Abstract
The ZNF198-FGFR1 fusion kinase is a constitutively activated tyrosine kinase associated with a specific atypical myeloproliferative disease. The chimeric protein localizes to the cytoplasm, unlike the wild type FGFR1 receptor kinase, and presumably inappropriately phosphorylates specific targets as part of the oncogenic signaling cascade. Other than known targets of the FGFR1 kinase itself, few specific targets of ZNF198-FGFR1 have been identified. Using a genetically engineered HEK 293 cell system, we have identified proteins that are specifically phosphorylated in the presence of the fusion kinase using anti-phosphotyrosine immunoprecipitation and mass spectrometry. Compared with 293 cells expressing exongenous wild type FGFR1, ZNF198-FGFR1 is associated with phosphorylation of several proteins including SSBP2, ABL, FLJ14235, CALM and TRIM4 proteins. The specificity of the phosphorylation events in the SSBP2 and ABL proteins, which have previously been implicated in leukemogenesis, were further confirmed independently using immunoprecipitation with protein-specific antibodies and Western blotting. The MS analysis also identified the phosphorylation events in the ZNF198 moiety in the chimeric protein that might be related to its function. These studies identify the intersection of several different leukemia-related pathways in the development of this myeloproliferative disorder and provide new insights into the substrates of FGFR1 under defined conditions.
Keywords: Mass spectrometry, Phosphopeptide fingerprinting, FGFR1 fusion kinase, Abl kinase, Myeloproliferative disease
INTRODUCTION
The fusion of the zinc finger domain of the ZNF198 gene with the kinase domain of the FGFR1 (fibroblast growth factor receptor-1) gene was originally defined (1–4) in an AMPD (atypical myeloproliferative disease) with poor prognosis (1–3;5;6). Since then, variant translocations have been described (7–13) where, although the partner gene fused with FGFR1 is different, in all cases analyzed so far, the specific partner gene is anticipated to facilitate dimerization of the FGFR1 kinase domain resulting in its constitutive activation (14–16). The chimeric kinase protein, unlike FGFR1, is no longer tethered to the cytoplasmic membrane but resides predominantly in the cytoplasm (14;17) and acts as a constitutive tyrosine kinase. Members of the STAT family of transcription factors represent one of the well characterized protein families phosphorylated in the presence of the ZNF198/FGFR1 fusion kinase and are constitutively activated in the presence of the various fusion kinases (14;18;19). In the BaF3 growth factor-dependent hematopoietic cells, the fusion kinases can confer growth factor independence, although when expressed in primary mouse hematopoietic stem cells, is apparently not sufficient in itself to cause cell transformation (20).
Various approaches have been taken to investigate the function of the FGFR1 fusion kinases more extensively. The constitutive dimerization of the wild type ZNF198 gene suggests that this event is important in the function of the protein. We have shown that the fusion kinase can dimerize with the endogenous wild type gene (14;17) and thereby potentially affect the wild type protein in a dominant negative way. Thus, one of the early observations was that ZNF198 was found in a protein complex with the UBE2 (HHR6) and RAD18 proteins (21), which are essential for the correct repair of UVB- induced chromosome breaks and cell survival. In the presence of the fusion kinase this function is compromised and cells were more sensitive to this DNA damaging agent. Mass spectrometry studies also demonstrated that ZNF198 existed in a complex with a number of proteins predicted to be involved in RNA processing and transcription (22), and that some of these interactions were lost in the presence of the fusion kinase. In normal cells, ZNF198 is localized to punctate structures in the nucleus of most cells (15;17). In the presence of the fusion kinase, these structures are lost, presumably because the cytoplasmic location of the kinase sequesters the wild type gene in the cytoplasm (17). We recently demonstrated that ZNF198 interacts with SUMO1 and is localized in PML bodies. Mutation of the sumoylation site in ZNF198 results in loss of PML bodies and a predominantly cytoplasmic membrane localization of ZNF198. In the presence of the fusion kinase, the PML bodies are also lost. Thus, from all of these observations suggest that the expression of the fusion kinase may interfere with important protein-protein interactions in the cell, which may contribute to the oncogenicity of this protein. Gene expression studies have also shown that the SERPINB2 and HSPA1A genes are upregulated by ZNF198/FGFR1 and their presence is required to maintain stability of the chimeric protein (23;24).
The observations that the STAT proteins are constitutively activated in the presence of the fusion kinase points to another of its potential transforming activities - the inappropriate activation of target genes in the cytoplasm through phosphorylation of critical tyrosine residues. Identifying these target proteins, which can be clearly identified in western blots from whole cell lysates expressing the kinase gene (H Baumann, Roswell Park Cancer Institute, pers comm.), has been difficult and largely restricted to candidate gene approaches. Several proteins activated by the FGFR1 kinase domain are also activated in cells expressing the fusion kinase, although it is presumably the proteins that are specifically activated by the chimeric protein which are likely involved in transformation of the leukemic stem cell. Recently, Rush et al (25) described an approach to specifically identify proteins undergoing tyrosine phosphorylation using a combination of IP (immunoprecipitation) and MS (mass spectrometry). Proteins from whole cell lysates expressing a chimeric tyrosine kinase were compared to cells not expressing the fusion kinase gene. IP of tryptic peptides using anti-phosphotytrosine antibodies were prepared from the different cells and the recovered peptides were analysed using linear ion trap MS. Proteins that were potential targets of the tyrosine kinases in question were identified in cells expressing the chimeric protein but not in cells that did not. Here we have applied this approach to identify proteins specifically phosphorylated by the ZNF198/FGFR1 protein in this model system. This analysis demonstrated that the ZNF198 moiety in the fusion gene is phosphorylated at multiple positions and we have identified the ABL kinase and SSBP2 genes as specific targets for phosphorylation. Both of these proteins have independently been shown to play important roles in leukemogenesis.
MATERIALS AND METHODS
Cell lines
The generation of HEK 293 cells stably expressing GFP (green fluorescent protein)-tagged ZNF198-FGFR1 fusion kinase were described previously (19). These cells show a cytoplasmic localization for the chimeric protein. The FGFR1 gene was amplified from 293 cells using RT-PCR, cloned into pcDNA3 and HEK 293 cells stably expressing the exogenous gene were isolated in 500ug/ml G418. Specific gene expression was confirmed using RT-PCR and protein expression was confirmed by western blotting using anti-FGFR1 antibodies. Addition of bFGF to these cells results in the constitutive activation of the exogenous gene. All cells were maintained in DMEM with 10% FBS in 5% CO2.
