sampling is the most successful area being developed in protein-ligand docking. Roughly, there are three types of ligand sampling algorithms: shape matching, systematic search, and stochastic algorithms.
3.1. Shape Matching
The shape matching method is one of the simplest sampling algorithms which is often used in the early stages of the docking process or in the first step of other more advanced ligand sampling methods. It places the ligand using the criterion that the molecular surface of the placed ligand must complement the molecular surface of the binding site on the protein. The six degrees of freedom (three translational and three rotational) of the ligand allow for many possible ligand binding orientations. Therefore, how to quickly place the ligand in the binding site with a good shape complementarity is the goal of the shape matching algorithm. Examples of docking programs that use shape matching include DOCK [1], FRED [44], FLOG [45], EUDOC [46], LigandFit [47], Surflex [48], MS-DOCK [49], and MDock [32,33]. The major advantage of shape matching is its computational efficiency. However, the conformation of the ligand is normally fixed during the shape matching process. Therefore, flexible-ligand docking with the shape matching method is usually performed by docking an ensemble of pre-generated ligand conformations into the protein [50], followed by merging/reranking of the docked poses from different docking runs according to their energy scores (see below).
3.2. Systematic Search
Systematic search algorithms are normally used for flexible-ligand docking, which generate all possible ligand binding conformations by exploring all degrees of freedom of the ligand. There are three types of systematic search methods: exhaustive search, fragmentation and conformational ensemble.
The most straightforward systematic algorithms are exhaustive search methods, in which flexible-ligand docking is performed by systematically rotating all possible rotatable bonds of the ligand at a given interval. Despite its sampling completeness for ligand conformations, the number of the combinations can be huge with the increase of the rotatable bonds. Therefore, to make the docking process practical, geometric/chemical constrains are normally applied to the initial screening of ligand poses, and the filtered ligand conformations are further subject to the more accurate refinement/optimization procedures. Glide [51,52] and FRED [44] are two typical examples of this type of hierarchical sampling methods.
In fragmentation methods, the ligand is first divided into different rigid parts/fragments. Then, the ligand binding conformation is incrementally grown by placing one fragment at a time in the binding site or by docking all the fragments into the binding site and linking them covalently. DOCK [53], LUDI [54], FlexX [55], ADAM [56], and eHiTs [57] are example docking programs that use fragmentation methods.
In conformational ensemble methods [50], ligand flexibility is represented by rigidly docking an ensemble of pre-generated ligand conformations with other programs such as OMEGA (OpenEye Scientific Inc, NM). Then, ligand binding modes from different docking runs are collected and ranked according to their binding energy scores. Examples of the conformational ensemble methods for docking include FLOG [45], DOCK3.5 [58], PhDOCK [59], MS-DOCK [49], MDock [32,33], and Q-Dock [60].
3.3. Stochastic Algorithms
In stochastic algorithms, ligand binding orientations and conformations are sampled by making random changes to the ligand at each step in both the conformational space and the translational/rotational space of the ligand, respectively. The random change will be accepted or rejected according to a probabilistic criterion. There are four types of stochastic algorithms: Monte Carlo (MC) methods, evolutionary algorithms (EA), Tabu search methods, and swarm optimization (SO) methods.
In a Monte Carlo method, the probability to accept a random change is calculated by using the following Boltzmann probability function:
| (1) |
where E0 and E1 stand for the energy scores of the ligand before and after the random change, respectively, kB is the Boltzmann constant, and T is the absolute temperature of the system. The docking programs that use the MC methods include DockVision [61], ICM [18], QXP [62], Prodock [63], and MCDOCK [64].
Evolutionary algorithms (EAs) search for the correct ligand binding mode based on the idea from the evolutionary process in biological systems. The most popular type of EAs is the genetic algorithms (GAs). GOLD [65,66], AutoDock [28], DIVALI [67], DARWIN [68], MolDock [69], PSI-DOCK [70], FLIPDock [42], Lead finder [71], and EADock [72] are the examples that have implemented evolution algorithms.
In Tabu search methods, the probability of acceptance depends on the previously explored areas in the conformational space of the ligand. The random change will be rejected if the RMSD between the current ligand binding conformation and any of the previously recorded solutions is less than a cutoff; otherwise, the random change will be accepted. Example docking programs are PRO_LEADS [73] and PSI-DOCK [70].
Swarm optimization (SO) algorithms attempt to find an optimal solution in a search space by modeling swarm intelligence. In the method, movements of a ligand mode through the search space are guided by the information of the best positions of its neighbors. Examples of docking programs that use swarm optimization algorithms include SODOCK [74], Tribe-PSO [75], PSO@Autodock [76], and PLANTS [77].
4. Scoring Functions
The scoring function is a key element of a protein-ligand docking algorithm, because it directly determines the accuracy of the algorithm [78–82]. Speed and accuracy are the two important aspects of a scoring function. An ideal scoring function would be both computationally efficient and reliable. Numerous scoring functions have been developed in the past decades and can be grouped into three basic categories according to their methods of derivation: force field, empirical, and knowledge-based scoring functions.
