Abstract
Neuronal mitochondria need to be transported and distributed in axons and dendrites in order to ensure an adequate energy supply and provide sufficient Ca2+ buffering in each portion of these highly extended cells. Errors in mitochondrial transport are implicated in neurodegenerative diseases. Here we present useful tools to analyze axonal transport of mitochondria both in vitro in cultured rat neurons and in vivo in Drosophila larval neurons. These methods enable investigators to take advantage of both systems to study the properties of mitochondrial motility under normal or pathological conditions.
1. INTRODUCTION
Neurons are exquisitely organized and compartmentalized. Their cytoskeleton and motor proteins are necessary to distribute essential components to axons, dendrites and synapses in sufficient supply and to return organelles and signals from the periphery to the cell soma. Mitochondria are one such important organelle that neurons need to allocate properly. Absent adequate mitochondrial movement, the distal portions of neurons, which in a human can be a meter distant from the cell body, may be deprived of sufficient ATP production and may be incapable of adequately buffering cytosolic Ca2+. Moreover, the involvement of mitochondria in generating reactive oxygen species and apoptotic signaling heightens the importance of keeping healthy mitochondria in the soma, axons, and dendrites.
Mitochondrial movement is therefore needed to deliver the organelles to the periphery. In addition, it is likely the movement from the periphery back to the soma is needed to provide for protein turnover, since most mitochondrial proteins are nuclear-encoded and peripheral proteins synthesis is likely to be limited. Damaged mitochondria can be highly destructive to the cell via the production of reactive oxygen species (Hoye et al., 2008). Therefore, turnover of the organelle and refreshment of peripheral mitochondria by fusion with newly minted mitochondria that arrive from the soma may be an important means of maximizing mitochondrial function and minimizing their deleterious potential. This may be one of the most significant reasons that 30-40% of the mitochondria in an axon or dendrite are in motion at any given time (Chen et al., 2007; Overly et al., 1996; Waters and Smith, 2003). Moreover, the optimal density of mitochondria differs from one neuron to another, even from one synapse from another within the same neuron, and is constantly changing (Hollenbeck and Saxton, 2005) as the activity of the neuron shifts the energetic needs of a subcellular domain. Activation of receptors and channels can also place an acute burden of Ca2+ influx on a subcellular region, and this too may require a shift in mitochondrial distribution. The necessity of tailoring mitochondrial density to changing needs may be an additional reason that mitochondria are so frequently in motion.
Mitochondria can congruently move a long distance without stopping, or travel a short distance and stop and start again. The majority of their movements are microtubule-based, either anterograde via kinesins, toward the (+)-ends of the microtubules such as in axonal terminal, or retrograde via dynein, toward the (−)-ends in the cell bodies (Chang et al., 2006; Hollenbeck and Saxton, 2005; Overly et al., 1996). It is not surprising, therefore, that the dynamic nature of mitochondria is significant for neurons to maintain neuronal function and plasticity, and the abnormalities in mitochondrial distribution have been found in many neurodegenerative disease models such as Charcot-Marie-Tooth (Baloh et al., 2007), Amyotrophic Lateral Sclerosis (De Vos et al., 2007), Alzheimer's (Pigino et al., 2003; Rui et al., 2006; Thies and Mandelkow, 2007), Huntington's (Trushina et al., 2004), and Hereditary Spastic Paraplegia (Ferreirinha et al., 2004; McDermott et al., 2003). To understand the means by which mitochondrial distribution is achieved and the dynamic regulation of that distribution, static imagery is insufficient. Only through live video microscopy can the mechanisms governing mitochondrial distribution be understood.
Although great progresses have been made in visualizing the transport of neuronal mitochondria, there remain significant challenges that include finding appropriate mitochondrial markers, an ideal perfusion/imaging system to keep neurons alive, and an amenable in vivo animal model. Offsetting these difficulties, neurons also offer advantages over other cell types for studying basic mechanisms of mitochondrial movement. The long parallel arrays of microtubules in neurites provide a simpler geometry in which long-distance movements of mitochondria can be followed. In addition, axons have largely uniform polarities of microtubules, with all (+)-ends oriented towards axon terminals. Therefore, the study of mitochondrial transport in axons provides the opportunity to distinguish kinesin-based movements from dynein-based. In dendrites, microtubule polarity is typically more mixed, but the analysis of dendritic motility offers the potential to examine how that motility is affected by the activation of postsynaptic receptors on the dendrite surface and the association of mitochondria with postsynaptic specializations.
Here we describe two systems that have been used successfully in our laboratory and elsewhere to study axonal transport of mitochondria: cultured rat hippocampal neurons and Drosophila neurons. Both systems permit detailed measurements of the parameters of mitochondrial movement and the manipulation of the mechanism either via transgene expression or classical genetics. Separately, they provide the complementary advantages, detailed below, of either in vitro tissue culture or imaging in a semi-intact in vivo environment.
