Abstract
Force-bearing linkages between the cytoskeleton and extracellular matrix are clearly important to normal cell viability – as is evident in a disease such as Duchenne Muscular Dystrophy (DMD) which arises in the absence of the linkage protein dystrophin. Therapeutic approaches to DMD include antisense-mediated skipping of exons to delete nonsense mutations while maintaining reading frame, but the structure and stability of the resulting proteins are generally unclear. Here we express and physically characterize dystrophin ‘nano’-constructs based on multi-exon deletions that might find use in a large percentage of DMD patients. The primary structure challenge is addressed first with Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) which can detect tryptic peptides from 53 of dystrophin’s 79 exons; for equivalent information from immunodetection, 53 different high-specificity antibodies would be required. Folding predictions for the nano-constructs reveal novel helical bundle domains arising out of exon-deleted ‘linkers’, while secondary structure studies confirm high helicity and also melting temperatures well above physiological. Extensional forces with an Atomic Force Microscope (AFM) nonetheless unfold the constructs, and the ensemble of unfolding trajectories reveal the number of folded domains, proving consistent with structure predictions. A mechanical cooperativity parameter for unfolding of tandem domains is also introduced as the best predictor of a multi-exon deletion that is asymptomatic in humans. The results thereby provide insight and confidence in exon-skipped designs.
Keywords: dystrophin, exon skipping, folding, AFM
Introduction
Dystrophin constitutes a critical link between the force-generating cytoskeleton in muscle and the extracellular matrix (Fig. 1A). Mutations that block expression of dystrophin lead to DMD, while mutations – including exon deletions – that shorten dystrophin’s central rod domain can lead to more mild forms of Becker muscular dystrophy (BMD) or even prove asymptomatic. Dystrophin’s rod domain normally consists of two-dozen three-helix bundle domains that are the repeated domains in ubiquitously expressed spectrin superfamily proteins. In red blood cells, these spectrin repeats have been shown to unfold when cells are mechanically sheared at physiological fluid shear stresses (Johnson 2007), and single molecule extension by Atomic Force Microscopy (AFM) has also shown that spectrin and dystrophin repeats unfold under similar forces (Bhasin 2005). However, the structure and stability of dystrophin deletants have been studied only to a limited extent and mostly in solution (Kahana 1997; Ruszczak 2009) although there is pressing need for deeper insight: emerging therapeutic efforts for DMD involve antisense-mediated deletion of multiple exons within the DMD gene’s 79 exons with the goal of skipping nonsense mutations while maintaining reading frame.
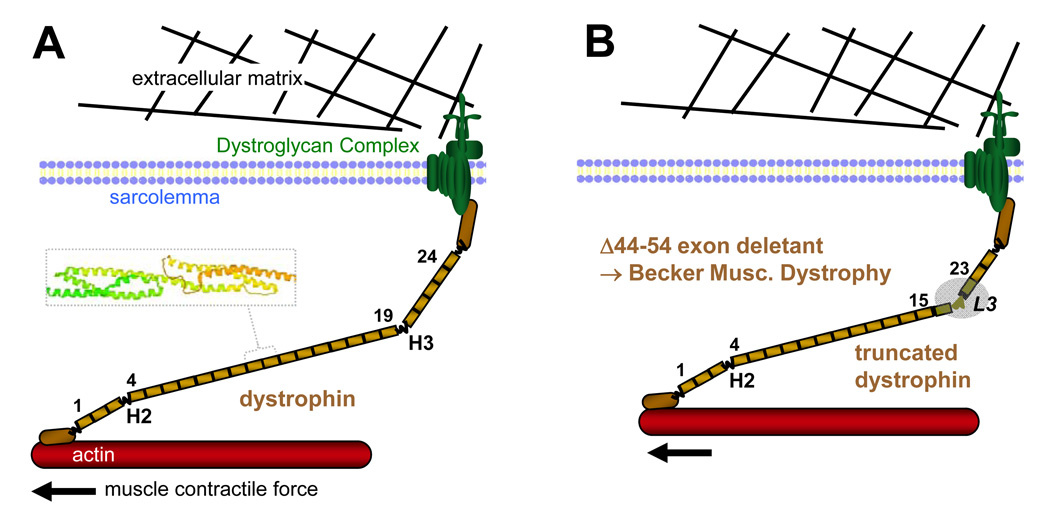
Figure 1. Full-length and representative truncated, Becker MD dystrophin in the dystrophin-glycoprotein complex.
Schematic of dystrophin and the dystroglycan complex (DGC). In normal skeletal muscle, the muscle cytoskeleton (actin) is linked by dystrophin to extracellular matrix via the cell membrane. Boxed inset illustrates crystal structure for tandem repeats 2–3 from actinin with triple helix repeats linked by a continuous and extended helix. (A) Wild-type and (B) truncated dystrophin corresponding to milder BMD phenotype with in-frame exon 17–40 deletion.
Combinations of antisense oligonucleotides (AONs) that are needed to target multiple exons (eg. Δ44–54, Fig.1B) can be problematic in application due in part to inefficient skipping (Aartsma-Rus 2006, 2007), but one such AON cocktail has been theorized to rescue up to 63% of DMD patients by transforming the DMD phenotype into an asymptomatic or mild BMD phenotype (Béroud 2007). Human clinical trials with a single AON designed to skip exon 51 (van Deutekom 2007) lead to modest levels of dystrophin expression (<35% of normal controls) using standard antibody approaches that remain state of the art in expression analysis (Freund 2007), but the limited availability of antibodies can be constraining if one needs to assess expression from every exon. Multi-exon deletion studies would clearly benefit from such assessments, and initial efforts at proteomic profiling of dystrophin in muscle suggest detectability (Lewis 2009). Here we address this issue of primary structure coverage with Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS), prior to focusing on secondary and tertiary structures of dystrophin deletants.
Exon deletions of potential interest for treating patients (Table 1) generally disrupt dystrophin’s spectrin-like repeats since exons and repeats possess distinct phasing (Fig. 2). Truncated dystrophins might nonetheless fold to form novel repeat domains that contribute protective properties of the full-length protein, and proper insight into what constitutes functional dystrophin structures could help guide AON-achievable designs. Proline-rich ‘hinge’ regions (H2, H3) appear unique to dystrophin when compared to spectrins and have been assumed to be unfolded, but there is evidence that H3 can lead to a mechanical coupling of repeats (Bhasin 2005). Such structure and stability issues have traditionally been studied in solution with chemical denaturants and non-physiological temperature changes, while single molecule AFM studies now allow one to apply more relevant mechanical forces. Most AFM studies in the past have focused on obtaining deeper understanding of known folded structures; the goal here is to obtain novel insight into unknown structures. Homology modeling based on spectrin-type repeats is used here to predict the folding of domains that result from key exon deletions, and then domain structures and stability are assessed by thermal denaturation and AFM, with the latter providing insight into the number of folded domains and linker or hinge regions. We examine truncated dystrophins that naturally occur in BMD plus a truncated dystrophin not yet found in any patients but predicted here to yield stably folded domains. Cooperative unfolding under force emerges as an important design parameter.
TABLE 1. Potential dystrophin deletants created by exon skipping.
| Mutations found in Duchenne MD patients |
Targeted exons for skipping |
Total exon deletions |
% DMD patients w/ • deletion (s) • point mutation in deleted exons |
MD phenotype |
|---|---|---|---|---|
| Deletion exons 45–54 |
Exon 44 | Δ44–54* | 66.6 20.4 |
Mild BMD |
| Deletion exon 45 |
Exon 46 | Δ45–46 | 32.9 3.6 |
Not reported |
| Deletion exons 45–50 |
Exon 51 | Δ45–51* | 58.9 13.2 |
Asymptomatic |
| Deletion exons 48–50 |
Exon 45 + Exon 51 | Δ45–51* | 58.9 13.2 |
Asymptomatic |
| Deletion exons 46–51 |
Exon 45 | Δ45–51* | 58.9 13.2 |
Asymptomatic |
| Deletion exons 46–50 |
Exon 45 + Exon 51 | Δ45–51* | 58.9 13.2 |
Asymptomatic |
| Deletion exons 48–50 |
Exon 51 | Δ48–51 | 22.4 8.3 |
Asymptomatic |
| Point Mutation exon 49 |
Exon 49 | Δ49* | 4.5 1.0 |
BMD |
| Deletion exon 50 |
Exon 51 | Δ50–51 | 9.0 4.9 |
BMD |
| Deletion exons 51–55 |
Exon 50 | Δ50–55 | 12.4 8.8 |
Not reported |
| Deletion exon 52 |
Exon 51 | Δ51–52 | 7.1 3.5 |
BMD |
| Deletion exon 52 |
Exon 51 + Exon 53 | Δ51–53 | 8.3 4.3 |
BMD |
| Deletion exon 52 |
Exon 53 | Δ52–53 | 3.0 1.7 |
BMD |
| Δ42–55* | 68.8 20.4 |
Not found in any patient |
Asterisk(*) indicates a basis for constructs studied here.
