Abstract
Electrical percolation based biosensing is a new technology. This is the first report of an electrical percolation-based biosensor for real-time detection. The label-free biosensor is based on electrical percolation through a single-walled carbon nanotubes (SWNTs)-antibody complex that forms a network functioning as a “Biological Semiconductor” (BSC). The conductivity of a BSC is directly related to the number of contacts facilitated by the antibody-antigen “connectors” within the SWNT network. BSCs are fabricated by immobilizing a pre-functionalized SWNTs-antibody complex directly on a poly(methyl methacrylate) (PMMA) and polycarbonate (PC) surface. Each BSC is connected via silver electrodes to a computerized ohmmeter, thereby enabling a continuous electronic measurement of molecular interactions (e.g., antibody-antigen binding) via the change in resistance. Using anti-Staphylococcal enterotoxin B (SEB) IgG to functionalize the BSC, we demonstrate that the biosensor was able to detect SEB at concentrations as low as 5 ng/ml at a signal to baseline (S/B) ratio of 2. Such measurements were performed on the chip in wet conditions.
The actuation of the chip by SEB is immediate, permitting real-time signal measurements. In addition to this “direct” label-free detection mode, a secondary antibody can be used to “label” the target molecule bound to the BSC in a manner analogous to an immunological sandwich “indirect” detection-type assay. Although a secondary antibody is not needed for direct detection, the indirect mode of detection may be useful as an additional measurement to verify or amplify signals from direct detection in clinical, food safety and other critical assays. The BSC was used to measure SEB both in buffer and in milk, a complex matrix, demonstrating the potential of electrical percolation-based biosensors for real-time label-free multi-analyte detection in clinical and complex samples. Assembly of BSCs is simple enough that multiple sensors can be fabricated on the same chip, thereby creating “Biological Central Processing Units (BCPUs)” capable of parallel processing and sorting out information on multiple analytes simultaneously which may be used for complex analysis and for point of care diagnostics.
Keywords: biosensor, semiconductor, carbon nanotubes, electrical percolation, antibody, point of care, personalized medicine
INTRODUCTION
Several types of direct detection biosensors such as Surface Plasmon Resonance (SPR), piezoelectric and cantilever sensors were developed for label-free detection of analytes. Single-walled carbon nanotubes (SWNTs) (Iijima 1991) have also been used for label-free detection in field effect transistor (FET) based sensors (Kong and Dai 2001; Kong et al. 2000). FETs sensors fabricated from single SWNTs grown by chemical vapor deposition (CVD) can measure biological interactions on the surface of SWNTs by measuring changes of electrical conductance in individual nanotubes. They have been used in chemical and biological sensors (Kong and Dai 2001; Kong et al. 2000; Tans et al. 1997). In addition to single SWNTs based sensors, submonolayer of SWNTs also fabricated by CVD (Chen et al. 2003) were shown to exhibit semiconductor-like behavior in which surface interactions of biomolecules were used for biosensing (Chen et al. 2003; Chen et al. 2001).
More recently, SWNTs percolation-based sensing has been carried out using non-porous material (Yang et al. 2010) in which SWNT-antibody complex was used to form a bio-nanocomposite network on plastic forming a “Biological Semiconductor” (BSC) for biodetection. In this SWNT-antibody complex, a recognition element which binds to a biological target was used to control the electrical conductivity of the bio-nanocomposite network via an electrical percolation principle.
In this model of electrical percolation-based system, the conductivity of the network is through the passage of current between the conductive ends of each SWNTs (the SWNTs ends within the network have to be connected for current to pass) and the overall conductivity dependent upon the continuity of the network (the number of connections). Binding of specific antigens to the SWNT-antibody complex disrupts the continuity by displacing the connected ends of the SWNT, resulting in increased tunneling distance and subsequent resistance. So the conductivity of the SWNT-antibody network increases with the increase in concentration of SWNT. At a specific SWNT concentration (the percolation transition point), the change in resistance begins to level off. Below this point, there is a still relatively low statistical distribution of “contacts” between the SWNT-antibody complexes in the network. Therefore, small changes in the SWNT-antibody complexes can lead to dramatic changes in conductivity.
Unlike the FET based sensors in which changes on the surface of s single carbon nanotubes result in changed conductivity, in BSC the change in the connectivity of the carbon nanotubes network results in a change in conductivity of the network.
Based on this model, we have shown (Yang et al. 2010) that for immunodetection, the bio-nanocomposite prepared with 1 mg/mL of SWNT will be the most sensitive to molecular interactions, other concentrations, such as 0.5 mg/mL, 1.5 mg/mL, 2 mg/mL, the response is much smaller. The best results have been obtained with 1 mg/mL of SWNT because it is near the percolation threshold. These measurements were conducted with dry SWNT-antibody complexes. However most biological interactions take place in fluids. In this work, we describe the adaptation of BSCs for measurements in fluids and the development of a real-time biosensor based on BSC technology for the detection of Staphylococcal enterotoxins (SEs).