4.1. Force Field Scoring Functions
Force field (FF) scoring functions [28,83,84] are based on decomposition of the ligand binding energy into individual interaction terms such as van der Waals (VDW) energies, electrostatic energies, bond stretching/bending/torsional energies, etc., using a set of derived force-field parameters such as AMBER [85] or CHARMM [86,87] force fields. One of the major challenges in FF scoring functions is how to account for the solvent effect. The simplest method is to use a distance-dependent dielectric constant ɛ (rij) such as the force field scoring function in DOCK [84]:
| (2) |
where rij stands for the distance between protein atom i and ligand atom j, Aij and Bij are the VDW parameters, and qi and qj are the atomic charges. ɛ (rij) is usually set to 4rij, reflecting the screening effect of water on electrostatic interactions.
The most rigorous FF methods are to treat water molecules explicitly such as FEP and TI (see [88] for review). However, these methods, together with their simplified approaches such as LIEPROFEC, and OWFEG are computationally expensive [88]. To reduce the computational expense, accelerated methods have been developed while preserving the reasonable accuracy by treating water as a continuum dielectric medium. The Poisson-Boltzmann/surface area (PB/SA) models [89–99] and the generalized-Born/surface area (GB/SA) models [100–111] are typical examples of such implicit solvent models.
In addition to the challenge on solvent effect, how to accurately account for entropic effect is an even more severe challenge for FF scoring functions. Moreover, whether the free energy of ligand binding can be decomposed into a linear combination of individual interaction terms without calculating the partition function (“ensemble average”) also remains in question, referred to as the nonadditive problem.
4.2. Empirical Scoring Functions
In empirical scoring functions, the binding energy score of a complex is calculated by summing up a set of weighted empirical energy terms such as VDW energy, electrostatic energy, hydrogen bonding energy, desolvation term, entropy term, hydrophobicity term, etc.
| (3) |
where {ΔGi} represent individual empirical energy terms, and the corresponding coefficients {Wi} are determined by reproducing the binding affinity data of a training set of protein-ligand complexes with known three-dimensional structures, using least squares fitting [112–121]. Compared to the force field scoring functions, the empirical scoring functions are normally much more computationally efficient due to their simple energy terms. However, the general applicability of an empirical scoring function depends on the training set due to the nature of its fitting to known binding affinities of its training set. With the rapid increase in the number of crystal structures of diverse protein-ligand complexes with known binding affinities, a general empirical scoring function could be developed by training on the binding constants of thousands of protein-ligand complexes. GlideScore [51,52], PLP [119,120], SYBYL/F-Score [55], LigScore [118], LUDI [115,117], SCORE [116], X-Score [121], ChemScore [114], MedusaScore [122], AIScore [123], and SFCscore [124] are examples of empirical scoring functions.
4.3. Knowledge-Based Scoring Functions
The potential parameters of knowledge-based scoring functions are directly derived from the structural information in experimentally determined protein-ligand complexes [125–128]. The principle behind knowledge-based scoring functions is the potential of mean force [129], which is defined by the inverse Boltzmann relation [130–133]
| (4) |
where kB is the Boltzmann constant, T is the absolute temperature of the system, ρ(r) is the number density of the protein-ligand atom pair at distance r in the training set, and ρ*(r) is the pair density in a reference state where the interatomic interactions are zero. After the potential parameters w(r) are derived, the energy of ligand binding for a given complex is simply the sum of the interaction terms for all the protein-ligand atom pairs in the complex. Based on the early idea of Tanaka and Scheraga [125], a number of knowledge-based scoring functions have been developed for protein-ligand interactions. Compared to the force field and empirical scoring functions, the knowledge-based scoring functions offer a good balance between accuracy and speed. Namely, because the potentials in Equation. (4) are extracted from a large number of structures rather than attempting to reproduce the known affinities by fitting, the knowledge-based scoring functions are relatively robust and general [134–137]. Their pairwise characteristic also enables the scoring process to be as fast as empirical scoring functions.
As the ideal reference state is inaccessible for complicated systems like proteins [132], one major challenge for knowledge-based scoring functions is the calculation for the afore-mentioned reference state; based on the methods knowledge-based scoring functions can be classified into three categories: traditional atom-randomized reference state, corrected reference state, and circumventing the reference state.
Traditional methods to approximate the reference state are randomization of the atoms in the training set. Examples include DrugScore [138,139], SMoG [140,141], BLEEP [142,143], GOLD/ASP [144], MScore [145], and KScore[146]. The disadvantage of the atom-randomization approximation is the neglection of the effects of excluded volume, interatomic connectivity, etc. [132]. Methods like PMF [134,135] and DFIRE [147] have introduced correction terms for the reference state. Yet, binding mode prediction and virtual database screening are main problems for most knowledge-based scoring functions as a result of the reference state problem. To circumvent the long-standing reference state problem, Huang and Zou have developed a physics-based iterative method and derived the ITScore scoring function, which has been extensively tested with multiple benchmarks for binding mode prediction, affinity prediction and virtual screening [136,137,148].
Other challenges for knowledge-based scoring functions include extension of the pairwise interactions to many-body interactions to account for hydrogen bonding and other directional interactions, development of an accurate method for entropic calculations [148], etc.