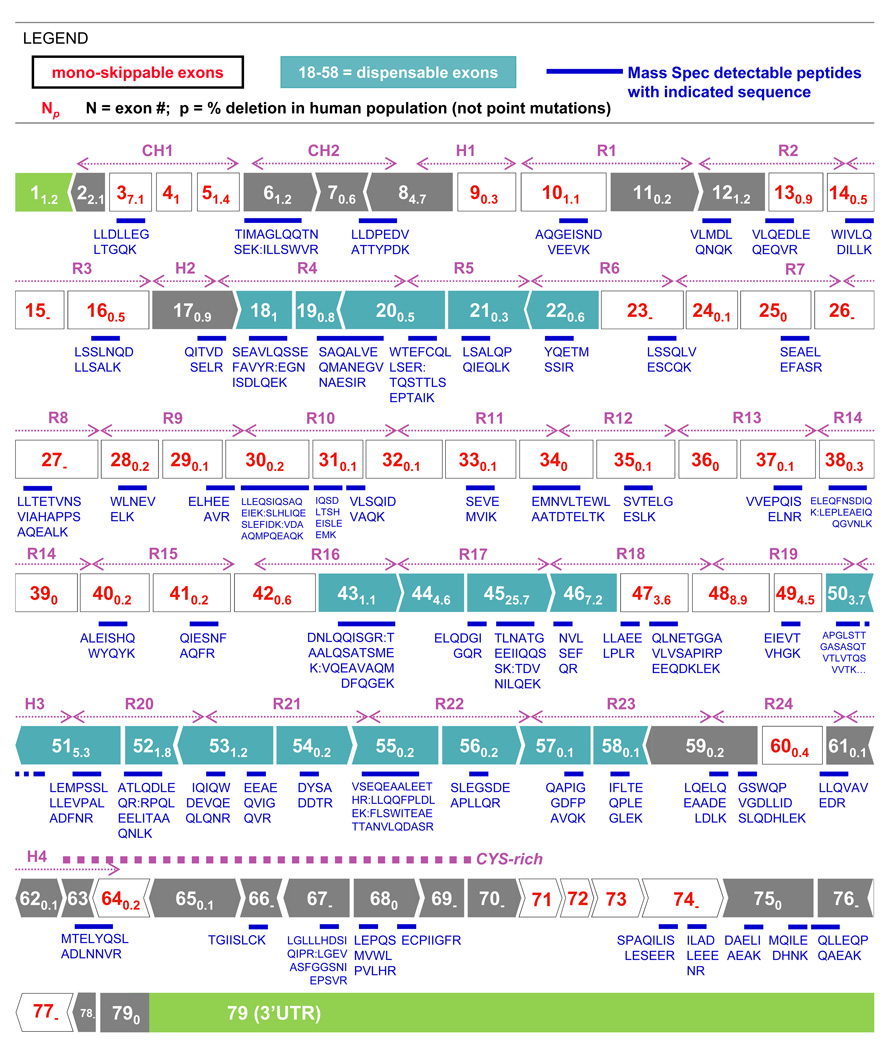
Figure 2. Phasing of DMD exons and protein domains with peptides detected by LC-MS/MS.
79 exons contribute to normal, full-length dystrophin, and. phasing of protein domains generally differs from exon boundaries. Each exon or repeat is numbered according to its distance from N-terminus, and the subscript indicates the percentage of DMD patients in the cited database (Fokkema 2005) with that particular exon deletion or duplication. For LC-MS/MS results, mouse and human dystrophin sequences were aligned, and the location of the human analog of the mouse peptide shown in blue, with the indicated sequence of the detected peptide. About half of the detected tryptic peptides are identical between human and mouse and should translate to analyses of patients.
Results
Mass Spectrometry Mapping of Dystrophin Primary Structure
As AON (and RNAi) therapies emerge, there is an increasing need to detect and even quantify proteins with complex splicing patterns. Evidence of changes in RNA from RT-PCR or other methods is simply not sufficient and antibody-based methods do not always yield the same results; in the cited human trials, for example, the efficiencies of exon skipping ranged from 49 to 90% for four patients while the amount of dystrophin in total protein extracts (with one antibody in Westerns) ranged from 3 to 12% of that found in a control specimen and from 17 to 35% for the immunofluorescent intensity ratio of dystrophin to laminin α2 relative to control (van Deutekom 2007). LC-MS/MS of tryptic peptides has emerged over the last decade as a powerful new method to map proteins, and it is used here to map dystrophin primary structure and perhaps provide some realism to detailed protein analyses of multi-exon deletions that are likely to be needed for most patients.
Multiple dystrophin peptides were detected in 4 of 5 LC-MS/MS studies of lysates from myotube cultures (see methods). Primary structure coverage of dystrophin in each run ranged from 3–20%, with a combined sequence coverage of 27%, and the 57 detectable peptides appear cumulatively scattered throughout the entire length of protein (Fig. 2). Indeed, when mapped back into the 79 exons that encode full-length dystrophin, 53 exons (or 67%) yield detectable tryptic peptides. These cumulated MS results could certainly supplement (or replace!) immunodetection of exon-deleted forms of dystrophin; for example, in the four deletion constructs considered next, all yield peptides from exons that are immediately upstream and downstream of the deletion or else just one exon removed, and all but two of the deleted exons also yield detectable peptides. Because multiple AON’s need to act on the same pre-mRNA in order to achieve the desired multi-exon skipping, the ability to detect (and even quantify) peptides from supposedly deleted exons is necessary to troubleshoot the fidelity of splicing and to know the primary structure of protein or, very possibly, mixtures of protein spliceoforms actually expressed in patients treated with multiple AONs.
Dystrophin nano-constructs and Structural predictions
DMD-causing mutations in a representative database (Fokkema 2005) are summarized in Table 1 together with exons that would be targeted if these specific patients were to undergo AON-therapy. Percentages of DMD patients that have mutations in the deleted exons and would therefore have the same truncated dystrophin are also shown together with an indication of predicted phenotype (eg. mild BMD), assuming that exon skipping works perfectly. There are many combinations of mutations and therapeutic deletions but only a handful of truncated dystrophins are functionally in-frame as illustrated with Δ45–51 which is the truncated dystrophin for four different DMD mutations. We designed nano-constructs of dystrophin (Fig. 3) based on four representative deletions: Δ49 has a typical BMD phenotype, Δ44–54 has a milder BMD phenotype, Δ45–51 is completely asymptomatic, and Δ42–55 has not been found in any patients so far but is predicted to be in-frame.