Immunoprecipitations and Western blotting
Cells were grown to 80% confluency and then washed twice with PBS. Lysis was achieved in RIPA buffer (50mM Tris containing 150mM NaCl 0.1% SDS 1% TritonX-100 1% sodium deoxycholate pH7.2) with 0.2% protease and phosphatase inhibitor cocktail (Sigma, MO) on ice for 10 min. Following centrifugation at 14,000 rpm for 20 min the supernatant was collected and used for IP (immunoprecipitation). Lysates were precleared using sepharose-coupled rabbit or mouse IgG and then the 500μl of supernatant was treated with 3μl of anti GFP rabbit polyclonal serum (Clontech) or 50μl of FGFR1 (Upstate Biotechnology, Lake Placid, NY), or SSBP2 and ABL (Cell Signaling) antibodies. The immune complexes were pulled down with Protein A- or Protein G-sepharose. The sepharose beads were washed five times with PBS and then proteins were eluted by incubation in SDS-sample buffer at 100°C for 3min and aliquots were subjected to SDS-PAGE (polyacrylamide gel electrophoresis) and western blot analysis. A polyclonal antibody against SSBP2 (24), a monoclonal antibody against GFP (Covance, New Jersey), and a rabbit polyclonal antibody against ABL were used for western blot analysis.
Mass Spectrometry
Lysates from 2 × 108 cells were recovered from each cell line and the phosphoproteins prepared as described previously (25;26). In short, cell lysates were precleared by centrifugation, reduced with DTT and alkylated with iodoacetamide. Proteins was then digested with trypsin and recovered by precipitation using trifluoroacetic acid (25). Individual fractions were then recovered from Sep-Pak columns using increasing concentrations of acetonitrile. Peptides from each fraction were immunoaffinity purified using anti-phospho-tyrosine P-Tyr-100 (Cell Signaling Technology) coupled resin and recovered using 0.1% TFA as described previously (25). Tandem mass spectra were collected in a data-dependent manner with an LCQ Deca XP Plus ion trap mass spectrometer (ThermoFinnigan, San Jose, CA), using a top-4 method, a dynamic exclusion repeat count of 1, and a repeat duration of 0.5 minute. Each biological sample was analyzed on two separate occasions and the data combined for analysis. TurboSequest (ThermoFinnigan) searches were performed against the National Center for Biotechnology Information (NCBI) human database released on March 4, 2008 containing 37, 742 protein entries allowing oxidized methionine (M_16) and phosphorylation (Y_80) as dynamic modifications.
False discoveries were controlled using the following stepwise processing of the data. Step 1: all small f assignments were first queried where phospho_num > 0 and rsp <= 10 and ((charge =1 and xcorr>=1.5) or (charge =2 and xcorr >=2.2) or (charge=3 and xcorr >=3.3)). Step 2: false positive assignments were removed according to; p = 0 and (z = 1, XCorr < 2.0 or z = 2, XCorr < 2.6 or z >= 3, Xcorr 3.6). Step 3: false negative assignments were then added where; CT (peptide count) >= 20 and fH >= 0.25 and Xcorr >= 1.5. In our experience this approach can limit the FDR to ~5%. The data from each independent analysis for each of the samples was combined and peptides identified in either analysis were compiled in the summary tables S1 and S2. A summary of the frequencies of the peptides identified in the combined analysis are also shown in supplemental table S1 and the associated technical/analytical details can be found in supplemental table S2.
RESULTS
The primary objective of this study was to identify proteins that are phosphorylated on tyrosine residues in the presence of the ZNF198-FGFR1 fusion kinase. Since there are no cell lines known to carry this specific fusion gene, or any of its common variants (see Discussion), we used a well characterized HEK293 model, involving a series of cell clones which express an exogenous ZNF198-FGFR1 transcript or an exogenous wild type FGFR1 transcript (14;22–24). Clones expressing the empty GFP vector were used as controls. This system provides a homogeneous genetic background on which the various exogenous genes that are expressed represent the only difference between the clones. The HEK293 cells expressing the fusion kinase have previously been shown to demonstrate activation of various members of the STAT family of transcription factors (14) and localize to the cytoplasm (17). These observations are consistent with other model cell systems used in the analysis of the FGFR1-chimeric kinases (19;27). We therefore used these cells in a comparison of peptides containing PY (phosphotyrosine) residues using the whole-proteome scan procedure described by Rush et al (25), involving IP and MS. Since the normal FGFR1 receptor, in the presence of bFGF, will also result in specific protein phosphorylation events, we also generated HEK293 cells stably expressing an exogenous wild type FGFR1 gene, which was shown to be activated by bFGF treatment, to identify proteins which are the natural substrates of the FGFR1 kinase in this system for comparison with the fusion kinase.
Total protein was isolated from lysates of the various cell clones and subjected to trypsin digestion. The tryptic digests were then mixed with anti-phosphotyrosine antibody-bound beads and the immunoprecipitated peptides were recovered and subjected to mass spectrometry. The data were then processed to reveal which PY events were common among the different clones and which were specific. The overall results from this analysis are summarized in tables 1 and 2. The full details are given in Supplemental tables S1 and S2. Although we were primarily interested in targets for ZNF198-FGFR1, this analysis also allowed us to review phosphorylation events related directly to FGFR1 activity under various conditions.
Table 1.
Summary of phosphorylated peptides seen specifically in 293 cells expressing FGFR1 before (F) and after induction by bFGF (FF) and those expressing ZNF198-FGFR1 (ZF). The identity of the peptides is given for each protein and the amino acid location of the modified tyrosine within that protein are shown.