SEs are a group of twenty-one heat stable toxins implicated in foodborne diseases (Archer and Young 1988; Bean et al. 1996; Bunning et al. 1997; Garthright et al. 1988; Olsen et al. 2000) and other diseases such as atopic eczema (Breuer et al. 2000; Bunikowski et al. 1999; Mempel et al. 2003), rheumatoid arthritis (Howell et al. 1991; Uematsu et al. 1991), and toxic shock syndrome (Herz et al. 1999). SEs are also recognized as potential bioweapons (Henghold 2004; Ler et al. 2006; Rosenbloom et al. 2002; Wiener 1996). Several immunological assays for SE detection have been described, including enzyme-linked immunosorbent assays (ELISA) (Bennett 2005), which generally use optical detection. Several different biosensors have been used for SEs detection (Homola et al. 2002; Nedelkov et al. 2000; Rasooly 2001; Rasooly and Herold 2006; Rasooly and Rasooly 1999; Sapsford et al. 2005; Shriver-Lake et al. 2003; Soelberg et al. 2005; Yu et al. 2005). In our previous work, we demonstrated that the sensitivity of sandwich immunoassays with a labeled secondary antibody detected with optical detectors could be enhanced by using single-walled carbon nanotubes (SWNTs) (Yang et al. 2008a, 2009; Yang et al. 2008b) SWNTs enhanced the sensitivity of biodetection because of their large surface area to volume ratio.
Here we present a BSC-based biosensor for SEs (and for other microbial toxins), which offers advantages such as a rapid real-time detection (monitoring conductivity changes as they happen) and high-throughput detection. The biosensor consists of an array of sixteen BSCs on a PMMA-PC substrate with a computerized ohmmeter connected and controlled by the PC and the data from the ohmmeter is sent to the PC for continuous electronic measurement of molecular interactions (e.g., antibody-antigen binding) via the change in resistance can be used for point of care diagnostics and personalized medicine.
2. Materials and Methods
2.1 Materials and Reagents
Staphylococcal enterotoxin B (SEB) and rabbit anti-SEB affinity purified IgG were purchased from Toxin Technology (Sarasota, FL). Poly(diallyldimethylammonium chloride) (PDDA) was purchased from Sigma-Aldrich (St. Louis, MO). Single-walled carbon nanotubes were obtained from Carbon Solutions Inc (Riverside, CA) and the food used for the analysis was purchased from a local grocery store.
2.2 Single wall carbon nanotube preparation
The single wall carbon nanotube solution was prepared as previously described (Yang et al. 2009; Yang et al. 2008b; Yang et al. 2010). SWNTs (30 mg) were oxidized by mixing with a concentrated sulfuric acid and nitric acid mixture (3:1 v/v) special care has to be taken care when handling sulphuric/nitric mixture. The SWNTs were shortened by sonication with a Fisher (FS-14) sonicator for 6 h, followed by extensive washing in water (100 ml) until neutralized (pH 7.0). The SWNTs were then dispersed in 100 ml 1M NaOH solution for 5 min to achieve net negative charged carboxylic acid groups and again washed with water (100 ml). The positively charged polycation (PDDA) was adsorbed by dispersing the SWNT in 50 ml of 1 mg/mL PDDA containing 0.5 M NaCl for 30 min followed by centrifugation (10,000 RPM) in a Beckman centrifuge for 15 minutes, then washed with 100 ml of water. The role of PDDA is to immobilize antibody onto SWNT surface through electrostatic adsorption.
To create the bio-nanocomposite material, the SWNTs were functionalized by dispersing in a rabbit anti-SEB IgG phosphate buffer solution (20 mM, pH 8.0) at a concentration of 0.01 mg/mL for 1 h at room temperature, so that the antibody was adsorbed onto the SWNT surface. After centrifugation (15 minutes) and extensive washing with water (10 ml), the SWNTs-antibody complex is ready to use.
2.3 Fabrication of BSC chips
The chip’s base (the non-conductive polymer) was fabricated as described in previous work (Sapsford et al. 2009; Sun et al. 2009; Yang et al. 2009; Yang et al. 2008b; Yang et al. 2010). The sixteen BSCs array used in this study were designed in CorelDraw 11 (Corel Corp. Ontario, Canada). The poly(methyl methacrylate) (PMMA) used for fabrication was micro-machined using a computer controlled Epilog Legend CO2 65W laser cutter (Epilog, Golden, CO). Before cutting, PMMA sheets were coated with 3M 9770 adhesive transfer double sided tape (Piedmont Plastics, Beltsville, MD), and the polycarbonate (PC) film, which serves as the bottom of the chip, was bonded to it to form a PMMA-PC chip. The recesses for the silver electrodes were engraved into the top PMMA layer and the surface contacts for the SWNT-antibody complex were cut through. The SWNTs-antibody complex (1 mg/mL) was applied to the chip surface by depositing pre-functionalized SWNTs with antibody to form a biological semiconductor layer into the PMMA-PC chip. For each BSC, 60 uL of SWNT complex solution was used.