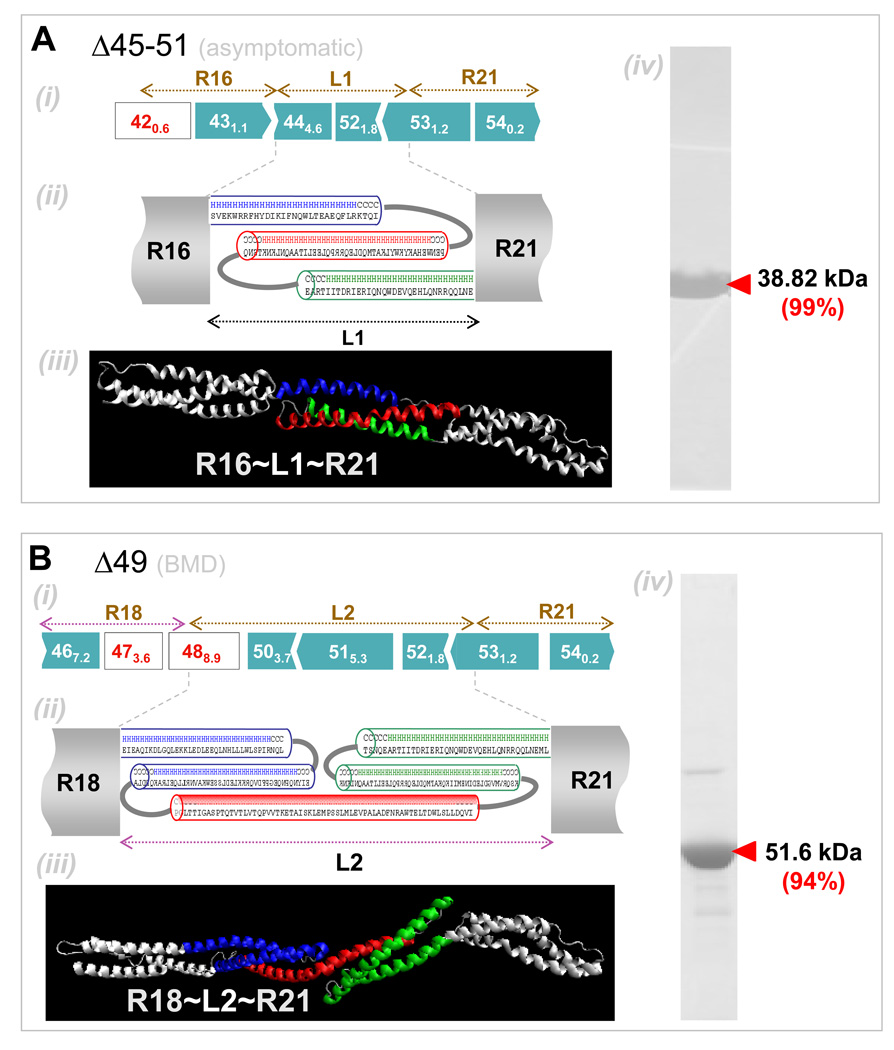
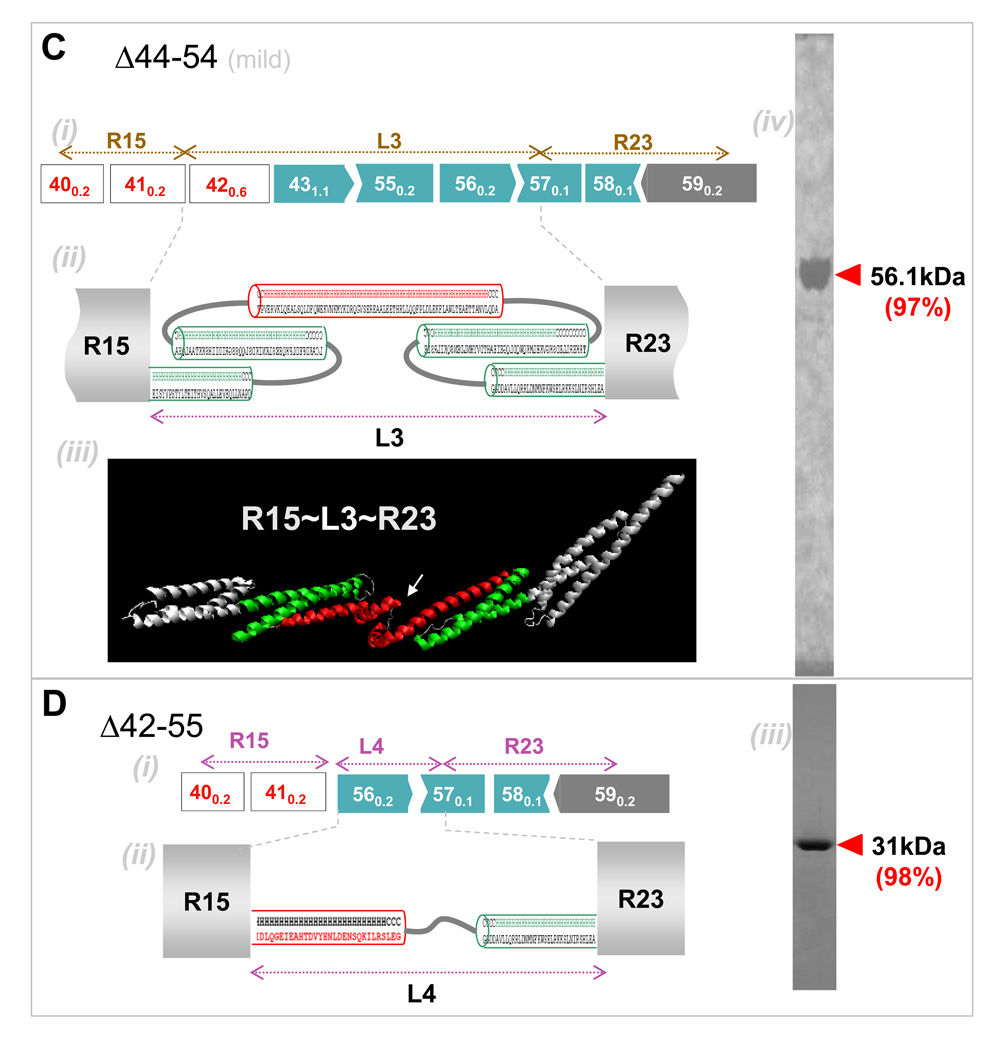
Figure 3. Secondary and tertiary structure prediction of BMD-inspired nano-dystrophin constructs.
(A) R16~L1~R21: (i) Exon map (ii) Secondary structure predictions for abnormal ‘linker’, L1, show this is mostly helical. Helical wheel predictions (not shown) indicate the presence of a hydrophobic core, necessitating packing of the helices into a spectrin-like domain. (iii) Homology model prediction for L1 with adjacent normal repeats shown in white. Color coding matches that of the structure prediction. (iv) Coomassie blue staining of purified nano-construct in SDS/PAGE, with purity in densitometry of 99%. (B) R18~L1~R21: (i) Exon map. (ii) Linker region L2 is again predicted to be mostly helical and to form two spectrin-like domains. (iii) Homology model for L3 also predicts folding into two repeat domains. (iv) Purified protein. (C) R15~L3~R23: (i) Exon map. (ii) Secondary structure predictions predict L3 to be mostly helical with two spectrin-like domains. (iii) Homology model prediction shows linker L3 might also contain another kink or hinge-like domain as indicated by a white arrow. (iv) Purified protein. (D) R15~L4~R23. (i) Exon map. (ii) The linker region was predicted to be helical, with an unstructured coil region in the middle. (iii) Purified protein.
In most types of structural studies of large proteins such as dystrophin, signals from the bulk of the protein dominate the signal from any key region of interest, and so nano-constructs were designed here to maintain phasing with at least one repeat on either side of the deletion-created ‘linker’ (Table 2). L1 in the nano-construct R16~L1~R21 results, for example, from deletion of exons 45–51 (Fig. 3A). We distinguish native wild-type sequences from non-native sequences, respectively, with ‘-’ versus ‘~’. Secondary structure and tertiary structure predictions were done on each linker region to assess whether: (i) the linker region can fold into a stable repeat domain, or (ii) folding of the linker domain might disrupt folding of native neighboring domains. In the former case, the number of amino acids seems important: the linker region in construct R16~L1~R21 (Fig. 3B) has a contour length well within the range of a single repeat domain, whereas L2 in construct R18~L2~R21 appears long enough to fold into two domains. Interestingly, L2 does not result from splicing together of two repeat domains but instead results from splicing of a single repeat to the hinge H3 (Fig. 1A), which has been shown to be helical (Bhasin 2005) but not necessarily folded into a stable tertiary structure. In contrast, linker L3 in construct R15~L3~R23 is too long for two repeat domains but falls short of forming three repeat domains, and L4 in R15~L4~R23 is much too short to generate even one repeat domain (Fig. 3C, D). This fact alone is not alarming because long helical linker regions between repeat domains are characteristic for this protein. In general, the helical content in the linker regions appears preserved, and tertiary folding through bundling of helices is driven by the need to shield hydrophobic patches. This occurs in L1, where one helix from repeat 17 easily substitutes for the missing helix in repeat 20. For simplicity, we refer to the various nano-constructs where appropriate as L1, L2, L3, or L4.