| Peptides only identified in bFGF induced FGFR1 cells (FF) | ||
| Abi-1 | Y197 | NTPyKTLEPVKPPTVPNDyMTSPAR |
| BUD31 | Y97 | QGyENLCCLR |
| COPB | Y354 | DMGSCEIyPQTIQHNPNGR |
| DCBLD2 | Y715 | ATGNQPPPLVGTyNTLLSR |
| eIF5B | Y134 | PKVEMySGSDDDDDFNKLPK |
| EphA2 | Y772 | VLEDDPEATyTTSGGKIPIR |
| FGFR1 | Y605; | DLVSCAyQVAR |
| FGFR3 | Y599 | DLVSCAyQVAR |
| FLJ34658 | Y123 | LMEIFGTQCSyLLSR |
| hnRNP A1 hnRNP |
Y357 | NQGGyGGSSSSSSYGSGR |
| A2/B1 | Y336 | NMGGPYGGGNyGPGGSGGSGGYGGR |
| HSFY1 | Y175 | LKFyyNPNFK |
| HSFY1 | Y176 | LKFyyNPNFK |
| IGF2BP2 | Y40 | SGyAFVDYPDQNWAIR |
| ITGB3 | Y773 | WDTANNPLyK |
| KCTD12 | Y119 | EAEyFELPELVR |
| LDH-B | Y239 | MVVESAyEVIK |
| LOC120364 | Y300 | NQGGyGGSSSSSSYGSGR |
| LOC441495 | Y83 | AGPRDLLPSAPTPyPPGPAPSPK |
| MIA3 | Y1569 | AVSAAEEVKTyKR |
| PLXNC1 | Y1350 | VKEMyLTKLLSTKVAIHSVLEK |
| POLR1A | Y1126 | MWyELDEESRR |
| RPL38 | Y40 | VRCSRyLYTLVITDKEK |
| SDCCAG1 | Y883 | MKKMKEKyKDQDEEDRELIMK |
| SHIP-2 | Y986 | NSFNNPAyYVLEGVPHQLLPPEPPSPAR |
| SIPA1L3 | Y1141 | RPVSFPETPyTVSPAGADR |
| TLN1 | Y70 | ALDyYMLR |
| TTC2 | Y50 | DYNEAYNyYTK |
| HDLBP | Y437 | MDyVEINIDHK |
| VIM | Y61 | SLYASSPGGVyATR |
| ZFR | Y610 | RHRLQyKKKVNPDLQVEVK |
| Peptides identified in FF and ZF | ||
| CDK5 | Y15 | IGEGTyGTVFK |
| DYRK2 | Y382 | VYTyIQSR |
| DYRK4 | Y264 | VYTyIQSR |
| ENO1 | Y286 | SFIKDyPVVSIEDPFDQDDWGAWQK |
| FGFR1 | Y583 | RPPGLEyCyNPSHNPEEQLSSK |
| FGFR1 | Y654 | DIHHIDyyK |
| FGFR1 | Y653 | DIHHIDyyKK |
| PTPRA | Y798 | VVQEYIDAFSDyANFK |
| RDLP0 | Y24 | IIQLLDDyPK |
| RPS27 | Y31 | LVQSPNSyFMDVK |
| Peptides only identified in cells overexpressing FGFR1 (F) | ||
| BRF1 | Y115 | QPGGGQVNSSRyK |
| C12orf63 | Y1080 | DSyLEMALLyFHLKK |
| C12orf63 | Y1080 | DSyLEMALLyFHLKK |
| CASKIN2 | Y253 | NTyNQTALDIVNQFTTSQASR |
| Cdc2 | Y19 | IGEGTYGVVyK |
| CDK2 | Y19 | IGEGTYGVVyK |
| CDK3 | Y19 | IGEGTYGVVyK |
| CdkL5 | Y171 | NLSEGNNANYTEyVATR |
| CDV-3 | Y190 | KTPQGPPEIySDTQFPSLQSTAK |
| CENTD1 | Y473 | HSYPLSSTSGNADSSAVSSQAISPyACFYGASAK |
| DENND2C | Y195 | SLENIySEPEGQECGPSINPLPKPR |
| Dok1 | Y315 | IAPCPSQDSLySDPLDSTSAQAGEGVQR |
| Dok1 | Y449 | SHNSALySQVQK |
| GRF-1 | Y1087 | SVSSSPWLPQDGFDPSDyAEPMDAVVKPR |
| IL1F6 | Y96 | DIMDLyNQPEPVK |
| MAML2 | Y513 | IPSPSFGQQTFSPQSSPMPGVAGGSGQSKVMANyMyK |
| OCLN | Y287 | SNILWDKEHIyDEQPPNVEEWVK |
| p38-delta | Y182 | HADAEMTGyVVTR |
| Pcdhb20 | Y202 | ALDyEQEAELRLTLTAVDGGSPPKSGTTLVLIK |
| PKP4 | Y487 | NNYALNTTATYAEPYRPIQyR |
| PLCG1 | Y506 | NGILYLEDPVNHEWyPHYFVLTSSK |
| PLCG1 | Y783 | IGTAEPDYGALYEGRNPGFyVEANPMPTFK |
| RDX | Y134 | yGDYNKEIHK |
| RPL12 | Y14 | VVyLRCTGGEVGATSALAPK |
| SPTY2D1 | Y640 | yMESSWKEQQKEEAKSLR |
| SSX2IP | Y231 | DKKIAMDILNyVGRADGKR |
| Syk | Y323 | QESTVSFNPyEPELAPWAADKGPQR |
| TRAF6 | Y326 | METQSMyVSELK |
| ZO2 | Y1118 | IEIAQKHPDIyAVPIK |
| Peptides identified in F and FF | ||
| ELMO2 | Y48 | EVCDGWSLPNPEyYTLR |
| EphA7 | Y791 | VIEDDPEAVyTTTGGKIPVR |
| PSF | Y488 | FAQHGTFEyEYSQR |
| Shc1 | Y427 | ELFDDPSyVNVQNLDK |
| SUGT1 | Y317 | LFQQIySDGSDEVKR |
| TSRC1 | Y159 | SRLRDPIKPGMFGyGR |
| DLG3 | Y673 | RDNEVDGQDyHFVVSR |
| FGFR1 | Y585 | RPPGLEyCyNPSHNPEEQLSSK |
| FGFR1 | Y463 | LSSSGTPMLAGVSEyELPEDPRWELPR |
| Hrs | Y216 | VCEPCyEQLNR |
| Hrs | Y132 | VVQDTyQIMK |
| PIN4 | Y122 | FGyHIIMVEGR |
| PLCG1 | Y977 | ACyRDMSSFPETK |
Table 2.
Summary of phosphorylated peptides that were only identified in cells ZNF198-FGFR1 and peptides that were specifically not identified in cells expressing the fusion kinase although were present in the parental 293 cells and cells expressing exogenous FGFR1.