To speed up the drying of the material, a simple evaporator with a variable speed 12V computer fan facing a Minco clear heater (Minco Minneapolis, MN) was fabricated (fig 2B). In this dryer, the chip is placed on the heater and a flow of air from the fan dry the material. After drying, electrodes were painted with silver contacts using “Silver Liquid” (Electron Microscopy Sciences (Hatfield, PA)) on both sides of the printed SWNT-antibody bio-nanocomposite.
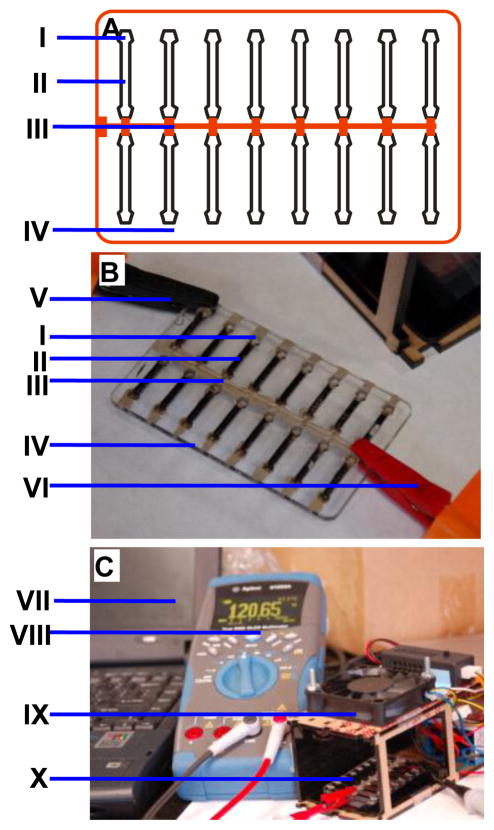
Figure 2. Electrical percolation based biosensor.
A. The schematic of a sensor with sixteen BSCs each with electrode specific (I). a ground connector common electrode (III) and space for the SWNT-antibody bio-nanocomposite gate (II) all printed on a PMMA board (IV). B. A photograph of the actual biosensor with the electrode specific connector to the ohmmeter (V), the BSC specific electrode (I), the SWNT-antibody complex gate (II), the common ground electrode (III) printed on PMMA board (IV) and the positive ground ohmmeter connector (VI). C. The electrical percolation based biosensor setup. The laptop computer (VII) is connected to a digital ohmmeter (VIII) via USB port. The BSC dryer (XI) and the BSC connected to the ohmmeter (X).
The distance between the 2 silver electrodes is 15mm, the thickness was not measured but empirically applied to cover completely the PMMA surface, the estimated thickness of the electrode is 50–200 uM.
2.4 Biosensor measurements
The resistance of the BSC was measured and recorded with a U1253A/001 Digital Multimeter (Agilent Technologies, Santa Clara CA) connected to a laptop computer via a USB port and used in the ohmmeter mode. The data generated was then imported into Microsoft Excel (Microsoft, Redmond, WA) for further analysis. The measurements were made in a wet chip so drying of the chip is not needed. The first step before applying SEB samples, a buffer or the sample fluid being tested (e.g. milk) without the toxin was added to the chip to establish the baseline resistance (R0) of the BSC measured with the digital multimeter. After establishing R0, different concentrations of SEB samples in phosphate buffer, milk or other food matrices were then added to the chip by directly pipetting the solutions (e.g. 5 ul) to the chip channel at room temperature (25 °C), ~ 8ul can be added before the cell overflows.
After the signal stabilized, the resistance (R1) was measured again. We tested the toxin in wet condition, so it is not necessary to dry after the addition of sample. The difference between the two readings (R1–R0) was used as the signal corresponding to different concentrations of SEB. Therefore, the relative value is taken under exactly the same conditions at each concentration of SEB. The value obtained with concentration of 0 ng/mL SEB was defined as background. The ratio of the signal to background (S/B) was further used to quantify the SEB concentration.
2.5 Food sample analysis
For the baby food analysis, 1 g of food sample was transferred to a centrifuge tube then buffer and different concentrations of SEB were added. The tubes containing the spiked food samples were vortexed briefly, centrifuged at 14,000 rpm for 2 min, and then the resulting supernatant transferred into a fresh tube. Loading buffer was added into the supernatant to reach a sample volume of 1 mL.
3.0 RESULTS AND DISCUSSION
3.1 Electrical percolation based biosensor
The electrical percolation-based biosensor consists of four main elements: (1) the “Biological Semiconductor” (BSC) which is a network of randomly distributed SWNTs-antibody complex that functions as the “gate” to control electrical current; (2) the silver electrodes used for measurement; (3) non-conductive material (PMMA) which is used as an insulator and base for fabrication of the semiconductor complex deposited on a polycarbonate (PC) layer bonded with adhesive to the PMMA; and (4) an ohmmeter or other digital multimeter device.