4.4. Consensus Scoring
Consensus scoring is not really a specific type of scoring function but a technique in protein-ligand docking [149]. It improves the probability of finding a correct solution by combining the scoring information from multiple scoring functions in hopes of balancing out the errors of the individual scoring functions. Therefore, the main issue in consensus scoring is how to make the combination rule for individual scores so that the true binders can be discriminated from others according to the consensus rule [150,151]. MultiScore and X-Cscore are two examples of consensus scoring methods [121,152].
4.5. Clustering and Entropy-Based Scoring Methods
In addition to consensus scoring, another technique to improve the performances of scoring functions is clustering-based scoring methods, which incorporate the entropic effects by dividing generated ligand binding modes into different clusters [169–171]. The entropic contribution in each cluster is measured by the configurational space covered by the ligand poses or the number of the ligand poses in the cluster. One restriction in clustering-based scoring methods is that its performance depends on the ligand sampling protocol that is used, i.e., it is docking program-dependent. These methods in combination with ligand conformational sampling using AutoDock have significantly improved binding mode prediction [148,169–171].
5. Conclusion and Discussions
We have reviewed three important aspects of protein-ligand docking: protein flexibility, ligand sampling, and scoring functions. Rapid advances in the last two decades have almost solved the ligand sampling issue. Although equal or even more efforts have been paid to scoring function development, entropy and desolvation effects remain the two major challenging issues for current scoring functions, particularly for the force field scoring functions. Speed and accuracy are the two important characteristics of a scoring function. Because of the rapid increase in computing power, how to improve the accuracy is the future direction for scoring function development. In contrast to ligand sampling and scoring functions which have been extensively studied for more than two decades, protein flexibility has only been addressed recently because of the difficulty resulting from the enormous degrees of freedom and the limitation of the computing power. The development of computational methods for protein flexibility is still in its infancy and thereby remains one of the major future directions in protein-ligand docking. Finally, how to evaluate different docking programs and scoring functions is another active area [153–157]. Although many comparison studies for docking and scoring have been published [158–166], publicly available docking benchmarks such as DUD (http://dud.docking.org/) [167,168] and CSAR (http://www.csardock.org/) are extremely valuable for systematic and consistent evaluation and improvement of new and existing docking algorithms.
Acknowledgments
We thank Sam Z. Grinter for critical reading of the manuscript. Support to XZ from OpenEye Scientific Software Inc. (Santa Fe, NM) and Tripos, Inc. (St. Louis, MO) is gratefully acknowledged. XZ is supported by NIH grant R21GM088517, Cystic Fibrosis Foundation grant ZOU07I0, the Research Board Award RB-07-32 and the Research Council Grant URC 09-004 of the University of Missouri. The work is also supported by Federal Earmark NASA Funds for Bioinformatics Consortium Equipment and additional financial support from Dell, SGI, Sun Microsystems, TimeLogic, and Intel.
References
- 1.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 2.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu. Rev. Biophys. Biomol. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 3.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: An overviewof search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 4.Shoichet BK, McGovern SL, Wei B, Irwin JJ. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 5.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004;3:935–948. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 6.Muegge I, Rarey M. Small molecule docking and scoring. Rev. Comput. Chem. 2001;17:1–60. [Google Scholar]
- 7.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: Current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 8.Kolb P, Ferreira RS, Irwin JJ, Shoichet BK. Docking and chemoinformatic screens for new ligands and targets. Curr. Opin. Biotech. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson HA. Protein flexibility is an important component of structure-based drug discovery. Curr. Pharm. Des. 2002;8:1571–1578. doi: 10.2174/1381612023394232. [DOI] [PubMed] [Google Scholar]
- 10.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000;57:213–218. [PubMed] [Google Scholar]
- 11.Teodoro ML, Kavraki LE. Conformational flexibility models for the receptor in structure based drug design. Curr. Pharm. Des. 2003;9:1635–1648. doi: 10.2174/1381612033454595. [DOI] [PubMed] [Google Scholar]
- 12.Teague SJ. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 13.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, et al. Target flexibility: An emerging consideration in drug discovery and design. J. Med. Chem. 2008;51:6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: A practical alternative. Curr. Opin. Struct. Biol. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang F, Kim SH. Soft docking: Matching of molecular surface cubes. J. Mol. Biol. 1991;219:79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari AM, Wei BQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. J. Med. Chem. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leach AR. Ligand docking to proteins with discrete side-chain flexibility. J. Mol. Biol. 1994;235:345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 18.Abagyan R, Totrov M, Kuznetzov D. ICM – a new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994;15:488–506. [Google Scholar]