| Peptides identified only in the ZNF198-FGFR1 expressing cells | ||
| Abl | Y393 | LMTGDTyTAHAGAK |
| Arg | Y439 | LMTGDTyTAHAGAK |
| ANXA11 | Y482 | SLyHDISGDTSGDYR |
| CALM1 | Y99 | VFDKDGNGyISAAELR |
| DNTT | Y477 | MILDNHALyDKTK |
| ERO1L | Y73 | LQKLLESDyFR |
| FLJ36208 | Y37 | KPHYIPRPWGKPYNyK |
| hCG2045503 | Y58 | STyKGSFSAAVLLYR |
| IL1RL1 | Y179 | AHKSFLVIDNVMTEDAGDyTCK |
| InsR | Y1185 | DIyETDyYRK |
| IGF1R | Y1161 | DIyETDyYRK |
| PCDHB13 | Y192 | DGRKyPELVLDK |
| PCDHB3 | Y191 | DGRKyPELVLDK |
| PCDHB5 | Y191 | DGRKyPELVLDK |
| PCNT | Y3139 | LYLHyLRAESFRK |
| PPARBP | Y224 | yyVSPSDLLDDK |
| PPP1R12B | Y549 | GNEIPQTIAPSTyVSTYLK |
| Ran | Y147 | NLQYyDISAK |
| RESP18 | Y192 | CSyGGLDMMQAPGPSK |
| SMEK1 | Y169 | EKLALALENEGyIK |
| SMEK2 | Y171 | EKLALALENEGyIK |
| SSBP2 | Y192 | QQGHPNMGGPMQRMTPPRGMVPLGPQNyGGAMR |
| TAF172 | Y415 | yALAVRQDVINTLLPK |
| TRIM4 | Y152 | RLIRLLLyHSK |
| UMPS | Y37 | SGLSSPIyIDLR |
| VIM | Y53 | SLyASSPGGVYATR |
| YB-1 | Y162 | NYQQNyQNSESGEKNEGSESAPEGQAQQR |
| ZNF198 | Y502 | FCCQSCVSEyK |
| ZNF198 | Y557 | FFDMTQCIGPNGyMEPYCSTACMNSHK |
| ZNF198 | Y595 | NSLPQyQATMPDGK |
| ZNF198 | Y595, Y605 | NSLPQyQATMPDGKLyNFCNSSCVAK |
| ZNF198 | Y680 | LHCIVTyCEYCQEEK; LHCIVTyCEyCQEEK |
| ZNF198 | Y683 | LHCIVTYCEyCQEEK; LHCIVTyCEyCQEEK |
| ZNF198 | Y729 | CVTCNyCSQLCK |
| Peptides not identified in ZNF198-FGFR1 expressing cells | ||
| ANXA2 | Y23 | LSLEGDHSTPPSAyGSVK |
| EphA3 | Y779 | VLEDDPEAAyTTR |
| EphA4 | Y833 | VLEDDPEAAyTTR |
| EphA5 | Y779 | VLEDDPEAAyTTR |
| Fyn | Y213 | KLDNGGyYITTR |
| Yes | Y221 | KLDNGGyYITTR |
| JNK2 | Y185 | TACTNFMMTPyVVTR |
| Tyk2 | Y292 | LLAQAEGEPCyIR |
Phosphorylation events involving FGFR1
The 293 cells used as hosts for the various exogenous genes produce tumors when injected into nude or SCID (severe combined immunodeficiency) mice (JKC unpublished observations). As such, these transformed cells showed constitutive phosphorylation (activation) of a wide range of proteins (Table S1), many of which have been described in different transformed cell systems (see below). Since the MS approach described here is only semi-quantitative, changes in the overall cellular levels of these phosphorylation events could not be accurately determined, but provide the background of phosphorylation events present in these cells. Proteins in this category include cell cycle control genes such as cdk2/3, Erk1/2 and src (table S2).
Constitutive overexpression of receptor tyrosine kinases frequently result in the low level auto-activation (phosphorylation) of the receptor, and consequent phosphorylation of down stream targets. Thus, in cells over expressing FGFR1 in the absence of ligand, the receptor was shown to be autophosphorylated at several sites (Y463 and Y585) as expected, and in addition to these sites others (Y599, Y605) were also phosphorylated in the presence of bFGF. Several phosphorylation events (Y583, Y653 and Y654) were also seen in both ligand stimulated and ZNF198-FGFR1 expressing cells. Since these events were not identified in parental, unstimulated 293 cells (Table 1), which express very low levels of FGFR1 constitutively, they apparently result from over expression of the receptor. When bFGF was added phosphorylation of Y583, Y605, Y653 and Y654 was detected. Combinations of PY events were seen alone and together involving Y653/654 and Y583/585. These data further demonstrate that of over expression of the FGFR1 receptor can result in its auto phosphorylation in the absence of ligand.
Since FGFR1 overexpression activates the receptor, it would be expected that downstream phosphorylation events would accompany this activation event. It was possible, therefore, to define a series of proteins which were apparently specifically phosphorylated in the presence of over-expressed FGFR1 receptor but in the absence of ligand (table 1). In some cases, these events constituted different sites on the same proteins that were phosphorylated elsewhere in other cell clones, such as PLCG1, a well characterized target for FGFR1 [26]. Addition of the ligand, however, super-activates the receptor, and the targets induced by this stimulus predominate in the lysates used for MS and so appear as either novel targets of ligand activation, or are more readily detected simply as a result of the increased levels of their phosphorylation (table 1). In none of these specific cases, however, were the same proteins apparently phosphorylated by the ZNF198-FGFR1 fusion kinase at the particular sites described. A small subset of proteins, downstream of the FGFR1 activation, were identified which were phosphorylated both in the presence of the overexpressed FGFR1 gene as well as expression of the fusion kinase gene. This series includes the large ribosomal protein RPLP0, enolase 1 and at least one member of the dual specificity tyrosine phosphorylation regulated kinase (DYRK) family. These proteins are considered important targets of the fusion kinase, as well as over-expressed FGFR1 kinase in presence of the ligand, since the endogenous FGFR1 gene is not active in the 293 cells over expressing ZNF198-FGFR1 and, therefore, these events identify proteins that are phosphorylated by the FGFR1 kinase domain, whether it is within the context of the over expressed wild type receptor or the ZNF198 fusion protein, unlike other proteins described in table 2. Also included in this category (table S1) are a small group of proteins which were not detected as constitutively phosphorylated in HEK293 cells, but which were present both in the cells expressing the FGFR1 gene (either with or without stimulation by bFGF) and the fusion kinase gene, including Ack, CDK5, DCBLD2, PRPRA and PKM2 (pyruvate kinase M).
In contrast, several other proteins (e.g. tyk2, ANAX2 and members of the JNK and EPH families) were not phosphorylated in the presence of the fusion kinase but were constitutively phosphorylated in 293 cells (table 2). These proteins may represent consequences of loss of specific functions as a result of the abnormal expression of the fusion kinase, or its localization in a different intracellular compartment (17). It is also possible that the fusion kinase may affect transcription of these genes as a result of reduced expression of upstream effectors. The same is possibly also the case for genes such as IGF1R, InsR and CDH18, which were only detected as phosphorylated proteins in 293 cells (table S1). Although phosphorylation of members of the STAT family of proteins have been shown to be increased in the presence of the fusion kinase, we did not identify STATs 1 and 5 in this analysis. The reason for this is unclear but possibly if the phosphopeptides derived from these proteins were too large or too small then they would not be detected. STAT3 was identified by a single peptide as phosphorylated in the parental 293 cells (table S1) and so did not meet the criteria to be included in the group of unique proteins phosphorylated by the fusion kinase, even though western blotting clearly demonstrates significantly increased levels of phosphoprotein.
Auto phosphorylation sites on the ZNF198 component of the chimeric protein
Our previous studies had suggested that the ZNF198/FGFR1 kinase was autophosphorylated on the ZNF198 moiety present in the fusion kinase (14). Analysis of the phosphorylated peptides in this series of cell lines demonstrates that phosphorylation of ZNF198 was not identified in the parental 293 cells, or in those clones overexpressing FGFR1, arguing against phosphorylation of the endogenous ZNF198 protein. Only cells constitutively expressing the fusion kinase showed evidence of tyrosine phosphorylation on ZNF198 (table 2). As such, the individual tyrosines involved have now been identified at positions; Y502, Y557, Y595, Y605, Y680, Y683 and Y729. The breakpoint in the ZNF198 gene that gives rise to the ZNF198/FGFR1 fusion protein, is located between amino acids 913–914 (2;14). The MS observations, therefore, are consistent with this interpretation since, only tyrosine residues in ZNF198 that are retained in the fusion protein are phosphorylated, further supporting the idea that it is the dimerized chimeric protein that is being auto phosphorylated rather than the endogenous wild type ZNF198 protein in these cells. In addition, there was a suggestion of differential phosphorylation at Y595, Y605, Y680 and Y683, since peptides which had single tyrosine phosphorylation events, as well as peptides with double phosphorylation sites, were identified.