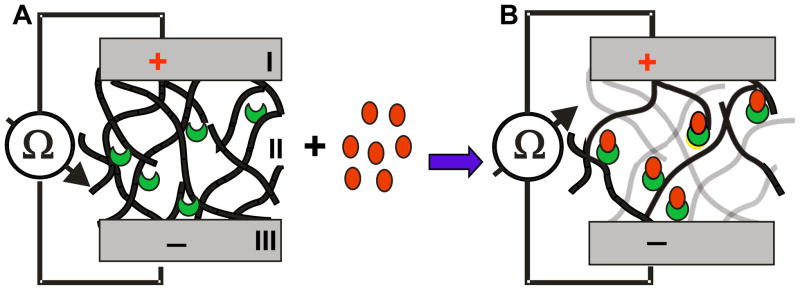
The model of the BSC’s operation is shown in figure 1. The sensor (figure 1A) is a unipolar device, with the electrodes (I and III) on both sides of the printed SWNT-antibody bio-nanocomposite gate (II). The SWNT-antibody complex gate is shown as an interconnected network of the BSC (black lines) with the antibodies (crescents). Binding of antigens (circles) to the antibody in the network results in disruption of the network (non-contact SWNTs are shown in grey), thus increasing electrical resistance (figure 1B).
Figure 1. A model of the Electrical percolation based biosensor.
Biological Semiconductor (BSC) with two electrodes (I and III) and a network (II) of randomly distributed SWNTs (black lines) with embedded antibodies (crescents). BSC is shown in low resistance mode (A) with no antigen present. In the presence of a specific antigen (B) which binds to the antibody (circles), the contacts between the SWNTs in the network (grey lines) are disturbed, resulting in a higher resistance.
The schematic of the actual sensor is shown in figure 2A. In this design, sixteen BSCs are fabricated in two rows with each BSC-specific electrode painted with silver contact paste on one of the sides (I), a ground electrode (III) common to all sixteen BSCs painted along the center between the two rows, and a space for the SWNT-antibody bio-nanocomposite gate (II) all printed on the PMMA board (IV). A photograph of the actual biosensor is shown in figure 2B where the assembled biological semiconductor is shown with the BSC-specific silver electrode (I), the SWNT-antibody complex gate (II), which is immobilized into the channel to form the network, and the common electrode filled with silver conducting solution (III) printed on the PMMA board (IV). The system also includes the ohmmeter connectors to the BSC-specific electrode (V) and the ground (VI). The device is unipolar so the polarity of the ohmmeter connectors is not important.
The electrical percolation-based biosensor setup is shown in figure 2C. Any ohmmeter can be used. An ohmmeter (VIII) that logs the data into a computer (VII) via a USB port can be used for continuous monitoring and analysis. The BSC dryer (IX), which speeds up the drying of SWNT-antibody complex on the PMMA surface is made with a simple 12V DC fan (blowing downward at low speed) and a thin heater. Measurements can be made inside the dryer (e.g. for operation in low external temperature or in fixed temperature). The BSC placed in the dryer is shown in figure 2C The SWNT-antibody complex is typically dried for 60 minutes, and is stable at room temperature for several days.
The electrical percolation BSC is operated by measuring the electrical resistance between the silver paste electrodes of each BSC and the common ground electrode. The use of a common electrode simplifies fabrication but introduces a constant difference of resistance among the 16 BSCs (based on their position relative to the measuring point). In our case, since we are measuring the change in resistance over time (Rt0 – Rt1), this constant difference has no effect.
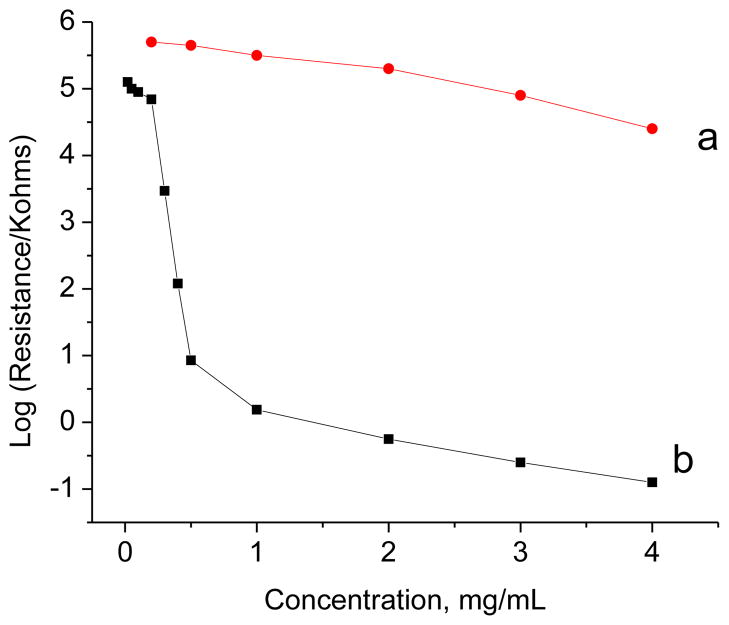
Simple graphite or conductive carbon has a lower surface area/volume ratio than SWNTs (Yang et al. 2008a, 2009; Yang et al. 2008b). Furthermore, graphite is relatively insensitive at the percolation threshold as shown in figure 3. As shown, the variation in film resistance is shown as a function of (a) graphite and (b) carbon nanotube concentrations. The resistance of graphite (a) changes relatively little with the change of graphite concentration while the resistance of carbon nanotubes (b) changes dramatically with the change of concentration especially in the 0.5–1 ng/ml range, which is utilized for BSC. These data indicate percolation of carbon nanotubes at less than 1 mg/ml but no percolation for graphite, which is a necessary condition for the biosensing principle.