- 19.Desmet J, Wilson IA, Joniau M, De Maeyer M, Lasters I. Computation of the binding of fully flexible peptides to proteins with flexible side-chains. FASEB J. 1997;11:164–172. doi: 10.1096/fasebj.11.2.9039959. [DOI] [PubMed] [Google Scholar]
- 20.Schaffer L, Verkhivker GM. Predicting structural effects in HIV-1 protease mutant complexes with flexible ligand docking and protein side-chain optimization. Proteins. 1998;33:295–310. doi: 10.1002/(sici)1097-0134(19981101)33:2<295::aid-prot12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 21.Schnecke V, Kuhn LA. Virtual screening with solvation and ligand-induced complementarity. Perspect. Drug Discov. Des. 2000;20:171–190. [Google Scholar]
- 22.Frimurer TM, Peters GH, Iversen LF, Andersen HS, Moller NP, Olsen OH. Ligand-induced conformational changes: Improved predictions of ligand binding conformations and affinities. Biophys. J. 2003;84:2273–2281. doi: 10.1016/S0006-3495(03)75033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meiler J, Baker D. ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–584. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 24.Nabuurs SB, Wagener M, de Vlieg J. A flexible approach to induced fit docking. J. Med. Chem. 2007;50:6507–6518. doi: 10.1021/jm070593p. [DOI] [PubMed] [Google Scholar]
- 25.Apostolakis J, Pluckthun A, Caflisch A. Docking small ligands in flexible binding sites. J. Comput. Chem. 1998;19:21–37. [Google Scholar]
- 26.Davis IW, Baker D. ROSETTALIGAND docking with full ligand and receptor flxibility. J. Mol. Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Knegtel RM, Kuntz ID, Oshiro CM. Molecular docking to ensembles of protein structures. J. Mol. Biol. 1997;266:424–440. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- 28.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 29.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: Incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 30.Claussen H, Buning C, Rarey M, Lengauer T. FlexE: Efficient molecular docking considering protein structure variations. J. Mol. Biol. 2001;308:377–395. doi: 10.1006/jmbi.2001.4551. [DOI] [PubMed] [Google Scholar]
- 31.Wei BQ, Weaver LH, Ferrari AM, Matthews BW, Shoichet BK. Testing a flexible-receptor docking algorithm in a model binding site. J. Mol. Biol. 2004;337:1161–1182. doi: 10.1016/j.jmb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Huang S-Y, Zou X. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 33.Huang S-Y, Zou X. Efficient molecular docking of NMR structures: Application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking: A fast and accurate account of discrete receptor flexibility in ligand docking. J. Med. Chem. 2009;52:397–406. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broughton HB. A method for including protein flexibility in protein-ligand docking: Improving tools for database mining and virtual screening. J. Mol. Graph. Model. 2000;18:247–257. doi: 10.1016/s1093-3263(00)00036-x. [DOI] [PubMed] [Google Scholar]
- 36.Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA. Developing a dynamic pharmacophore model for HIV-1 integrase. J. Med. Chem. 2000;43:2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- 37.Meagher KL, Carlson HA. Incorporating protein flexibility in structure-based drug discovery: Using HIV-1 protease as test case. J. Am. Chem. Soc. 2004;126:13276–13281. doi: 10.1021/ja0469378. [DOI] [PubMed] [Google Scholar]
- 38.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: The relaxed complex scheme. J. Am. Chem. Soc. 2002;124:5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 39.Zavodszky MI, Lei M, Thorpe MF, Day AR, Kuhn LA. Modeling correlated main-chain motions in proteins for flexible molecular recognition. Proteins. 2004;57:243–261. doi: 10.1002/prot.20179. [DOI] [PubMed] [Google Scholar]
- 40.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Sanner MF. FLIPDock: Docking flexible ligands into flexible receptors. Proteins. 2007;68:726–737. doi: 10.1002/prot.21423. [DOI] [PubMed] [Google Scholar]
- 43.May A, Zacharias M. Protein-ligand docking accounting for receptor side chain and global flexibility in normal modes: Evaluation on kinase inhibitor cross docking. J. Med. Chem. 2008;51:3499–3506. doi: 10.1021/jm800071v. [DOI] [PubMed] [Google Scholar]
- 44.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers. 2003;68:76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 45.Miller MD, Kearsley SK, Underwood DJ, Sheridan RP. FLOG: A system to select quasi-flexible ligands complementary to a receptor of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8:153–174. doi: 10.1007/BF00119865. [DOI] [PubMed] [Google Scholar]
- 46.Pang YP, Perola E, Xu K, Prendergast FG. EUDOC: A computer program for identification of drug interaction sites in macromolecules and drug leads from chemical databases. J. Comput. Chem. 2001;22:1750–1771. doi: 10.1002/jcc.1129. [DOI] [PubMed] [Google Scholar]