Proteins phosphorylated in the presence of ZNF198-FGFR1
Analysis of the MS data as described above, identified peptides showing tyrosine PY (phosphorylation) events that were seen exclusively in cells expressing the ZNF198-FGFR1 fusion kinase (table 2). Interestingly, proteins such as cadherin 18 and the insulin receptor were only identified in the parental 293 cells, which may simply reflect that induction of other protein phosphorylation events overwhelm the detection system in this study. Within this series of experiments, we identified phosphorylation events that were only seen in cells expressing the fusion kinase and these are defined in table 2. The TRIM4 (isoform 1) protein, had not been known to show PY until this study (see discussion). This gene is a member of the tetraspannin containing gene family of membrane bound proteins. The suppressor of MEK signaling like gene (SMEK1/2) is a relatively uncharacterized protein, with no known function, but its extensive homology to other members of this family implies it probably has a similar function. Two other proteins, ABL and SSBP2, which were identified as specifically phosphorylated in cells expressing the fusion kinase, however, have been previously implicated in leukemogenesis. The PY-peptide identified in ABL kinase is also present in the C-truncated isoform of this protein, as well as the closely related Arg protein, which has parallel function with ABL (28). It was not possible from the MS analysis alone, therefore, to determine directly which of these two genes was phosphorylated by ZNF198-FGFR1, or whether both of them were (see below). The SSBP2 protein was shown to be phosphorylated only at Y192 in this analysis. Another protein apparently showing specific phosphorylation in the cells expressing the fusion kinase was calmodulin, at Y99, which is a target for other receptor and non receptor kinases in different cell systems (29).
Proteins specifically not phosphorylated in the presence of ZNF198-FGFR1
In this analysis, a number of proteins were identified that were apparently specifically phosphorylated by over expression and ligand induction of FGFR1 (table 1), other proteins that were constitutively expressed in the parental 293 cells as well as being phosphorylated in cells carrying the fusion kinase. Another group of proteins, however, although showing phosphorylation in both the parental cells, and cells expressing FGFR1, were not detected in cells expressing the fusion kinase (table 2). This series of proteins included members of the EPHA family (again the high homology of the peptides within family members could not distinguish between specific family members) of receptor tyrosine kinases and specific tyrosine residues in the FYN and YES proteins as well as JNK and ANXA2, which is a major cellular substrate of the Src gene.
SSBP2 is specifically tyrosine phosphorylated in ZNF198-FGFR1 expressing 293 cells
The MS data has revealed proteins that are associated with FGFR1 activation as well as some that are associated with the function of the ZNF198-FGFR1 chimeric kinase. The demonstration that SSBP2 was a member of this group of proteins was particularly relevant because of its prior implication in leukemogenesis (30). To independently confirm the MS observation, we used an SSBP2-specific antibody (30) to IP this protein from cell lysates from various clones described above. These proteins were then analyzed using an anti-phosphotyrosine antibody. As shown in figure 1, the SSBP2 protein, with a molecular weight of ~40 kDa, can be readily identified in the input lysates. When SSBP2 was immunoprecipitated from these lysates and western blot analyses performed using an anti-PY antibody, only the IPs from cells expressing the fusion kinase showed a reactive protein with the molecular weight indicative of wild type SSBP2. In these experiments, the K562 myeloid cell-derived line was used as the positive control, since SSBP2 has previously been shown to be constitutively expressed in these cells (30). Importantly, this analysis confirms the observation from the MS data, that SSBP2 could be a specific target for ZNF198-FGFR1.
Figure 1.
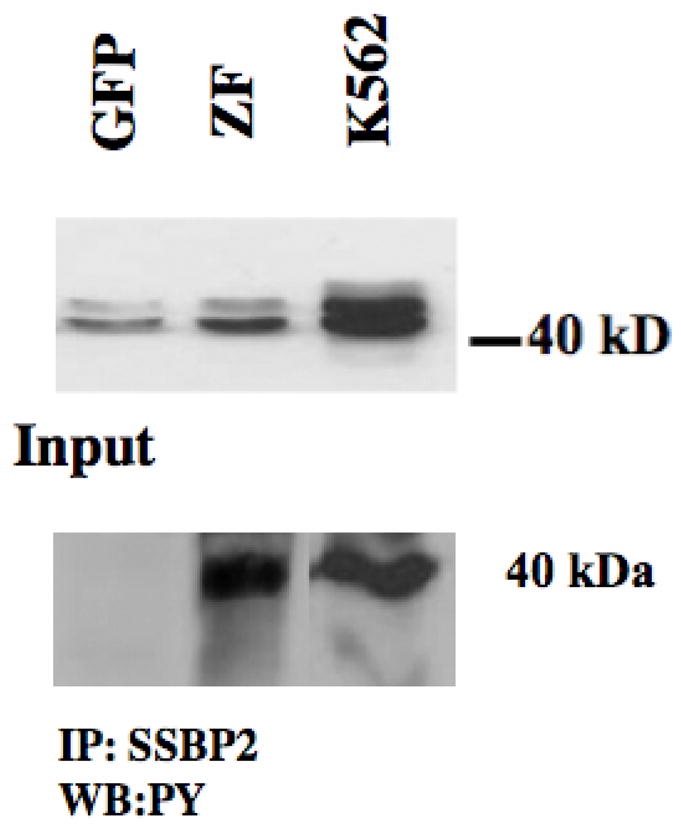
Analysis of SSBP2 protein phosphorylation in ZNF198-FGFR1 expressing cells. Western blot analysis of 293 cells shows a doublet at 40kDa in cells expressing the empty vector and the fusion kinase (ZF). High level expression is seen in K562 cells which is included as a positive control. When lysates from the same cell lines are used in IP studies using anti-SSBP2 antibodies and probed with anti-phosphotyrosine, the 40 kDa protein is seen only in the ZF cells and K562 control cells and not 293 cells expressing the empty vector.
ABL is specifically phosphorylated by ZNF198-FGFR1
The ABL kinase has also been strongly implicated in the development of leukemia (28), where the observation is that it is constitutively activated either by fusion with the dimerization domain of the BCR gene, or through constitutive phosphorylation of the wild type protein. In parental HEK 293 cells, the levels of phospho-ABL were below the levels of detection using western blotting which has been well described. We therefore assayed ABL phosphorylation following transient transfection of the wild type gene into the various cell lines (figure 2). HEK 293 cells stably expressing exogenous GFP (control) showed high levels of ABL protein in transiently transfected cells. However, when ABL was then immunoprecipitated from these cells and probed with an antiphosphotyrosine antibody PY (phosphotyrosine), its phosphorylation was below the levels of detection. When the same experiment was performed using lysates from cells expressing the GFP-tagged ZNF198-FGFR1, the ABL protein is clearly shown to be phosphorylated (figure 2) supporting the suggestion that the fusion kinase phosphorylates ABL. Phosphorylated ZNF198-FGFR1 was also seen in the ABL IP, demonstrating an interaction between the two proteins. When the cells expressing the ZNF198-FGFR1 kinase were transiently transfected with ABL, as described for the control cells, increased levels of ABL protein were detected as expected, although there appears to be relatively little additional phosphorylated ABL, suggesting a steady state has already been achieved in the fusion kinase expressing cells. This series of experiments demonstrates that ABL and ZNF198-FGFR1 co-exist in a protein complex and that ABL kinase is phosphorylated in this complex, but only apparently in the presence of ZNF198-FGFR1.