Figure 3. Comparison of the resistance for graphite and carbon nanotube films.
Film resistance as a function of graphite (a) and carbon nanotube (b) concentration
Binding of the specific antigen to the antibody disrupts the network (figure 1B) and increases the resistance when measured by an ohmmeter. The change in resistance depends on the amount of bound antigen to the SWNTs-antibody complex.
3.2 Electrical percolation based biosensor for real time direct detection of SEB
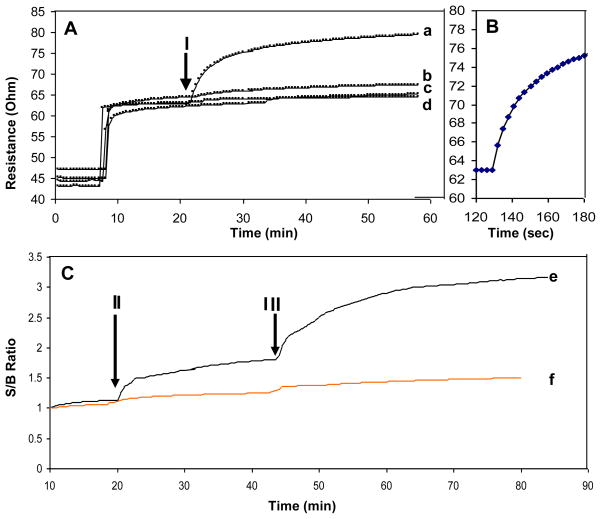
One of the main advantages of label-free biological detection is the speed of detection. As a direct biosensor, the assay involves a single reaction: binding of the analyte to the primary antibody detected as change in conductivity as a real-time detection (monitoring conductivity changes as they happen) In contrast, indirect (sandwich based) assays require two antibody-binding reactions: primary antibody to analyte, and then binding of the labeled secondary antibody to the primary antibody. To demonstrate the speed of the BSC response, 100 ng/ml of SEB in buffer was added to the chip (figure 4A arrow I) after establishing a stable baseline (R0) with buffer. As shown in figure 4A, the signal increased rapidly upon adding the toxin (figure 4A–a) and the measurable response time is ~10 seconds.” (figure 4B). The time is consistent with that previously observed for detection of a similar enterotoxin with a Surface Plasmon Resonance biosensor (Rasooly 2001).
Figure 4. Real-time detection of SEB.
A. BSC Continuous monitoring mode. B. Enlarged section of the period after SEB injection and C. sandwich immunoassay detection of the captured SEB on the BSC. In these experiments the samples were applied to BSC (arrow I) composed of 1 mg/ml SWNT with immobilized anti-SEB IgG and the resistance was plotted. For curve a, 100 ng/mL SEB; (b), 1 ug/mL BSA; (c) 1 ug/mL lysozyme; (d) 1 ug/mL IgG were applied to the sensor. To show the rapid response of BSC, one minute monitoring is shown in B. To demonstrate that SEB is actually bound to the BSC, after SEB injection (II) a second anti-SEB IgG was injected (III) and the signal to baseline (S/B) was plotted. In the presence of SEB (e) a signal is detected. Such signal not detected in the control (no SEB) (f).
It is important to note that the speed of detection depend on many factors, including the affinity of the antibody for the analyte, the analyte’s concentration, assay conditions, the design of the microfluidics and chip surface area, the response of the sensor, contaminants in the sample, etc. The measurement shown here only demonstrates the potential of the “electronic” response of the detector under conditions that minimize the effect of other factors.
To measure the specificity of the signal, non-specific antigens were tested. Solutions containing either 1 ug/mL BSA (figure 4A–b), 1 ug/mL lysozyme (figure 4A–c) or 1 ug/mL IgG (figure 4A–d) were applied to the sensor. All produced a low signal, demonstrating the specificity of the SEB measurements.
As an additional control to demonstrate that SEB is actually bound to the BSC, a second antibody was used to bind and “label” the captured SEB on the BSC in a manner analogous to immunological “sandwich” assays. According to our model, the binding of such antibody to the captured antigen should further disrupt the SWNT network and increase resistance for these measurements (figure 4C). In plots of the signal to baseline ratio (S/B), SEB injection (marked with arrow II in figure 4C) caused increased resistance (figure 4-e), and addition of a second anti-SEB IgG (marked with arrow III in figure 4C) amplified the signal. To show the specificity of the signal, control experiments were performed without SEB (figure 4C–f), and the injection of the antibody did not increase resistance. The signal change resulting from the secondary antibody injection demonstrates that SEB was captured by BSC. Although a secondary antibody is not needed for direct detection by the BSC, this indirect mode of detection can be used to verify that the signal from the direct detection is specific. Use of a secondary antibody in this way can increase confidence in the results in clinical and food safety assays and amplify the signal.