- 47.Venkatachalam CM, Jiang X, Oldfield T, Waldan M. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 48.Jain AN. Surflex: Fully automatic molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 49.Sauton N, Lagorce D, Villoutreix B, Miteva M. MS-DOCK: Accurate multiple conformation generator and rigid docking protocol for multi-step virtual ligand screening. BMC Bioinformatics. 2008;9:184. doi: 10.1186/1471-2105-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorber DM, Shoichet BK. Hierarchical docking of databases of multiple ligand conformations. Curr. Top. Med. Chem. 2005;5:739–749. doi: 10.2174/1568026054637683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 52.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 53.Ewing TJA, Kuntz ID. Critical evaluation of search algorithms for automated molecular docking and database screening. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- 54.Bohm HJ. The computer program LUDI: A new method for the de novo design of enzyme inhibitors. J. Comput. Aided Mol. Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 55.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 56.Mizutani MY, Tomioka N, Itai A. Rational automatic search method for stable docking models of protein and ligand. J. Mol. Biol. 1994;243:310–326. doi: 10.1006/jmbi.1994.1656. [DOI] [PubMed] [Google Scholar]
- 57.Zsoldos Z, Reid D, Simon A, Sadjad BS, Johnson AP. eHiTS: An innovative approach to the docking and scoring function problems. Curr. Protein Pept. Sci. 2006;7:421–435. doi: 10.2174/138920306778559412. [DOI] [PubMed] [Google Scholar]
- 58.Lorber DM, Shoichet BK. Flexible ligand docking using conformational ensembles. Protein Sci. 1998;7:938–950. doi: 10.1002/pro.5560070411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joseph-McCarthy D, Thomas BEIV, Belmarsh M, Moustakas D, Alvarez JC. Pharmacophore-based molecular docking to account for ligand flexibility. Proteins. 2003;51:172–188. doi: 10.1002/prot.10266. [DOI] [PubMed] [Google Scholar]
- 60.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J. Comput. Chem. 2008;29:1574–1588. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hart TN, Read RJ. A multiple-start Monte Carlo docking method. Proteins. 1992;13:206–222. doi: 10.1002/prot.340130304. [DOI] [PubMed] [Google Scholar]
- 62.McMartin C, Bohacek RS. QXP: Powerful, rapid computer algorithms for structure-based drug design. J. Comput. Aided Mol. Des. 1997;11:333–344. doi: 10.1023/a:1007907728892. [DOI] [PubMed] [Google Scholar]
- 63.Trosset JY, Scheraga HA. Prodock: Software package for protein modeling and docking. J. Comput. Chem. 1999;20:412–427. [Google Scholar]
- 64.Liu M, Wang S. MCDOCK: A Monte Carlo simulation approach to the molecular docking problem. J. Comput. Aided Mol. Des. 1999;13:435–451. doi: 10.1023/a:1008005918983. [DOI] [PubMed] [Google Scholar]
- 65.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 66.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 67.Clark KP. Flexible ligand docking without parameter adjust-ment across four ligand-receptor complexes. J. Comput. Chem. 1995;16:1210–1226. [Google Scholar]
- 68.Taylor JS, Burnett RM. DARWIN: A program for docking flexible molecules. Proteins. 2000;41:173–191. [PubMed] [Google Scholar]
- 69.Thomsen R, Christensen MH. MolDock: A new technique for highaccuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 70.Pei J, Wang Q, Liu Z, Li Q, Yang KL, Lai L. PSI-DOCK: Towards highly efficient and accurate flexible ligand docking. Proteins. 2006;62:934–946. doi: 10.1002/prot.20790. [DOI] [PubMed] [Google Scholar]
- 71.Stroganov OV, Novikov FN, Stroylov VS, Kulkov V, Chilov GG. Lead finder: An approach to improve accuracy of protein-ligand docking, binding energy estimation, and virtual screening. J. Chem. Inf. Model. 2008;48:2371–2385. doi: 10.1021/ci800166p. [DOI] [PubMed] [Google Scholar]
- 72.Grosdidier A, Zoete V, Michielin O. EADock: Docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins. 2007;67:1010–1025. doi: 10.1002/prot.21367. [DOI] [PubMed] [Google Scholar]
- 73.Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD. Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins. 1998;33:367–382. [PubMed] [Google Scholar]
- 74.Chen H-M, Liu B-F, Huang H-L, Hwang S-F, Ho S-Y. SODOCK: Swarm optimization for highly flexible protein-ligand docking. J. Comput. Chem. 2007;28:612–623. doi: 10.1002/jcc.20542. [DOI] [PubMed] [Google Scholar]
- 75.Chen K, Li T, Cao T. Tribe-PSO: A novel global optimization algorithm and its application in molecular docking. Chemom. Intell. Lab. Syst. 2006;82:248–259. [Google Scholar]
- 76.Namasivayam V, Gunther R. PSO@Autodock: A fast flexible molecular docking program based on swarm intelligence. Chem. Biol. Drug. Des. 2007;70:475–484. doi: 10.1111/j.1747-0285.2007.00588.x. [DOI] [PubMed] [Google Scholar]
- 77.Korb O, Stutzle T, Exner TE. PLANTS: Application of ant colony optimization to structure-based drug design. Ant Colony Optimization and Swarm Intelligence, 5th International Workshop; Brussels, Belgium. 4–7 September, 2006; pp. 247–258. [Google Scholar]
- 78.Gohlke H, Klebe G. Statistical potentials and scoring functions applied to protein-ligand binding. Curr. Opin. Struct. Biol. 2001;11:231–235. doi: 10.1016/s0959-440x(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 79.Schulz-Gasch T, Stahl M. Scoring functions for protein-ligand interactions: A critical perspective. Drug Discov. Today: Tech. 2004;1:231–239. doi: 10.1016/j.ddtec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Jain AN. Scoring functions for protein-ligand docking. Curr. Protein Pept. Sci. 2006;7:407–420. doi: 10.2174/138920306778559395. [DOI] [PubMed] [Google Scholar]