TABLE 2. Predicted unfolding lengths of domains in dystrophin nano-constructs.
| Exon deletions (human phenotype) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Domain | Δ45–51 (Asymptomatic) |
Δ49 (BMD) |
Δ44–54 (mild) |
Δ42–55 (n/a) |
||||
| R16~L1~R21 | R18~L2~R21 | R15~L3~R23 | R15~L4~R23 | |||||
| name | lc (nm) | name | lc (nm) | name | lc (nm) | Name | lc (nm) | |
| D1 | R16 | 40.6 | R18 | 40.6 | R15 | 36.5 | R15 | 36.5 |
| D2 | L1 | 39.9 | L2a | 42.0 | L3a | 42.4 | L4 | 22.9 |
| D3 | R21 | 40.2 | L2b | 41.0 | L3b | 10.7 | R23 | 47.6 |
| D4 | -- | -- | R21 | 40.2 | L3c | 39.0 | -- | -- |
| D5 | -- | -- | -- | -- | R23 | 47.6 | -- | -- |
| Avg ±S.D. |
40.2 ± 0.4 |
41.0 ± 0.8 |
35.2 ± 14.3 |
35.7 ± 12.4 |
||||
Domain length predictions for dystrophin nano-constructs.
L2 proved an interesting case where the tertiary structure prediction shows warping or twisting in one of the novel repeat domains, most likely due to the flexibility of the hinge region. Despite, or perhaps because of, the flexibility in the hinge region, L2 was able to fold into two repeat domains. L3 was also able to fold into two repeat domains as well as a smaller, helical domain. The smaller domain formed between the two non-native repeat domains and is the residual of repeat 21. Its tertiary structure shields the hydrophobic patches that were exposed after removal of the majority of repeat 21. L4 had a similar problem where it was not long enough to form a full repeat domain. Only two of the three helices from repeat 22 remained. According to the “in-frame” model, repeats 15 and 23 in R15~L4~R23 were still in phase and should remain stable. One of the drawbacks of the structure predictions is that, due to the pre-existing helical nature of the peptide, the predictions will be biased towards giving the linker region a helical structure. However, doing so exposes hydrophobic patches on the helices, and whether that perturbation is strong enough to create a novel domain, as shown in L3, and whether that domain is stable under force must be determined experimentally.
Solution studies by Circular Dichroism
CD was used to measure thermal stability of the nano-constructs and to then estimate their helical content (eg. Fig. S1). Melting temperature (Tm) provides a simple metric of thermal stability even though physiological temperatures rarely differ from 37°C, and we find that the melting temperatures of R16~L1~R21 and R18~L2~R21 (Table 3) are both very close to the Tm reported for the full-length rod domain (Kahana 1997). R15~L3~R23 and R15~L4~R23 had slightly lower Tm, but all constructs should be highly stable at physiological 37°C.
TABLE 3. Thermal denaturation of dystrophin nano-constructs by circular dichroism (CD) and by Tryptophan fluorescence.
| Nano-Construct (human phenotype) |
CD Tm(°C) |
%-Helicity, 37°C | Tryptophan fluorescence |
|
|---|---|---|---|---|
| in PBS | w/TFE | Tm (°C) | ||
|
R16~L1~R21 (Δ45–51) (Asymptomatic) |
60 | 54 | 85 | 55 |
|
R18~L2~R21 (Δ49) (BMD) |
65 | 60 | 70 | 62 |
|
R15~L3~R23 (Δ44–54) (Mild) |
55 | 58 | 75 | 35 |
|
R15~L4~R23 (Δ42–55) (N/A) |
55 | 37 | n/a | n/a |
BMD denotes Becker MD, and n/a denotes not addressed.
For helicity determinations at 37°C, CD was done in PBS with or without trifluoroethanol (TFE), which tends to stabilize helical structure to a maximum value less than 100% helicity. One hypothesis was that linker structures might not be especially stable in PBS at physiological temperature, and TFE might therefore increase %-helicity. Three of the four nano-constructs showed %-helicity well above 50%, consistent with a mostly helical bundle structure. With TFE, the asymptomatic nano-construct R16~L1~R21 showed the greatest helicity and the biggest change relative to PBS, but the fact that this deletant is asymptomatic in humans suggests this physically measurable difference in secondary structure is not critically important to structure-function. The R15~L4~R23 showed the lowest %-helicity overall, but %-helicity in TFE could not be determined because of TFE-induced aggregation; whether the low helicity was due to L4 being a random coil thus could not be determined.
Tryptophan fluorescence can be used to assess changes in solvated structure prior to or during complete unfolding. With R15~L4~R23, aggregation was again a problem. Two of the other three constructs showed Tm similar to those found by CD, but for R15~L3~R23 the Tm appeared near physiological temperature, indicating that for a significant fraction of this construct’s ten Trp’s solvent exposure changes at physiological temperature. Two Trp’s at the ends of repeats are predicted in modeling to be solvent exposed (thus constant signal). Four Trp’s located in L3 (Fig. 3C) likely account for 50% of changes in signal intensity, and we speculate that these Trp’s become exposed to solvent at lower temperature than the hydrophobic tryptophans sequestered in wild type dystrophin repeats 15 and 23 because the latter repeats have been recently expressed and reported to unfold with Tm ~ 65°C (Mirza 2010). CD and Trp fluorescence measurements therefore suggest that wild-type repeat domains induce helicity in L3 but these adjacent bundles have little influence on bundling into a physiologically stable tertiary structure.
Domain counts and lengths by Forced extension
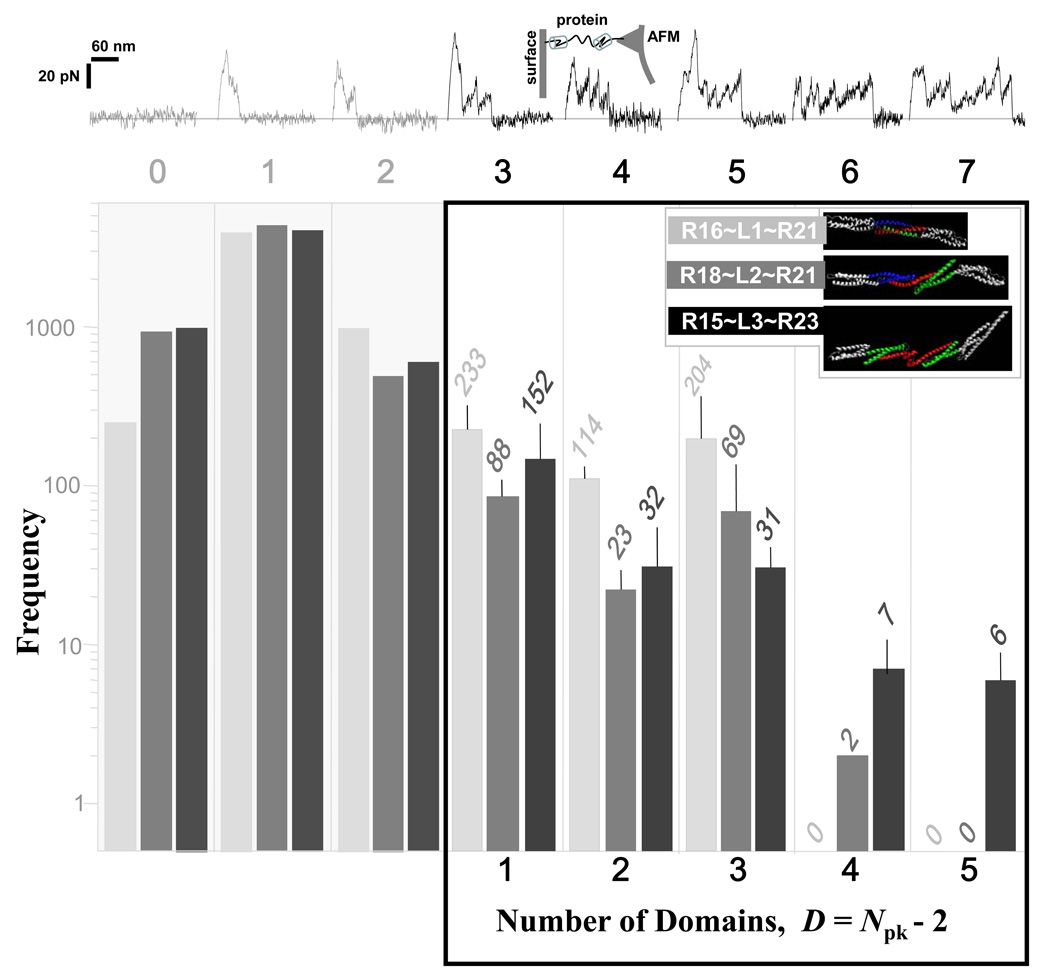
AFM was used to determine the number of unfoldable domains as well as the mechanical forces required to unfold stably folded domains. In such single molecule forced extension studies of spectrin family helical bundle proteins (Rief 1999; Law 2003; Bhasin 2005), domains unfold in a stochastic fashion with force spectrograms appearing as sawtooth patterns (Fig. 4, top). Such sawtooth patterns from tip retractions are unique to protein unfolding since other materials such as carbohydrates (Marszalek 2003) and synthetic polymers (Sun 2005) all reportedly show simpler, monotonic, stretching patterns. Thousands of pulling events here provide statistical fingerprints of mechanical unfolding for each nano-construct. If all of the predicted domains are stable, then the maximum number of peaks, max(Npk), seen in a large ensemble of force spectra would match the number of predicted domains plus two (Law 2003); the ‘two’ accounts for tip desorption events at the beginning and end of a force pull and is based on systematic studies of multiple 2, 3, and 4 spectrin repeat constructs. For the deletion constructs of dystrophin tested here (Fig. 4, bottom), the AFM data thus indicates three domains in R16~L1~R21, four domains in R18~L2~R21, and five domains in R15~L3~R23 – which are all in remarkable agreement with the tertiary structure predictions.