3.3 Direct BSC detection of SEB in food
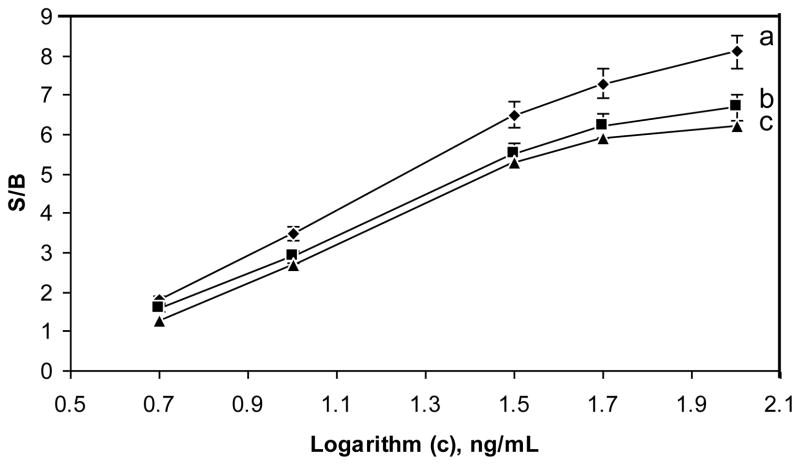
One of the challenges in biodetection is measuring analytes in a complex matrix (blood, food, soil, etc.). Although sample preparation and purification of the target analyte can reduce background noise and improve detection, such steps prolong and complicate the analysis and may result in some loss of the target analytes. Thus, direct measurement with minimal manipulation of samples is preferable. Staphylococcal enterotoxins (SEs) are a significant cause of food poisoning and detection of these toxins is important for public health and for food safety applications. In order to evaluate the SWNT sensor for detection in complex matrix, we assayed different food samples spiked with SEB and used BSC for detection. The measurement of SEB in food was compared to the measurement in buffer (Figure 5). The effect of endogenous substances inducing change in conductivity like salts or electroactive compounds is measured and accounted for in the control e.g. sample with no toxin (the first measurements in figure 5). In buffer, the biosensor was able to detect SEB at concentrations of 5 ng/ml at a S/B ratio of 2, as seen in Figure 5b. The BSC’s sensitivity with samples of SEB in milk is about 20% lower than for samples of SEB in buffer, while for samples of SEB in baby food (Figure 5c), the sensitivity was about 30% lower. The linear range of the response is from 5–100 ng/mL, for reproducibility, five measurements of 10 ng/mL of SEB with an RSD of 11.5% were obtained. These measurements were done without any sample prep of the sort used in previous work (Yang et al. 2008a, 2009; Yang et al. 2008b), suggesting that direct measurement of the toxin in food matrix is possible without sample prep.
Figure 5. Detection of SEB in various matrixes.
SEB samples in buffer (a), milk (b), and baby food (c) were detected with the electrical percolation based biosensor and the signal to baseline (S/B) was plotted.
The amount of enterotoxin necessary to cause intoxication is very small. The emetic dose in a monkey assay is 5–20 ug/animal ((Bergdoll 1967). An estimation of human dose of staphylococcal enterotoxin A to cause intoxication in a large outbreak of staphylococcal food poisoning involving chocolate milk determine that the mean amount of SEA in the 280-ml container in this outbreak was 144 ng (Evenson et al. 1988). So, detection of 5ng/ml may be for high and moderate level of contamination.
The sensitivity of all immunological assays depends on the speed of the assay: the more time allowed for antibody-antigen binding, the more sensitive and slow is the assay, real time rapid detectors are inherently less sensitive than slower assays. In addition, many food testing assays involve a sample preparation step which purifies and concentrates the sample and thus increase assay’s sensitivity. For example, to detect SEB in food samples, a partial sample purification with carboxymethylcellulose (CM) was used to lower the assay background by reducing cross-reaction of the antibodies with other components of the food matrix (Yang et al. 2008a, 2009; Yang et al. 2008b). Adding a CM sample purification step to the assay described in this paper, is likely to improve the sensitivity by 5–10 fold and make it comparable to ELISA based assays.
In comparisons with already published methods, ELISA is the most common method for detection of SEs with sensitivity of 1.56 ng/ml for colorimetric ELISA(Guglielmo-Viret et al. 2005). The addition of sample preparation and applying SWNT to improve the binding of the antibody increased the sensitivity of ELISA assay to 0.1 ng/mL (Yang et al. 2008b). To increase the sensitivity of the ELISA detection of purified SEB, gold nanoparticle-based enhanced chemiluminescence (ECL) immunosensor enable the detection of approximately 0.01 ng/mL (Yang et al. 2009). However, these assays required SEB purification step and ELISA is relatively slow assay, which may require several hours so it is not suitable for rapid detection. In previous work Electrical percolation was used for detection of SEB at concentrations of 1 ng/ml (Yang et al. 2010), which is different than the approach described here. The previous assay requires drying the BSC, which slowed the assay and is incompatible with the continuous monitoring of the toxin described here. So, while the sensitivity of the assay described here is lower, the advantage that this assay offers is rapid detection.