- 81.Rajamani R, Good AC. Ranking poses in structure-based lead discovery and optimization: Current trends in scoring function development. Curr. Opin. Drug. Discov. Devel. 2007;10:308–315. [PubMed] [Google Scholar]
- 82.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 83.Huang N, Kalyanaraman C, Irwin JJ, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. J. Chem. Inf. Model. 2006;46:243–253. doi: 10.1021/ci0502855. [DOI] [PubMed] [Google Scholar]
- 84.Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy approach to macromolecule-ligand interactions. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- 85.Weiner PK, Kollman PA. AMBER – assisted model building with energy refinementla general program for modeling molecules and their interactions. J. Comput. Chem. 1981;2:287–303. [Google Scholar]
- 86.Nilsson L, Karplus M. Empirical energy functions for energy minimization and dynamics of nucleic acids. J. Comput. Chem. 1986;7:591–616. [Google Scholar]
- 87.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM – a programm for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 88.Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: Recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu. Rev. Biophys. Biomol. Struct. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 89.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: Applications to the molecular systems and geometric objects. J. Comput. Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 90.Grant JA, Pickup BT, Nicholls A. A smooth permittivity function for Poisson-Boltzmann solvation methods. J. Comput. Chem. 2001;22:608–640. [Google Scholar]
- 91.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei BQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 93.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 94.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. Validation and use of the MM-PBSA approach for drug discovery. J. Med. Chem. 2005;48:4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- 95.Kuhn B, Kollman PA. Binding of a diverse set of ligands to avidin and strepavidin: An accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J. Med. Chem. 2000;43:3786–3791. doi: 10.1021/jm000241h. [DOI] [PubMed] [Google Scholar]
- 96.Pearlman DA. Evaluating the molecular mechanics poisson-boltzmann surface area free energy method using a congeneric series of ligands to p38 MAP kinase. J. Med. Chem. 2005;48:7796–7807. doi: 10.1021/jm050306m. [DOI] [PubMed] [Google Scholar]
- 97.Sims PA, Wong CF, McCammon JA. A computational model of binding thermodynamics: The deisgn of cyclin-dependent kinase 2 inhibitors. J. Med. Chem. 2003;46:3314–3325. doi: 10.1021/jm0205043. [DOI] [PubMed] [Google Scholar]
- 98.Huang D, Caflisch A. Efficient evaluation of binding free energy using continuum electrostatics solvation. J. Med. Chem. 2004;47:5791–5797. doi: 10.1021/jm049726m. [DOI] [PubMed] [Google Scholar]
- 99.Thompson DC, Humblet C, Joseph-McCarthy D. Investigation of MM-PBSA rescoring of docking poses. J. Chem. Inf. Model. 2008;48:1081–1091. doi: 10.1021/ci700470c. [DOI] [PubMed] [Google Scholar]
- 100.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 101.Zou X, Sun Y, Kuntz ID. Inclusion of solvation in ligand binding free energy calculations using the generalized-Born model. J. Am. Chem. Soc. 1999;121:8033–8043. [Google Scholar]
- 102.Liu H-Y, Kuntz ID, Zou X. Pairwise GB/SA scoring function for structure-based drug design. J. Phys. Chem. B. 2004;108:5453–5462. [Google Scholar]
- 103.Liu H-Y, Zou X. Electrostatics of ligand binding: Parametrization of the generalized born model and comparison with the Poisson-Boltzmann approach. J. Phys. Chem. B. 2006;110:9304–9313. doi: 10.1021/jp060334w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu H-Y, Grinter SZ, Zou X. Multiscale generalized born modeling of ligand binding energies for virtual database screening. J. Phys. Chem. B. 2009;113:11793–11799. doi: 10.1021/jp901212t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Majeux N, Scarsi M, Apostolakis J, Ehrhardt C, Caflisch A. Exhaustive docking of molecular fragments with electrostatic solvation. Proteins. 1999;37:88–105. [PubMed] [Google Scholar]
- 106.Cecchini M, Kolb P, Majeux N, Caflisch A. Automated docking of highly flexible ligands by genetic algorithms: A critical assessment. J. Comput. Chem. 2004;25:412–422. doi: 10.1002/jcc.10384. [DOI] [PubMed] [Google Scholar]
- 107.Huang D, Luthi U, Kolb P, Edler K, Cecchini M, Audetat S, Barberis A, Caflisch A. Discovery of cell-permeable non-peptide inhibitors of beta-secretase by high-throughput docking and continuum electrostatics calculations. J. Med. Chem. 2005;48:5108–5111. doi: 10.1021/jm050499d. [DOI] [PubMed] [Google Scholar]
- 108.Cho AE, Wendel JA, Vaidehi N, Kekenes-Huskey PM, Floriano WB, Maiti PK, Goddard WA., II The MPSim-Dock hierarchical docking algorithm: Application to the eight trypsin inhibitor cocrystals. J. Comput. Chem. 2005;26:48–71. doi: 10.1002/jcc.20118. [DOI] [PubMed] [Google Scholar]
- 109.Ghosh A, Rapp CS, Friesner RA. Generalized Born model based on a surface integral formulation. J. Phys. Chem. B. 1998;102:10983–10990. [Google Scholar]
- 110.Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J. Med. Chem. 2006;49:4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 111.Guimaraes CRW, Cardozo M. MM-GB/SA rescoring of docking poses in structure-based lead optimization. J. Chem. Inf. Model. 2008;48:958–970. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- 112.Jain AN. Scoring noncovalent protein-ligand interactions: A continuous differentiable function tuned to compute binding affinities. J. Comput.-Aided Mol. Des. 1996;10:427–440. doi: 10.1007/BF00124474. [DOI] [PubMed] [Google Scholar]