Figure 2.
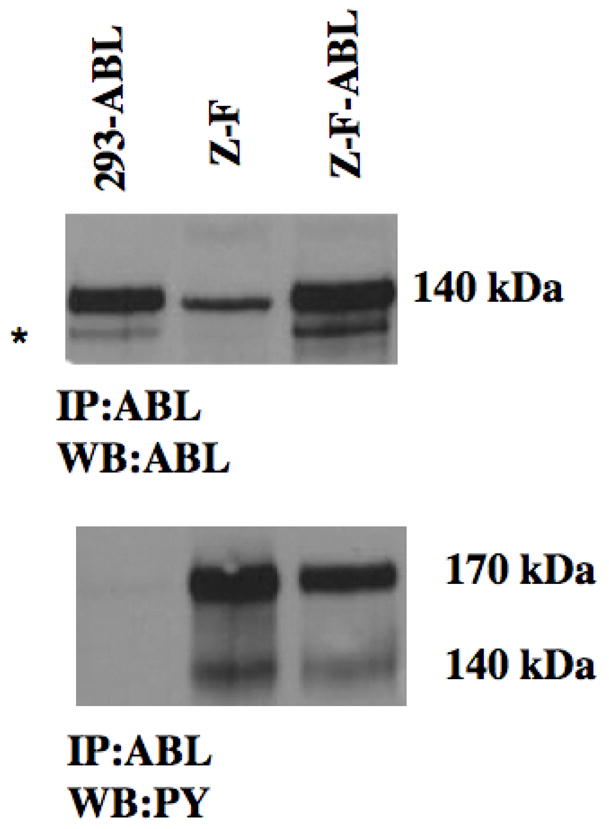
Lysates from 293 cells transfected with exogenous ABL show high level of protein expression. 293 cells expressing the ZNF198-FGFR1 fusion kinase express ABL endogenously and increased expression is seen in these cells following transient expression of exogenous ABL (top). Non Specific bands are shown (*). Immunoprecipitation of ABL from transiently transfected 293 cells, when probed with antiphosphotyrosine antibodies, fails to show phospho-ABL despite high levels of ABL protein expression. In 293 cells expressing the ZNF198-FGFR1 kinase the 140 kDa Phospho-ABL is clearly seen in the host cells and after transient transfection with exogenous ABL. The 170 kDa fusion kinase is also seen demonstrating that ABL is in a complex with this protein.
DISCUSSION
One of the challenges related to a better understanding of the function of novel fusion kinase genes has been to identify the substrate proteins that are inappropriately activated by them, since it is likely that these events are important in the transforming ability of the fusion kinase. Usually these studies have been largely guided by a prior knowledge of the proteins that are known to be activated by the wild type kinase domain involved in the chromosome translocation. In this case, for example, the role of PLCG in downstream signaling of FGFR1 has been extensively investigated and was clearly identified in this study. The recent development of an unbiased way of screening for tyrosine phosphorylation events in cells expressing these chimeric kinases, however, has provided new opportunities to define the role of these fusion genes in cancer development (31;32).
We have identified a change in the profile of tyrosine phosphorylation events that appear to be specific to various circumstances of FGFR1 expression, as well as specifically related to the presence of the ZNF198-FGFR1 oncogene. It is important to understand at the outset, however, that the analysis of tyrosine phosphorylation events in these experiments is not comprehensive and was not designed to be quantitative. By virtue of the technology, not all phosphorylated peptides will be identified in a given experiment, and it is likely that the more abundant proteins will be identified preferentially, although this does not necessarily mean that low abundance proteins will be overlooked. This concept was clearly seen in the comparison between phosphorylation events resulting from over expression of wild type FGFR1 and ligand-induced FGFR1 activity. In this comparison, although common targets were identified, a spectrum of differentially phosphorylated proteins were also identified in each case. Thus, within any given analysis, the ability to identify any particular phosphorylated protein is a balance between the presence/absence of the event and the frequency of that event in the cells being studied. Thus, the analysis does not represent the sum total of events in each cell line but provides clues about the dominant events that are associated with the presence of a specific type of kinase activity. In our experimental design, we selected proteins that were apparently exclusively phosphorylated in cells expressing the fusion kinase, although valuable information about FGFR1 targets was also obtained.
The function of the FGFR1-fusion proteins that are associated with atypical myeloproliferative disorder have been largely studied in experimental cell systems, because of the lack of cell lines that specifically carry the common variants of this chromosome rearrangement. In a recent survey of leukemia cell lines, however, Gu et al (33) identified an AML derived line, KG-1, that apparently carried a rare variant, FGFR1OP2, as the FGFR1 fusion gene partner. We have used the model 293 cell system which, although not specifically from a hematopoietic lineage, shows activation of the STAT and PLCG proteins by the fusion kinase as well as induction of SERPINB2, which was also seen in BaF3 hematopoietic cells expressing the ZNF198-FGFR1 gene (23). In this case, therefore, many of the known events demonstrated a normally functioning kinase. The advantage of the 293 system, is that we can specifically investigate novel events that are the consequences of the expression of ZNF198-FGFR1, since these cells can be compared directly to parental cells which differ only by the fact that they do not express the fusion kinase. In keeping with this concept, the ZNF198 component of the fusion kinase gene was only shown to be autophosphorylated in this cell system, demonstrating the specificity of this event. Thus, these cells provide an easily manipulated substrate in which to investigate downstream targets of the fusion kinase with the caveat that this survey is not comprehensive, since some lineage specific proteins may not be represented. Despite this potential limitation, we have successfully identified several proteins which appear to be specific targets of ZNF198-FGFR1, as well as obtaining some insights into the targets of FGFR1. The MS/PY approach has the added advantage that the specific phosphorylation sites within the substrate proteins are also identified, which allowed us to determine which of the tyrosines in the ZNF198 moiety in the chimeric protein are phosphorylated. It will now be possible, through site directed mutagenesis, to alter these sites to determine whether they have any effect on the function of the fusion kinase, specifically the dimerization event that is facilitated by the zinc finger motifs retained in the chimeric protein which are essential for the activation of the kinase domain (14).