Figure 4. Single molecule extension by AFM reveals the unfolding forces and number of protein domains.
(top) Representative sawtooth patterns for domain-by-domain unfolding when extended at 1 nm/msec. (bottom) Number of peaks, Npk, per extension with pre-adsorbed nano-dystrophin constructs R18~L1~R21, R16~L2~R21 and R15~L3~R23. Each experiment involved at least 6000 contacts between AFM tip and proteinaceous substrate. For PBS alone, there were only 0–1 peaks ever seen. The first peak and the last peak are ignored as desorption events, so that the number of mechanically detectable domains in is obtained by subtracting these 2 peaks from Npk.
The force to a unfold a protein domain depends in general on the log of the extension rate, and rates of muscle sarcomere contraction of order ~µm/sec suggest an extension rate of this magnitude is physiologically reasonable to apply. Past studies of forces to unfold spectrin repeats have revealed unfolding forces in the range of ~20–40 pN (Rief 1999; Law 2003; Bhasin 2005), and the results here (Fig. 4, top) appear very similar in magnitude.
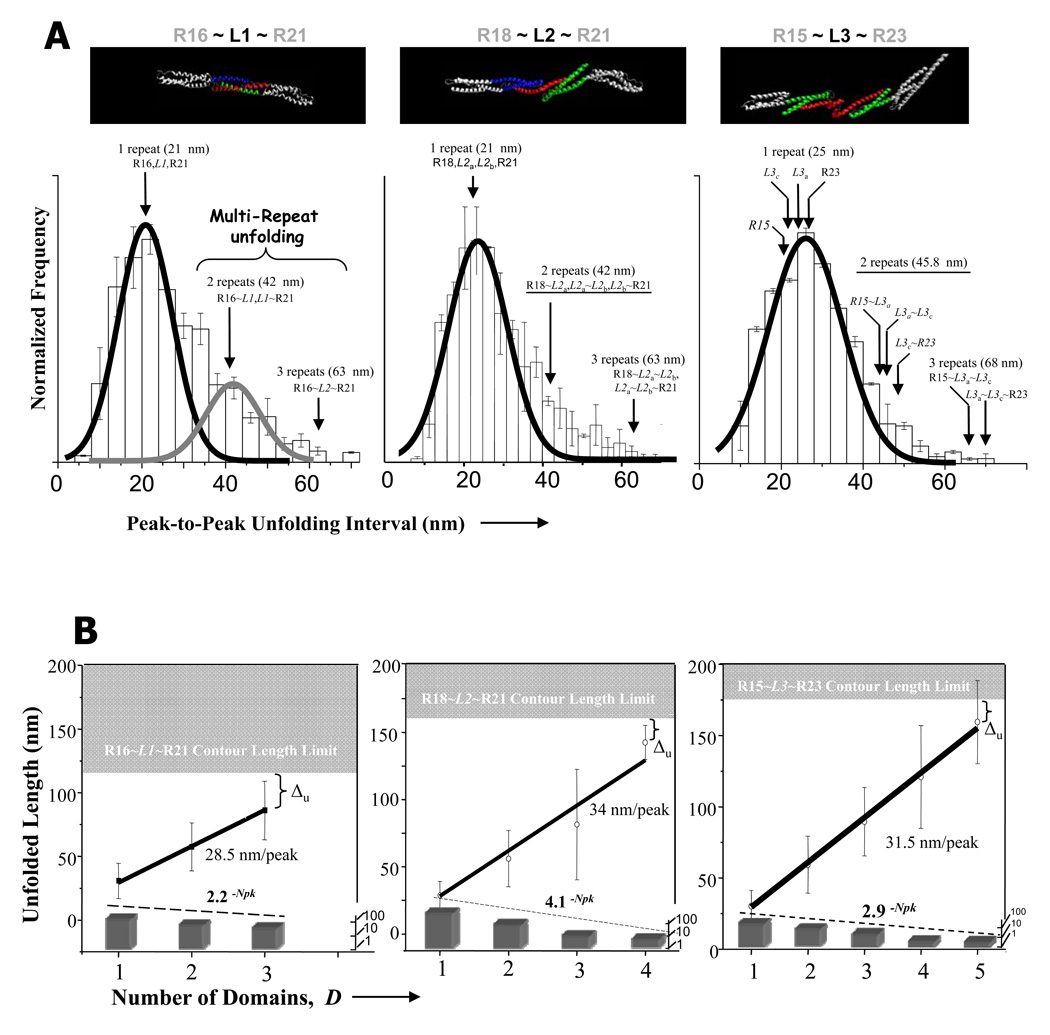
The peak-to-peak length (lpk-pk) represents the extension of an unfolded domain before the next domain is stressed to unfold and it therefore defines a domain unfolding length. Furthermore, adjacent domains might be sufficiently coupled to sometimes unfold in tandem – as documented with doubled lengths for various constructs of spectrins (Law 2003) as well as an R8-R15 dystrophin (Bhasin 2005). R16~L1~R21 was the one nano-construct here in which tandem unfolding pathways appear distinct as well as frequent: the cumulated histogram of lpk-pk (Fig. 5A) shows a significant number of events for this deletion construct in which unfolding lengths appear at twice the unfolding length of ~20 nm that is typical of a single repeat domain (Law 2003; Bhasin 2005). The widths of all distributions here are noticeably broader than those cited for single repeats. The increased widths highlight the greater diversity of pathways for extending these constructs with possibilities for unfolding ‘linker’ domains separately or in tandem with canonical repeats.
Figure 5. Unfolding length statistics by AFM for dystrophin nano-constructs.
(A) Histograms for peak-to-peak unfolding lengths for R18~L1~R21, R16~L2~R21 and R15~L3~R23 all show a major peak that was fit with a Gaussian for single domain unfolding centered near ~20 nm. Peaks at ~2× and ~3× the major peak value reflect simultaneous unfolding of multiple repeats. (B) After the 6000–8000 contacts of tip to surface, the total unfolding length (avg. ± S.D.) beyond the second peak is obtained for each sawtooth, which is categorized by Npk. The upper gray regions indicate the extension limits calculated from Table 2, and the slope of the best-fit line through all of the data (including zero total unfolding length at Npk = 2) is the average distance between peaks, which accounts for both single and multi-domain unfolding‥ The lower bargraph shows frequency distributions of Npk fitted to exponential decays of the form m−Npk with 3 < Npk < max(Npk) and the indicated values for m.