- 113.Head RD, Smythe ML, Oprea TI, Waller CL, Green SM, Marshall GR. Validate a new method for the receptor-based prediction of binding affinities of novel ligands. J. Am. Chem. Soc. 1996;118:3959–3969. [Google Scholar]
- 114.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 115.Böhm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput.-Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 116.Wang R, Liu L, Lai L, Tang Y. SCORE: A new empirical method for estimating the binding affinity of a protein-ligand complex. J. Mol. Model. 1998;4:379–394. [Google Scholar]
- 117.Böhm HJ. Prediction of binding constants of ptotein ligands: A fast method for the polarization of hits obtained from de novo design or 3D database search programs. J. Comput.-Aided Mol. Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 118.Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M. LigScore: A novel scoring function for predicting binding affinities. J. Mol. Graph. Model. 2005;23:395–407. doi: 10.1016/j.jmgm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 119.Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Freer ST. Molecular recognition of the inhibitor AG-1343 by HIV-1 Protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 1995;2:317–324. doi: 10.1016/1074-5521(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 120.Gehlhaar DK, Bouzida D, Rejto PA. In: Rational Drug Design: Novel Methodology and Practical Applications. Parrill L, Reddy MR, editors. Vol. 719. American Chemical Society; Washington, DC, USA: 1999. pp. 292–311. [Google Scholar]
- 121.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 122.Yin S, Biedermannova L, Vondrasek J, Dokholyan NV. MedusaScore: An accurate force-field based scoring function for virtual drug screening. J. Chem. Inf. Model. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raub S, Steffen A, Kämper A, Marian CM. AIScore – Chemically diverse empirical scoring function employing quantum chemical binding energies of hydrogen-bonded complexes. J. Chem. Inf. Model. 2008;48:1492–1510. doi: 10.1021/ci7004669. [DOI] [PubMed] [Google Scholar]
- 124.Sotriffer CA, Sanschagrin P, Matter H, Klebe G. SFCscore: Scoring functions for affinity prediction of protein-ligand complexes. Proteins. 2008;73:395–419. doi: 10.1002/prot.22058. [DOI] [PubMed] [Google Scholar]
- 125.Tanaka S, Scheraga HA. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules. 1976;9:945–950. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- 126.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: Quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
- 127.Sippl MJ. Calculation of conformational ensembles from potentials of mean force. J. Mol. Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 128.Verkhivker G, Appelt K, Freer ST, Villafranca JE. Empirical free energy calculations of ligand-protein crystallographic complexes. I. Knowledge-based ligand-protein interaction potentials applied to the prediction of human immunodeficiency virus 1 protease binding affinity. Protein Eng. 1995;8:677–691. doi: 10.1093/protein/8.7.677. [DOI] [PubMed] [Google Scholar]
- 129.Huang S-Y, Zou X. Mean-force scoring functions for protein-ligand binding. Annu. Rep. Comput. Chem. 2010;6:281–296. [Google Scholar]
- 130.Thomas PD, Dill KA. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. USA. 1996;93:11628–11633. doi: 10.1073/pnas.93.21.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koppensteiner WA, Sippl MJ. Knowledge-based potentials – Back to the roots. Biochemistry (Moscow) 1998;63:247–252. [PubMed] [Google Scholar]
- 132.Thomas PD, Dill KA. Statistical potentials extracted from protein structures: How accurate are they? J. Mol. Biol. 1996;257:457–469. doi: 10.1006/jmbi.1996.0175. [DOI] [PubMed] [Google Scholar]
- 133.McQuarrie DA. Statistical Mechanics. Harper Collins Publishers; New York, NY, USA: 1976. [Google Scholar]
- 134.Muegge I, Martin YC. A general and fast scoring function for protein-ligand interactions: A simplified potential approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 135.Muegge I. PMF scoring revisited. J. Med. Chem. 2006;49:5895–5902. doi: 10.1021/jm050038s. [DOI] [PubMed] [Google Scholar]
- 136.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: I. Derivation of interaction potentials. J. Comput. Chem. 2006;27:1866–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 137.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: II. Validation of the scoring function. J. Comput. Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- 138.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 139.Velec HFG, Gohlke H, Klebe G. DrugScoreCSD-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J. Med. Chem. 2005;48:6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- 140.DeWitte RS, Shakhnovich EI. SMoG: de Novo design method based on simple, fast, and accutate free energy estimate. 1. Methodology and supporting evidence. J. Am. Chem. Soc. 1996;118:11733–11744. [Google Scholar]
- 141.Ishchenko AV, Shakhnovich EI. Small molecule growth 2001 (SMoG2001): An improved knowledge-based scoring function for protein-ligand interactions. J. Med. Chem. 2002;45:2770–2780. doi: 10.1021/jm0105833. [DOI] [PubMed] [Google Scholar]
- 142.Mitchell JBO, Laskowski RA, Alex A, Thornton JM. BLEEP – Potential of mean force describing protein-ligand interactions: I. Generating potential. J. Comput. Chem. 1999;20:1165–1176. [Google Scholar]