In addition to ELISA which is indirect method of detection, Surface Plasmon Resonance (SPR) direct detection was used for label-free detection of SEs with sensitivity ranging from 0.5 ng/ml (Homola et al. 2002), 1 ng/ml (Nedelkov et al. 2000) and approximately 10 ng/ml (Rasooly 2001) which is the same range as the BSC detector described here.
4.0 CONCLUSION
We describe here an electrical percolation-based label free biosensor with 16 BSCs capable of continuous measurement of SEB in various matrices. In the model for our sensor, antigen binding to the gate leads to rearrangement of the SWNT-antibody network, resulting in physical depletion of electron carriers in the bulk of the SWNT-antibody bio-nanocomposite through changes in contact between the SWNTs in the BSC network.
One of the most attractive features of the approach described here is the simplicity of the preparation (screen-printing) and the simplicity of measurement (ohmmeter). In contrast, FETs based biosensors (Kong and Dai 2001; Kong et al. 2000) are often fabricated using chemical vapor deposition (CVD) and require a high-tech infrastructure for microfabrication of solid-state semiconductor components while electrical percolation BSCs described here can be printed on any nonconductive material to create biosensors capable of detecting a variety of molecules.
In our previous work Electrical percolation was used for detection of SEB at solid state (Yang et al. 2010). The previous assay requires drying the BSC, which slowed the assay and is incompatible with the continuous monitoring of the toxin described here but solid state measurements enable detection of SEB at concentrations of 1 ng/ml, while the sensitivity of the assay described here is lower (~5 ng/ml), the advantage that this assay offers is rapid detection. Although we demonstrated here the application of electrical percolation for immunodetection of SEB, the selectivity of the BSC can be achieved by printing with different specific biological “dopants” (e.g., various antibodies, DNA, receptors, aptamers, etc.).
Because FET devices fabricated by CVD are hard to fabricate and it is difficult to selectively functionalize individual wires, most current FET-based biosensor applications are not scalable to perform multi-analyte detection. In contrast, the data prented here demonstrate that numerous BSCs can be easily fabricated on the same chip for multi-analyte detection. Using BSCs, it is possible to fabricate miniaturized “Biological Central Processing Units (BCPUs)” with multiple biological elements, capable of processing and sorting out information on multiple analytes simultaneously. In combination with computer algorithms, it may be possible to automatically perform multi-analyte detection and make decisions analogous to the way a silicon chip processes digital information to make decisions. Such a “Biological CPU” can provide immediate decision-making capability, which is useful for many biomedical and clinical applications including, direct biodetection of microbial pathogens and their toxins, cancer, cardiovascular, or other rapid point of care diagnostics and personalized medicine. The technology may also be used as biological switches for the regulation of implantable biomedical devices such as insulin pumps.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer DL, Young FE. Clin Microbiol Rev. 1988;1:377–398. doi: 10.1128/cmr.1.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean NH, Goulding JS, Lao C, Angulo FJ. Mor Mortal Wkly Rep CDC Surveill Summ. 1996;45:1–66. [PubMed] [Google Scholar]
- Bennett RW. J Food Prot. 2005;68:1264–1270. doi: 10.4315/0362-028x-68.6.1264. [DOI] [PubMed] [Google Scholar]
- Bergdoll MS. In: Biochemistry of some foodborne microbial toxins. Mateles RI, Wogan GN, editors. MIT press; Cambridge: 1967. pp. 1–25. [Google Scholar]
- Breuer K, Wittmann M, Bosche B, Kapp A, Werfel T. Allergy. 2000;55:551–555. doi: 10.1034/j.1398-9995.2000.00432.x. [DOI] [PubMed] [Google Scholar]
- Bunikowski R, Mielke M, Skarabis H, Herz U, Bergmann RL, Wahn U, Renz H. J Allergy Clin Immunol. 1999;103:119–124. doi: 10.1016/s0091-6749(99)70535-x. [DOI] [PubMed] [Google Scholar]
- Bunning VK, Lindsay JA, Archer DL. World Health Stat Q. 1997;50:51–56. [PubMed] [Google Scholar]
- Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NW, Shim M, Li Y, Kim W, Utz PJ, Dai H. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RJ, Zhang Y, Wang D, Dai H. Journal of the American Chemical Society. 2001;123:3838–3839. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- Evenson ML, Hinds MW, Bernstein RS, Bergdoll MS. Int J Food Microbiol. 1988;7:311–316. doi: 10.1016/0168-1605(88)90057-8. [DOI] [PubMed] [Google Scholar]
- Garthright WE, Archer DL, Kvenberg JE. Public Health Rep. 1988;103:107–115. [PMC free article] [PubMed] [Google Scholar]
- Guglielmo–Viret V, Attree O, Blanco-Gros V, Thullier P. J Immunol Methods. 2005;301:164–172. doi: 10.1016/j.jim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Henghold WB., 2nd Dermatol Clin. 2004;22:257–262. v. doi: 10.1016/j.det.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Herz U, Bunikowski R, Mielke M, Renz H. Int Arch Allergy Immunol. 1999;118:240–241. doi: 10.1159/000024085. [DOI] [PubMed] [Google Scholar]
- Homola J, Dostalek J, Chen S, Rasooly A, Jiang S, Yee SS. International journal of food microbiology. 2002;75:61–69. doi: 10.1016/s0168-1605(02)00010-7. [DOI] [PubMed] [Google Scholar]
- Howell MD, Diveley JP, Lundeen KA, Esty A, Winters ST, Carlo DJ, Brostoff SW. Proc Natl Acad Sci U S A. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima S. Nature. 1991;354:56–58. [Google Scholar]
- Kong J, Dai HJ. J Phys Chem B. 2001;105:2890–2893. [Google Scholar]
- Kong J, Franklin NR, Zhou C, Chapline MG, Peng S, Cho K, Dai H. Science (New York, NY. 2000;287:622–625. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- Ler SG, Lee FK, Gopalakrishnakone P. J Chromatogr A. 2006;1133:1–12. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]
- Mempel M, Lina G, Hojka M, Schnopp C, Seidl HP, Schafer T, Ring J, Vandenesch F, Abeck D. Eur J Clin Microbiol Infect Dis. 2003;22:306–309. doi: 10.1007/s10096-003-0928-0. [DOI] [PubMed] [Google Scholar]
- Nedelkov D, Rasooly A, Nelson RW. International journal of food microbiology. 2000;60:1–13. doi: 10.1016/s0168-1605(00)00328-7. [DOI] [PubMed] [Google Scholar]
- Olsen SJ, MacKinnon LC, Goulding JS, Bean NH, Slutsker L. Mor Mortal Wkly Rep CDC Surveill Summ. 2000;49:1–62. [PubMed] [Google Scholar]
- Rasooly A. J Food Prot. 2001;64:37–43. doi: 10.4315/0362-028x-64.1.37. [DOI] [PubMed] [Google Scholar]
- Rasooly A, Herold KE. J AOAC Int. 2006;89:873–883. [PubMed] [Google Scholar]
- Rasooly L, Rasooly A. Int J Food Microbiol. 1999;49:119–127. doi: 10.1016/s0168-1605(99)00053-7. [DOI] [PubMed] [Google Scholar]
- Rosenbloom M, Leikin JB, Vogel SN, Chaudry ZA. Am J Ther. 2002;9:5–14. doi: 10.1097/00045391-200201000-00003. [DOI] [PubMed] [Google Scholar]
- Sapsford KE, Francis J, Sun S, Kostov Y, Rasooly A. Anal Bioanal Chem. 2009;394:499–505. doi: 10.1007/s00216-009-2730-z. [DOI] [PubMed] [Google Scholar]
- Sapsford KE, Taitt CR, Loo N, Ligler FS. Appl Environ Microbiol. 2005;71:5590–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver-Lake LC, Shubin YS, Ligler FS. J Food Prot. 2003;66:1851–1856. doi: 10.4315/0362-028x-66.10.1851. [DOI] [PubMed] [Google Scholar]
- Soelberg SD, Chinowsky T, Geiss G, Spinelli CB, Stevens R, Near S, Kauffman P, Yee S, Furlong CE. J Ind Microbiol Biotechnol. 2005;32:669–674. doi: 10.1007/s10295-005-0044-5. [DOI] [PubMed] [Google Scholar]
- Sun S, Ossandon M, Kostov Y, Rasooly A. Lab Chip. 2009;9:3275–3281. doi: 10.1039/b912097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans SJ, Devoret MH, Dai HJ, Thess A, Smalley RE, Geerligs LJ, Dekker C. Nature. 1997;386:474–477. [Google Scholar]
- Uematsu Y, Wege H, Straus A, Ott M, Bannwarth W, Lanchbury J, Panayi G, Steinmetz M. Proc Natl Acad Sci U S A. 1991;88:8534–8538. doi: 10.1073/pnas.88.19.8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener SL. Mil Med. 1996;161:251–256. [PubMed] [Google Scholar]
- Yang M, Kostov Y, Bruck HA, Rasooly A. Anal Chem. 2008a;80:8532–8537. doi: 10.1021/ac801418n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Kostov Y, Bruck HA, Rasooly A. International journal of food microbiology. 2009;133:265–271. doi: 10.1016/j.ijfoodmicro.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Kostov Y, Rasooly A. International journal of food microbiology. 2008b;127:78–83. doi: 10.1016/j.ijfoodmicro.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Yang M, Sun S, Kostov Y, Bruck HA, Rasooly A. Anal Chem. 2010 doi: 10.1021/ac902644z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kim SN, Papadimitrakopoulos F, Rusling JF. Mol Biosyst. 2005;1:70–78. doi: 10.1039/b502124c. [DOI] [PubMed] [Google Scholar]