The greater the number of peaks observed in a sawtooth pattern, the longer the total extension (Fig. 4A, top; Fig. 5B), even though sawtooths with very large Npk are rare. The length limits calculated as contour lengths from the number of residues (gray regions in Fig. 5B; sums in Table 2) are approached as expected only for extension curves at the max(Npk). The difference, Δu, between the maximum unfolded length and the contour length is just 15–25 nm and is much smaller than the Δu ≈ 50 nm reported previously for extension of constructs with dystrophin’s native hinge regions (Bhasin 2005). In the latter case, we concluded hinges extended before sawtooth-type unfolding of the repeats, whereas here Δu is so small that it likely reflects fraying or unwinding at the ends of domains.
The best-fit slope of the total unfolded length in a sawtooth versus the number of domains (Fig. 5B) shows for both R16~L1~R21 and R18~L2~R21 that the unfolding length per peak is significantly lower than the average domain contour lengths listed in Table 2. These slopes reflect an averaging of single and multi-domain unfolding events and understandably exceed the ~20 nm length for the main peak seen in the lpk-pk distributions. R15~L3~R23 was the only construct for which unfolding length per peak (31.5 nm/peak) and average domain contour length (Table 2) were within one standard deviation (35.2 ± 14.3 nm domain).
The synthetic construct, R15~L4~R23, was shown above in solution studies to be structurally less stable than the asymptomatic and BMD-inspired constructs. While AFM extension results are not shown in additional detail here, the maximum peak number for R15~L4~R23 was found to be 3, indicating that not even the two flanking wild-type repeats were stable. Additionally, the unfolding length per domain was only 17 nm, which shows that very few amino acids were in stable helical structures. This seemingly ‘negative’ construct highlights the potential complexity that can result from deletion of multiple exons in dystrophin.
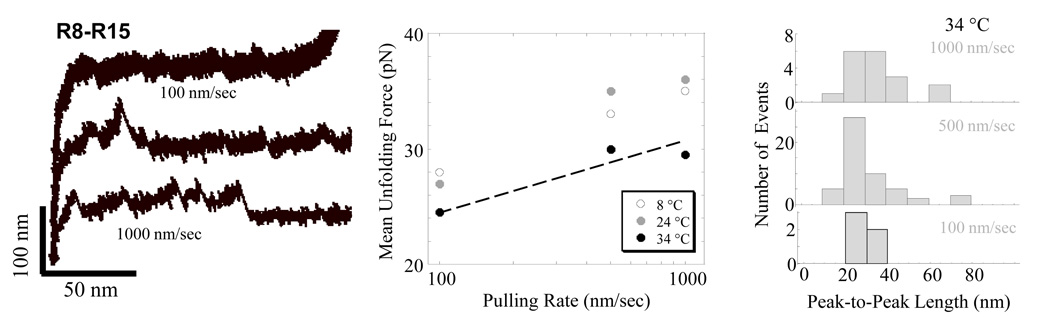
For completeness, we have also examined the rate dependence for forced unfolding of a fragment in dystrophin’s rod domain, R8–R15 (Fig. 6), up to near-physiological temperature (34 °C). Past studies at room temperature have suggested this construct’s 8 canonical spectrin repeats are interconnected by helical linkers that can propagate unfolding of up to 4–5 repeats as a single event – although such processes are exponentially more rare compared to single and tandem repeat unfolding (Bhasin 2005). The mean forces to unfold these canonical repeats increase only slightly with rate of pulling, consistent with reports for spectrin (Rief 1999), and the forces decrease slightly with temperature up to near-physiological. Multi-repeat unfolding was also evident at all temperatures, including near-physiological, with higher rates of pulling perhaps exhibiting more propagation of cooperative unfolding. The implications for rates of muscle contraction and cell pathophysiology are intruiging.
Figure 6. Forced unfolding of native dystrophin R8–R15 at different pulling rates and temperatures.
While the effects of pulling rate are weak, they do suggest the expected log-dependence on pulling rate as well as a slight weakening of the interactions at the highest temperature, 34°C. Tandem unfolding events, with peak-to-peak unfolding lengths exceeding ~20 nm per Fig.5A, are generally evident even at 34°C.
Discussion
Multi-exon deletions in both asymptomatic and BMD patients (Table 1, Fig.1B) lead to novel domain structures (Fig. 3, Table 2) with physical features both similar to and distinct from normal dystrophin. With DMD patients, similar structures might one day be generated by antisense-mediated skipping of exons – perhaps assessed at the primary structure level with Mass Spectrometry based mapping of exon expression (Fig. 2), but all such efforts seem likely to benefit from studies of secondary structure in solution combined with single molecule forced extension that reveals aspects of tertiary structure.
Helicity in solution together with melting temperatures are averaged properties that vary significantly here between nano-constructs (Table 3) just as they do for isolated single repeats and tandem repeats (Rusczak 2010). Such measurements might or might not lend insight into the function and mechanics of exon-deleted domains as suggested by the finding here that the one construct that represents an asymptomatic deletion (R16~L1~R21) exhibited the third lowest helicity and also showed the greatest disparity with TFE-induced helicity (Table 3). On the other hand, solution studies did quickly demonstrate that R15~L4~R23, with its low helicity, tends to aggregate and might therefore be a poor design for multi-exon skipping therapy.
Forced extension experiments (Figs. 4–5) agree with tertiary structure predictions in terms of the number of unfoldable domains, D, that one can determine from an ensemble of extension curves: D = max(Npk) - 2. The homology modeling here that also predicts D accounts for both helicity (i.e. hydrogen bonding) and packing of helices to sequester hydrophobic residues. We have previously simulated forced extension of tandem repeat spectrins by fully hydrated, atomistic molecular dynamics and found that helical linkers between domains can either (i) concentrate the strain and stochastically decouple domains or else (ii) remain intact and facilitate unwinding and unfolding of the tandem (Ortiz 2005). The pulling experiments test the strength of the hydrophobic and hydrogen bonds within a domain, and ensemble-level analyses of such processes reveal the number of domains together with the range of forces holding the structures together. Unfolding forces of 20–40 pN are detected here (Fig. 4-top and Fig. 6) seemingly independent of native domain or linker-derived domain and also independent of single or tandem unfolding, pulling rate or temperature (up to near-physiological), and such forces are typical of unfolding 3-helix bundle types of spectrin repeats (Rief 1999; Law 2003; Bhasin 2005). Myosin molecules individually generate peak forces on F-actin of about 6 pN at similar transition rates as here (Ishijima 1996), which suggests that the collective action of a half-dozen myosin molecules is sufficient to extensibly unfold dystrophin in muscle. Spectrin in red blood cells has indeed been shown to unfold during fluid-shearing of these cells through the application of a novel ‘Cysteine Shotgun Mass Spectrometry’ method in which in situ labeling of buried Cys is followed by Mass Spectrometry mapping of all labeled sites in lysates (Johnson 2007). Among muscle proteins, titin’s repeat domains are known to unfold at much higher forces than spectrin repeats and calculations suggest that the probability of unfolding of some titin domains is low, but not zero in muscle (Li 2002).
The predominant unfolding length per peak of ~20 nm for the nano-constructs (Fig. 5A) is also the same as that measured for wild-type dystrophin (Bhasin 2005), and the additional distinct peak at twice this length for R16~L1~R21 provides clear evidence of cooperative tandem domain unfolding. With the other two constructs shown, the peak-to-peak length distributions are broad but do encompass all of the other expected domain transitions when scaled from the full contour length. Mixed modes of unfolding are also clear by comparing the ~20 nm histogram values to the significantly higher ‘mean unfolding length’ determined from the slopes of total unfolded length versus number of domains, D (Fig. 5B). One additional metric of cooperative unfolding is revealed upon fitting the exponential decays of the Npk distributions with
which yields values for m > 1. We have previously shown in systematic studies of short and long spectrin repeat constructs (Law 2003) that m provides a simple measure of the m-fold fewer ways (i.e. degrees of freedom) for achieving one additional unfolded domain (single and tandem) in a protein that spans the gap between AFM tip and surface. For example, with D increasing from 2 to 4 in studies of both α- and β-spectrin, m decreases exponentially from ~5 to ~3; theoretically, for D = 2, there are at most three unfolding pathways inasmuch as unfolding can involve one tandem repeat event and two single repeat events, whereas with D = 4, there are four possible single repeat events plus a combination of these with at least three tandem repeat events. Kinetic Monte Carlo simulations model this behavior with reasonable accuracy (Fig. S2). For the dystrophin nano-constructs here, m ≈ 2 for the highly cooperative and asymptomatic R16~L1~R21 while m ≈ 4 for the minimally cooperative BMD construct R18~L2~R21 and m is in between for the mild phenotype R15~L3~R23. Because of the inverse relationship, we introduce 1/m as a mechanical cooperativity metric in forced unfolding; 1/m ranges from 0 to 1 and the higher the value, the greater the mechanical cooperativity.