Within this overall cell system we also demonstrated that some of the signaling events associated with FGFR1 could be readily detected, such as phosphorylation of PLC-gamma as well as autophosphorylation events at Y653/654 in FGFR1, which occur within the kinase domain and which are essential for its activation (34). It was interesting that the Y766 phosphorylation event was not observed in any of these cell clones, since this is an essential event for binding to PLC-gamma and efficient endocytosis of FGFR1, although not critical for mitogenic responses (34). Endocytosis, however, is not relevant for the chimeric kinase which, unlike wild type FGFR1, is located constitutively within the cytoplasm and is no longer tethered to the membrane (17).
Two genes identified by MS analysis as apparently specifically activated by the ZNF198-FGFR1 kinase, have previously been implicated in leukemia biology. The specificity of the phosphorylation events in both of these cases was subsequently independently confirmed. Thus, the SSBP2 gene, encodes a protein presumed to function as a single stranded DNA binding protein that can regulate gene expression and, by modulating LIM domain proteins, affects growth and differentiation of hematopoietic cells by interacting with LIM binding protein 1 (30;35–37). SSBP2 was found in a multiprotein complex with the SCL/Tal1 transcription factor (38) and was also shown to be downregulated in AML (39). Recently it has also been suggested that SSBP2 is involved in prostate cancer development where it is preferentially inactivated in advanced stage tumors and reexpression of the gene in prostate cells induces cell cycle arrest (40). More importantly in HEK 293 cells endogenous SSBP2 localizes to aggresomes, due its direct interaction with the adenoviral oncoprotein E1b55K (41). Differential phosphorylation of SSBP2 by the fusion kinase despite the altered subcellular localization of SSBP2 suggests a potentially important role for this interaction. The implications from our analysis is that, in cells expressing ZNF198-FGFR1, the phosphorylation of SSBP2 is important in a signaling cascade that is initiated by the chimeric kinase. One of the potential predicted functions of SSBP2 is to interact with the cytoskeleton, mediated through the FISH domain in this protein, which is characteristic of proteins that interact with the cytoskeleton.
Interestingly, the other protein that was identified and independently confirmed as a target of the ZNF198-FGFR1 kinase, ABL, has also been implicated in cytoskeleton reorganization, although its primary role has been defined as a constitutively active kinase that also affects downstream targets. In the ZNF198-FGFR1 expressing cells, ABL is phosphorylated at Y412, which is well known to be required for full activation of the kinase function.
The suggestion from our MS data is that, through their involvement as downstream targets of ZNF198-FGFR1, SSBP2 and ABL demonstrates the intersection of different pathways known to be implicated in leukemogenesis in the development of this stem cell leukemia. Further evidence of this intersection comes from observations implicating ZNF198-FGFR1 in the regulation of PML body formation (17). The formation of PML bodies requires sumoylation of the PML protein and we have previously demonstrated that the wild type ZNF198 protein is essential for PML body formation (23). The presence of the ZNF198-FGFR1 protein in the cytoplasm in some way inhibits SUMO1 localization to the nucleus and is associated with absence of PML bodies. It is possible, therefore, that in addition to activating key signal transduction pathways, ZNF198-FGFR1 also acts in a manner similar to the PML-RARA fusion protein in promyelocytic leukemia by preventing PML body formation which contributes to the leukemogenesis process.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (Grant CA076167-JC; HL074449-LN).
Abbreviations
- FGFR1
fibroblast growth factor receptor-1
- AMPD
atypical myeloproliferative disease
- IP
immunoprecipitation
- MS
mass spectrometry
- GFP
green fluorescent protein
- PAGE
polyacrylamide gel electrophoresis
- PY
phosphotyrosine
- SCID
severe combined immunodeficiency
- PML
promyelocytic leukemia
References
- 1.Xiao S, Nalabolu SR, Aster JC, Ma JL, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nature Genetics. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 2.Still IH, Cowell JK. The t(8;13) atypical myeloproliferative disorder: further analysis of the ZNF198 gene and lack of evidence for multiple genes disrupted on chromosome 13. Blood. 1998;92:1456–1458. [PubMed] [Google Scholar]
- 3.Smedley D, Hamoudi R, Clark J, Warren W, Abdul-Rauf M, Somers G, Venter D, Fagan K, Cooper C, Shipley J. The t(8;3)(p11;q11–12) rearrangement associated with an atypical myeloproliferative disorder fuses the fibroblast growth factor receptor 1 gene to a novel gene RAMP. Human Molecular Genetics. 1998;7:637–642. doi: 10.1093/hmg/7.4.637. [DOI] [PubMed] [Google Scholar]
- 4.Popovici C, Adelaide J, Ollendorff V, Chaffanet M, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is used to FIM in stem-cell myeloproliferative disorder with t(8;13)(p12;q12) Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abruzzo LV, Jaffe ES, Cotelingam JD, Whangpeng J, Delduca V, Medeiros LJ. T-Cell Lymphoblastic Lymphoma with Eosinophilia Associated with Subsequent Myeloid Malignancy. American Journal of Surgical Pathology. 1992;16:236–245. doi: 10.1097/00000478-199203000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Popovici C, Adelaide J, Ollendorff V, Chaffanet M, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is used to FIM in stem-cell myeloproliferative disorder with t(8;13)(p12;q12) Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walz C, Chase A, Schoch C, Weisser A, Schlegel F, Hochhaus A, Fuchs R, Schmitt-Graff A, Hehlmann R, Cross NCP, Reiter A. The t(8;17)(p11;q23) in the 8p11 myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia. 2005;19:1005–1009. doi: 10.1038/sj.leu.2403712. [DOI] [PubMed] [Google Scholar]
- 8.Popovici C, Zhang B, Gregoire MJ, Jonveaux P, Lafage-Pochitaloff M, Birnbaum D, Pebusque MJ. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood. 1999;93:1381–1389. [PubMed] [Google Scholar]
- 9.Guasch G, Mack GJ, Popovici C, Dastugue N, Birnbaum D, Rattner JB, Pebusque MJ. FGFR1 is fused to the centrosome-associated protein CEP110 in the 8p12 stem cell myeloproliferative disorder with t(8;9)(p12;q33) Blood. 2000;95:1788–1796. [PubMed] [Google Scholar]
- 10.Guasch G, Popovici C, Mugneret F, Chaffanet M, Pontarotti P, Birnbaum D, Pebusque MJ. Endogenous retroviral sequence is fused to FGFR1 kinase in the 8p12 stem-cell myeloproliferative disorder with t(8;19)(p12;q13.3) Blood. 2003;101:286–288. doi: 10.1182/blood-2002-02-0577. [DOI] [PubMed] [Google Scholar]