- 143.Mitchell JBO, Laskowski RA, Alex A, Forster MJ, Thornton JM. BLEEP – Potential of mean force describing protein-ligand interactions: II. Calculation of binding energies and comparison with experimental data. J. Comput. Chem. 1999;20:1177–1185. [Google Scholar]
- 144.Mooij WT, Verdonk ML. General and targeted statistical potentials for protein-ligand interactions. Proteins. 2005;61:272–287. doi: 10.1002/prot.20588. [DOI] [PubMed] [Google Scholar]
- 145.Yang CY, Wang RX, Wang SM. M-score: A knowledge-based potential scoring function accounting for protein atom mobility. J. Med. Chem. 2006;49:5903–5911. doi: 10.1021/jm050043w. [DOI] [PubMed] [Google Scholar]
- 146.Zhao X, Liu X, Wang Y, Chen Z, Kang L, Zhang H, Luo X, Zhu W, Chen K, Li H, Wang X, Jiang H. An improved PMF scoring function for universally predicting the interactions of a ligand with protein, DNA, and RNA. J. Chem. Inf. Model. 2008;48:1438–1447. doi: 10.1021/ci7004719. [DOI] [PubMed] [Google Scholar]
- 147.Zhang C, Liu S, Zhu Q, Zhou Y. A knowledge-based energy function for protein-ligand, protein-protein, and protein-DNA complexes. J. Med. Chem. 2005;48:2325–2335. doi: 10.1021/jm049314d. [DOI] [PubMed] [Google Scholar]
- 148.Huang S-Y, Zou X. Inclusion of solvation and entropy in the knowledge-based scoring function for protein-ligand interactions. J. Chem. Inf. Model. 2010;50:262–273. doi: 10.1021/ci9002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Charifson PS, Corkery JJ, Murcko MA, Walters WP. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J. Med. Chem. 1999;42:5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 150.Wang R, Wang S. How does consensus scoring work for virtual library screening? An idealized computer experiment. J. Chem. Inf. Comput. Sci. 2001;41:1422–1426. doi: 10.1021/ci010025x. [DOI] [PubMed] [Google Scholar]
- 151.Clark RD, Strizhev A, Leonard JM, Blake JF, Matthew JB. Consensus scoring for ligand/protein interactions. J. Mol. Graph. Model. 2002;20:281–295. doi: 10.1016/s1093-3263(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 152.Terp GE, Johansen BE, Christensen IT, Jorgensen FS. A new concept for multidimensional selection of ligand conformations (MultiSelect) and multidimensional scoring (MultiScore) of protein-ligand binding affinities. J. Med. Chem. 2001;44:2333–2343. doi: 10.1021/jm001090l. [DOI] [PubMed] [Google Scholar]
- 153.Cole JC, Murray CW, Nissink JWM, Taylor RD, Taylor R. Comparing protein-ligand docking programs is difficult. Proteins. 2005;60:325–332. doi: 10.1002/prot.20497. [DOI] [PubMed] [Google Scholar]
- 154.Chen H, Lyne PD, Giordanetto F, Lovell T, Li J. On evaluating molecular-docking methods for pose prediction and enrichment factors. J. Chem. Inf. Model. 2006;46:401–415. doi: 10.1021/ci0503255. [DOI] [PubMed] [Google Scholar]
- 155.Jain AN, Nicholls A. Recommendations for evaluation of computational methods. J. Comput.-Aided Mol. Des. 2008;22:133–139. doi: 10.1007/s10822-008-9196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hawkins PCD, Warren GL, Skillman AG, Nicholls A. How to do an evaluation: Pitfalls and traps. J. Comput.-Aided Mol. Des. 2008;22:179–190. doi: 10.1007/s10822-007-9166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kirchmair J, Markt P, Distinto S, Wolber G, Langer T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selectionsWhat can we learn from earlier mistakes. J. Comput.-Aided Mol. Des. 2008;22:213–228. doi: 10.1007/s10822-007-9163-6. [DOI] [PubMed] [Google Scholar]
- 158.Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 159.Stahl M, Rarey M. Detailed analysis of scoring functions for virtual screening. J. Med. Chem. 2001;44:1035–1042. doi: 10.1021/jm0003992. [DOI] [PubMed] [Google Scholar]
- 160.Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL., III Assessing scoring functions for protein-ligand interactions. J. Med. Chem. 2004;47:3032–3047. doi: 10.1021/jm030489h. [DOI] [PubMed] [Google Scholar]
- 161.Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000;43:4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 162.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56:235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 163.Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 164.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 165.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., III Comparative study of several algorithms for flexible ligand docking. J. Comput. Aided Mol. Des. 2003;17:755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 166.Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J. Comput. Chem. 2008;29:1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J. Med. Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: A feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ruvinsky AM. Calculations of protein-ligand binding entropy of relative and overall molecular motions. J. Comput.-Aided Mol. Des. 2007;21:361–370. doi: 10.1007/s10822-007-9116-0. [DOI] [PubMed] [Google Scholar]
- 170.Chang MW, Belew RK, Carroll KS, Olson AJ, Goodsell DS. Empirical entropic contributions in computational docking: Evaluation in APS reductase complexes. J. Comput. Chem. 2008;29:1753–1761. doi: 10.1002/jcc.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee J, Seok C. A statistical rescoring scheme for protein-ligand docking: Consideration of entropic effect. Proteins. 2008;70:1074–1083. doi: 10.1002/prot.21844. [DOI] [PubMed] [Google Scholar]