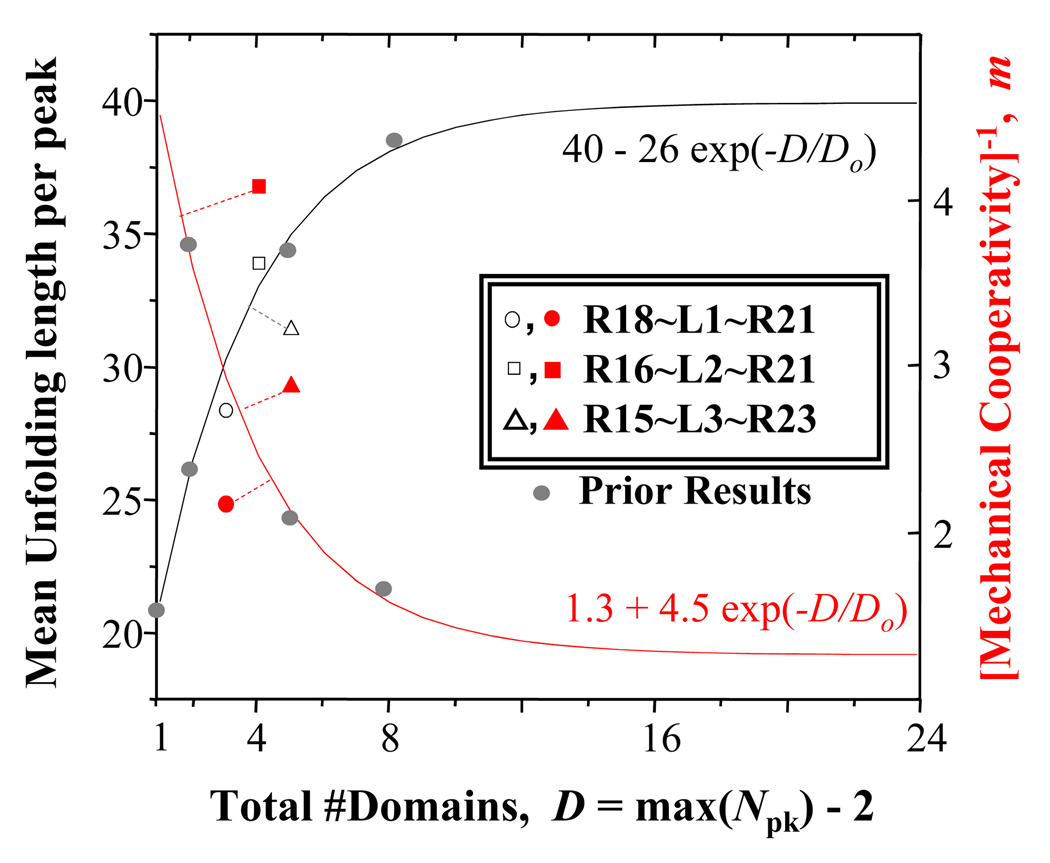
Past AFM results with the dystrophin rod domain R8–R15 and a mild BMD construct (H1-R1-2~H3~R22-24-H4) have indicated how both the Mean Unfolding Length per peak (i.e. slope) and the metric 1/m of cooperativity – as recognized here – vary with total domain number D (curves in Fig. 7), and the present results show that all of the nano-constructs here come close to previous results. Remarkably, both unfolding length and cooperativity are best preserved in the ‘asymptomatic’ construct that deviates the least from past results. Cooperativity deviates the most for the ‘BMD’ nano-construct, whereas unfolding length deviates the most for the ‘Mild’ nano-construct. Mechanical cooperativity might be crucial to dystrophin function and might be the reason that the R16~L1~R21 deletion does not lead to BMD. Forced unfolding of multiple repeats all at once produces – in an instant – an extra long linker between contracting muscle and matrix, thereby maintaining cell anchorage.
Figure 7. Comparison of AFM measures of unfolding lengths and mechanical cooperativity for dystrophin nano-constructs here to prior results for native and mild-BMD constructs (Bhasin 2005).
The exponential fits are for the prior results, and the dashed lines indicate deviation of results here from the prior results.
Conclusions
Single molecule AFM studies here together with the various bio-informatics analyses clarify the structure and mechanical cooperativity of repeat domains in exon-skipped dystrophins. While solution studies provide basic information on secondary structure and stability, the single molecule mechanical studies reveal tertiary structures with helical domains formed by exon-deleted ‘linkers’. AFM extension experiments further suggest that mechanical cooperativity with tandem domain unfolding is particularly key to dystrophin function.
Materials and Methods
Mass Spectrometry
C2C12 myoblasts were cultured by established methods (eg. Engler 2004) and differentiated for 14 days. C2C12 lysates were prepared for standard 1D-SDS-PAGE followed by in-gel trypsinization on the 460 kDa band that was then submitted for MS/MS following standard methods (Engler 2008).
Structural predictions
The helical content of nano-constructs was predicted using psi pred as described previously (Bhasin 2005). The hydrophobic packing of the helices was determined manually with helical wheel predictions that often indicated a hydrophobic core upon packing of the helices into a spectrin-like helical bundle domain. Homology prediction in conjunction with secondary structure predictions suggest that linking regions are purely helical and may fold into respective modular “molten” globules.
Plasmid Construction and Protein Preparation
Using native dystrophin as a guide, the dystrophin nano-constructs here were made by PCR per the designs of Fig. 2, 3 and Table 2. A start codon-embedded NdeI site, an additional stop codon, and an EcoRI site were engineered into PCR primers such that the PCR product could be subcloned into expression vector pMW172. Non-native splice sites in each construct are indicated by ‘~’, and sequencing verified that: construct R16~L1~R21 consisted of dystrophin residues Glu2012-Lys2146~Ala2515-Glu2686; construct R18~L2~R21, residues Gln2208-Gln2366~Arg2401-Glu2686; construct R15~L3~R23, residues Ile1874-Phe1966~Ala1972-Gly2096~Val2677-Glu2931; and construct R15~L4~R23, a hexaHis-tag followed by residues Ile1874-Phe1966~Ala1972-Ile1974~Asp2740-Glu2931.
Escherichia coli strain BL21 (Star)DE3 (Invitrogen) was transformed with the above plasmids. Bacteria were grown at 37°C in Luria-Bertani medium with 200 µg/ml of ampicillin, to an A600nm of 0.8–1.0, and expression was induced by addition of 1 mM isopropyl-β-d-thiogalactoside. After 3–4 h, cells were harvested by centrifugation at 5000g for 10 min and frozen in liquid N2. The frozen cell pellet (~10 ml, from 2 L of culture) was thawed for 10 min at 25°C and lysed by resuspension in 40 ml of Cellytic B (Sigma-Aldrich) containing 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml of leupeptin, and 10 mg/ml of aprotinin. After 15 min at 25°C, cell lysate was clarified by centrifugation at 45,000g for 15 min; centrifugation and all subsequent steps were performed at 4°C. For constructs R16~L1~R21, R18~L2~R21, and R15~L3~R23, the supernatant was loaded onto a DEAE-cellulose column (Whatman DE-52, 2.5 cm × 30 cm) pre-equilibrated with 20 mM Hepes (pH 7.0), 1 mM dithiothreitol, 1 mM sodium azide, and eluted at 35 ml/h with a 325 ml × 325 ml gradient of 0–0.5 M sodium chloride in Hepes buffer; fractions were collected at 12 min intervals. The expressed protein, identified by SDS-PAGE, eluted about halfway through the gradient. Solid ammonium sulfate was added to the pooled peak fractions to a final concentration of 1.6 M, and the material was applied to a 1.5 cm × 30 cm column of Butyl ToyoPearl 650 M (Tosoh Bioscience) in 1.6 M ammonium sulfate, 20 mM Hepes (pH 7.0), 1 mM dithiothreitol, 1 mM sodium azide. The column was eluted at 10 ml/h with a descending gradient of 1.6 M–0 M ammonium sulfate in 150 ml. Fractions were collected at 10 min intervals, and fractions containing the purified protein were identified by SDS-PAGE, pooled, and dialyzed against 100 volumes of phosphate buffered saline, with two changes. For construct R15~L4~R23 a final concentration of 6 M urea was added to crude lysate, and the cell pellet was resuspened in 6 M urea and centrifuged at 45,000g for 15 min 3×. Crude lysate and urea washes were combined and applied to a Ni-NTA Superflow column. Protein was eluted according to manufacturer’s instructions. Concentration was determined from the A280 nm using an extinction coefficient of 1.313 at 1 mg/ml, calculated from the amino acid composition.