- 11.Grand EK, Grand FH, Chase AJ, Ross FM, Corcoran MM, Oscier DG, Cross NCP. Identification of a novel gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes & Cancer. 2004;40:78–83. doi: 10.1002/gcc.20023. [DOI] [PubMed] [Google Scholar]
- 12.Demiroglu A, Steer EJ, Heath C, Taylor K, Bentley M, Allen SL, Koduru P, Brody JP, Hawson G, Rodwell R, Doody ML, Carnicero F, Reiter A, Goldman JM, Melo JV, Cross NCP. The t(8;22) in chronic myeloid leukemia fuses BCR to FGFR1: transforming activity and specific inhibition of FGFR1 fusion proteins. Blood. 2001;98:3778–3783. doi: 10.1182/blood.v98.13.3778. [DOI] [PubMed] [Google Scholar]
- 13.Belloni E, Trubia M, Gasparini P, Micucci C, Tapinassi C, Confalonieri S, Nuciforo P, Martino B, Lo-Coco F, Pelicci PG, Di Fiore PP. 8pII myeloproliferative syndrome with a novel t(7;8) translocation leading to fusion of the FGFRI and TIFI genes. Genes Chromosomes & Cancer. 2005;42:320–325. doi: 10.1002/gcc.20144. [DOI] [PubMed] [Google Scholar]
- 14.Baumann H, Kunapuli P, Tracy E, Cowell JK. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. Journal of Biological Chemistry. 2003;278:16198–16208. doi: 10.1074/jbc.M300018200. [DOI] [PubMed] [Google Scholar]
- 15.Ollendorff V, Guasch G, Isnardon D, Galindo R, Birnbaum D, Pebusque MJ. Characterization of FIM-FGFR1, the fusion product of the myeloproliferative disorder-associated t(8;13) translocation. Journal of Biological Chemistry. 1999;274:26922–26930. doi: 10.1074/jbc.274.38.26922. [DOI] [PubMed] [Google Scholar]
- 16.Smedley D, Demiroglu A, Abdul-Rauf M, Heath C, Cooper C, Shipley J, Cross NC. ZNF198-FGFR1 transforms Ba/F3 cells to growth factor independence and results in high level tyrosine phosphorylation of STATS 1 and 5. Neoplasia. 1999;1:349–355. doi: 10.1038/sj.neo.7900035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunapuli P, Kasyapa CS, Chin SF, Caldas C, Cowell JK. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp Cell Res. 2006;312:3739–3751. doi: 10.1016/j.yexcr.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Smedley D, Demiroglu A, Abdul-Rauf M, Heath C, Cooper C, Shipley J, Cross NC. ZNF198-FGFR1 transforms Ba/F3 cells to growth factor independence and results in high level tyrosine phosphorylation of STATS 1 and 5. Neoplasia. 1999;1:349–355. doi: 10.1038/sj.neo.7900035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath C, Cross NCP. Critical role of STAT5 activation in transformation mediated by ZNF198-FGFR1. Journal of Biological Chemistry. 2004;279:6666–6673. doi: 10.1074/jbc.M308743200. [DOI] [PubMed] [Google Scholar]
- 20.Roumiantsev S, Krause DS, Neumann CA, Dimitri CA, Asiedu F, Cross NCP, Van Etten RA. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. 2004;5:287–298. doi: 10.1016/s1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 21.Kunapuli P, Somerville R, Still IH, Cowell JK. ZNF198 protein, involved in rearrangement in myeloproliferative disease, forms complexes with the DNA repair-associated HHR6A/6B and RAD18 proteins. Oncogene. 2003;22:3417–3423. doi: 10.1038/sj.onc.1206408. [DOI] [PubMed] [Google Scholar]
- 22.Kasyapa CS, Kunapuli P, Cowell JK. Mass spectroscopy identifies the splicing-associated proteins, PSF, hnRNP H3, hnRNP A2/B1, and TLS/FUS as interacting partners of the ZNF198 protein associated with rearrangement in myeloproliferative disease. Exp Cell Res. 2005;309:78–85. doi: 10.1016/j.yexcr.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 23.Kasyapa CS, Kunapuli P, Hawthorn L, Cowell JK. Induction of the plasminogen activator inhibitor-2 in cells expressing the ZNF198/FGFR1 fusion kinase that is involved in atypical myeloproliferative disease. Blood. 2006;107:3693–3699. doi: 10.1182/blood-2005-04-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasyapa CS, Kunapuli P, Cowell JK. HSPA1A is an important regulator of the stability and function of ZNF198 and its oncogenic derivative, ZNF198-FGFR1. J Cell Biochem. 2007;102:1308–1317. doi: 10.1002/jcb.21362. [DOI] [PubMed] [Google Scholar]
- 25.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nature Biotechnology. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 26.Goss VL, Lee KA, Moritz A, Nardone J, Spek EJ, MacNeill J, Rush J, Comb MJ, Polakiewicz RD. A common phosphotyrosine signature for the Bcr-Abl kinase. Blood. 2006;107:4888–4897. doi: 10.1182/blood-2005-08-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smedley D, Demiroglu A, Abdul-Rauf M, Heath C, Cooper C, Shipley J, Cross NC. ZNF198-FGFR1 transforms Ba/F3 cells to growth factor independence and results in high level tyrosine phosphorylation of STATS 1 and 5. Neoplasia. 1999;1:349–355. doi: 10.1038/sj.neo.7900035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendergast AM. The Abl family kinases: Mechanisms of regulation and signaling. Advances in Cancer Research. 2002;8585:51–100. doi: 10.1016/s0065-230x(02)85003-5. [DOI] [PubMed] [Google Scholar]
- 29.Benaim G, Villalobo A. Phosphorylation of calmodulin - Functional implications. European Journal of Biochemistry. 2002;269:3619–3631. doi: 10.1046/j.1432-1033.2002.03038.x. [DOI] [PubMed] [Google Scholar]
- 30.Liang H, Samanta S, Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24:2625–2634. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- 31.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan ZP, Stokes M, Sullivan L, Mitchell J, Wetzel R, MacNeill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 32.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu TL, Goss VL, Reeves C, Popova L, Nardone J, MacNeill J, Walters DK, Wang Y, Rush J, Comb MJ, Druker BJ, Polakiewicz RD. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108:4202–4204. doi: 10.1182/blood-2006-06-026666. [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Molecular and Cellular Biology. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu ZX, Meng XZ, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes & Development. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, Ogryzko VV, Dawid IB, Westphal H. Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, Porcher C. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Molecular and Cellular Biology. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian ZJ, Fernald AA, Godley LA, Larson RA, Le Beau MM. Expression profiling of CD34(+) hematopoietic stem/progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu JW, Nagpal JK, Sun W, Lee J, Kim MS, Ostrow KL, Zhou S, Jeronimo C, Henrique R, Van Criekinge W, Moon CS, Califano JA, Trink B, Sidransky D. ssDNA-binding protein 2 is frequently hypermethylated and suppresses cell growth in human prostate cancer. Clinical Cancer Research. 2008;14:3754–3760. doi: 10.1158/1078-0432.CCR-07-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleisig HB, Orazio N, Liang H, Tyler AF, Adams HP, Weitzman MD, Nagarajan L. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene. 2007;26:4797–4805. doi: 10.1038/sj.onc.1210281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