Stable monomer was ultimately purified by gel permeation chromatography in phosphate-buffered saline (PBS) and kept on ice for AFM studies. Immediately before use, any protein aggregates were removed by centrifugation 2°C for 1 hour; and monodispersity was verified by dynamic light scattering.
Circular Dichroism Measurements
Circular dichroism spectra were measured and analyzed as before (Bhasin 2005) at a number of temperatures using a 1 mm path length cell on a Jasco J715 spectropolarimeter in the same buffer (PBS, pH 7.4) as that used in the AFM experiments. The instrument was calibrated with d-10-camphorsulfonic acid. Samples were equilibrated at each temperature for 20 min before taking measurements. Each spectrum was the average of three measurements. Circular dichroism spectra for each nano-construct were also measured at 25 °C with 40% v/v TFE.
Fluorescence Spectroscopy
All tryptophan fluorescence experiments were carried out as described elsewhere (Johnson 2007b) using a UV fluorometer from Photon Technology Instruments with a set excitation wavelength of 275 nm. The emission spectrum was monitored between 310–370 nm for each scan. Samples consisted of 480uL of PBS buffer (pH=7.5) and 20 µL of 1 mg/mL protein utilizing a 1 mL cuvette with 1cm path length. Temperature scans from 21–80 °C were collected at 2°C increments with appropriate time to thermoequilibriate at each temperature. A blank buffer sample spectrum was also collected for background subtraction. Emission intensities at 350 nm were analyzed using the previously reported methods.
Atomic Force Spectroscopy
50 µl of 0.1 mg/ml protein solution was adsorbed on freshly cleaved mica for 30 min at room temperature per (Bhasin 2005). All measurements were carried out in PBS using a Nanoscope-E Multimode AFM (Digital Instruments, Santa Barbara, CA) equipped with a liquid cell. Sharpened silicon nitride (Si3N4) cantilevers (Park Scientific, Sunnyvale, CA) of nominal spring constant kC = 10 pN/nm were typically used, with equivalent results obtained using 30 pN/nm cantilevers. kC was measured for each cantilever using the manufacturer’s directions at each temperature, and additional calibrations were performed as described previously. Experiments were done at imposed displacement rates of 1 nm/msec = 1 µm/sec. Thousands of surface to tip contacts were collected and analyzed with the aid of a semi-automated, visual analysis program custom written in C++. Since protein unfolding events are stochastic and the experiment intrinsically random in many ways, collecting and analyzing thousands of peaks is necessary to provide an accurate statistical survey of extensible unfolding. Initial results at the beginning of a many hour experiment were similar to those obtained at the end of the experiment. Force spectrograms, or sawtooth patterns for each construct were analyzed by categorizing each according to the number of peaks (Npk) as well as peak-to-peak lengths (lpk-pk).
Supplementary Material
Acknowledgement
Support from the NIH (NIAMS, NHLBI, and NIBIB), Muscular Dystrophy Association (MDA), and the NSF (including Penn’s NSEC - NanoBio Interface Center) is very gratefully acknowledged. Experiments on the dystrophin rod domain at different temperatures and pulling rates were conducted by A.E.X. Brown as a visitor to the Laboratory of Prof. Matthias Rief (Technical University of Munich, Physics), and we sincerely acknowledge the opportunity to use their instruments and all of the assistance.
References
- Aartsma-Rus A, Kaman WE, Weij R, den Dunnen JT, van Ommen GJ, van Deutekom JC. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, van Ommen GJ, van Deutekom JC. Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med Genet. 2007;8:43–51. doi: 10.1186/1471-2350-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béroud C, Tuffery-Giraud S, Matsuo M, Hamroun D, Humbertclaude V, Monnier N, Moizard MP, Voelckel MA, Calemard LM, Boisseau P, Blayau M, Philippe C, Cossée M, Pagès M, Rivier F, Danos O, Garcia L, Claustres M. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat. 2007;28:196–202. doi: 10.1002/humu.20428. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Law R, Liao G, Safer D, Ellmer J, Discher BM, Sweeney HL, Discher DE. Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J Mol Biol. 2005;352:795–806. doi: 10.1016/j.jmb.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Engler A, Sheehan A, Sen S, Bonnemann C, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications of stiff or soft microenvironments. Journal of Cell Biology. 2004;166(6):877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A, Carag C, Johnson C, Raab M, Tang H-Y, Speicher D, Sanger J, Sanger J, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. Journal of Cell Science. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema IF, den Dunnen JT, Taschner PE. LOVD: easy creation of a locus-specific sequence variation database using an "LSDB-in-a-box" approach. Hum Mutat. 2005 Aug;26(2):63–68. doi: 10.1002/humu.20201. [DOI] [PubMed] [Google Scholar]
- Freund AA, Scola RH, Arndt RC, Lorenzoni PJ, Kay CK, Werneck LC. Duchenne and Becker muscular dystrophy: a molecular and immunohistochemical approach. Arq Neuropsiquiatr. 2007;65:73–76. doi: 10.1590/s0004-282x2007000100016. [DOI] [PubMed] [Google Scholar]
- Ishijima A, Kojima H, Higuchi H, Harada Y, Funatsu T, Yanagida T. Multiple- and single-molecule analysis of the actomyosin motor by nanometer-piconewton manipulation with a microneedle: unitary steps and forces. Biophys. J. 1996;70:383–400. doi: 10.1016/S0006-3495(96)79582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Massimiliano G, Ortiz V, Bhasin N, Harper S, Speicher D, Gallagher P, Discher DE. A pathogenic proline mutation in the linker between spectrin repeats: Disease due to spectrin unfolding. Blood. 2007b;109(8):3538–3543. doi: 10.1182/blood-2006-07-038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana E, Flood G, Gratzer WB. Physical properties of dystrophin rod domain. Cell Motil Cytoskeleton. 1997;36:246–252. doi: 10.1002/(SICI)1097-0169(1997)36:3<246::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C, Carberry S, Ohlendieck K. Proteomic profiling of x-linked muscular dystrophy. J Muscle Res Cell Motil. 2009;30:267–269. doi: 10.1007/s10974-009-9197-6. [DOI] [PubMed] [Google Scholar]
- Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Marszalek PE, Oberhauser AF, Li H, Fernandez JM. The force-driven conformations of heparin studied with single molecule force microscopy. Biophys J. 2003;85:2696–2704. doi: 10.1016/s0006-3495(03)74692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A, Sagathevan M, Sahni N, Choi L, Menhart N. A biophysical map of the dystrophin rod. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbapap.2010.03.009. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Ortiz V, Nielsen SO, Klein ML, Discher DE. Unfolding a linker between helical repeats. J Mol Biol. 2005;349:638–647. doi: 10.1016/j.jmb.2005.03.086. [DOI] [PubMed] [Google Scholar]
- Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- Ruszczak C, Mirza A, Menhart N. Differential stabilities of alternative exon-skipped rod motifs of dystrophin. Biochim Biophys Acta. 2009;1794:921–928. doi: 10.1016/j.bbapap.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Walker GC. Viscoelastic response of poly(dimethylsiloxane) in the adhesive interaction with AFM tips. Langmuir. 2005;21:8694–8702. doi: 10.1021/la050448j. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, de Kimpe SJ, Ekhart PF, Venneker EH, Platenburg GJ, Verschuuren JJ, van Ommen GJ. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.