1.1 Imaging Mitochondrial Movement in Cultured Rat Hippocampal Neurons
Embryonic hippocampal neuronal cultures have been widely used to study protein distribution, neuronal function and neuronal morphogenesis. Their utility derives from their elegant extension of neurites, their expression of key features of in vivo hippocampal neurons, and their suitability for transfection with transgenes (Kaech and Banker, 2006). Neurons dissociated from rat embryonic hippocampi contain few glial cells and can be cultured in vitro for up to a month. After 3-4 days in vitro, axons have differentiated from dendrites and exhibit unique morphological characteristics, such as long, thin and uniform diameters, a lack of branching, and obvious growth cones. The identification of axons can be enhanced by transfecting the cells with a fluorescent axonal marker such as synaptophysin-YFP (Chang et al., 2006), or marking dendrites with a marker such as PSD95-GFP (Chang et al., 2006).
The axons of cultured hippocampal neurons can reach a few mm in length, enabling the observation of long-range movements. In these neuronal cultures, it is also easy to apply extracellular agonists, antagonists, toxins, or ionophores to regulate intracellular signaling, change intracellular ion balance, trigger neuronal cell death, or mimic a pathological cellular effect. The consequences of these manipulations on mitochondrial movements can be monitored acutely. These advantages have been exploited extensively for the study of mitochondrial motility (Chada and Hollenbeck, 2003; Chada and Hollenbeck, 2004; Chen et al., 2007; Overly et al., 1996; Waters and Smith, 2003).
In the second section, we will describe the protocols to analyze mitochondrial movement in cultured rat hippocampal neurons. For optimal tracking of individual mitochondria, we have found it desirable to use cultures that were sparsely transfected with a construct encoding a fluorescent protein fused to a mitochondrial targeting sequence (RFP-mito). Under these circumstances, the processes of a single transfected neuron can be examined and thus the orientation of movement towards or away from the soma can be determined unambiguously. In densely transfected cultures or when membrane permeant dyes are used to image mitochondria, the bundling of neurites makes it impossible to determine the cell body or origin and the direction of movement. We and others have also found that some mitochondrial dyes may decrease movement of the organelles.
1.2 Imaging Mitochondrial Movement in Drosophila Larval Axons
As a model system for the genetic analysis of neurons, Drosophila remains unsurpassed. Existing mutant lines are available for dissecting many cell biological processes and ongoing mutant screens continue to identify additional proteins of interest. Despite progress with RNAi in mammalian systems, a null allele remains the best guaranty of complete loss of function. Large collections of RNAi lines are also available in the fly at low cost with which to screen candidate genes (http://stockcenter.vdrc.at/control/main). Additionally, the ease of combining different mutants and transgenes in an individual organism enables studies of double-mutants and phenotypic rescue with modified transgenes in an intact organism. Moreover, many mutations whose early lethality would prevent their analysis in a knockout mouse, can be studied in the fly through genetic tricks to make mutations homozygous or express RNAi selectively in small subsets of neurons. In consequence, Drosophila has been used to establish many human neurodegenerative disease models, an endeavor justified by the high degree of similarity of their genome to our own and similarity of the resultant cellular phenotypes to human neurodegeneration (Bolino et al., 2004; Clark et al., 2006; Gunawardena et al., 2003; Wang et al., 2007). Overexpression of a normal or a disease-related allele can be accomplished by crossing fly strains with cell type or tissue-specific GAL4 drivers to the fly strains harboring the gene-of-interest downstream of a GAL4 binding site (UAS) (Brand and Perrimon, 1993).
For live imaging studies, the opaque cuticle of the adult fly poses an optical challenge and makes dissections more difficult. Therefore, most imaging studies have chosen the third-instar larval stage for analysis. These larvae are easy to dissect and many mutants and transgenic lines of interest survive to this stage. Mutations that severely impair the transport of mitochondria may die before this stage, however; milton null alleles, for example, die in the first instar (Stowers et al., 2002). This need not preclude live imaging of mitochondria, however, though it does make the dissection more challenging and imaging in intact specimens through the cuticle may be preferred by those who have not mastered the dissection.
In Drosophila larvae, the cell bodies of central nervous system neurons form the cortex of the brain lobes and ventral nerve cord (VNC), and are bilaterally symmetrical. In the core of the VNC two parallel zones of neuropil lie to either side of the midline. It is in this neuropil where the axonal and dendritic projections of the cell bodies comingle and communicate. The complexity of the neuropil has, to date, made it unattractive for imaging studies of axonal transport. Instead, laboratories have focused on the nerves that issue from the VNC and extend to the body wall in each segment. These nerves include the axons of motorneurons that emanate from the VNC and terminate in well-characterized neuromuscular junctions on larval body walls. These segmental nerves also contain sensory axons whose cell bodies reside in the body wall and whose axons extend into the VNC, with the opposite orientation of the motorneurons. Therefore, to know the polarity of mitochondrial movement in these nerves, it is necessary to selectively label a subset of neurons of either the sensory or motorneuron class. To date, this has principally been done by the selective expression in motorneurons of a fusion of GFP with a mitochondrial import signal (Miller et al., 2005; Piling et al., 2006). In the third section, we will introduce the protocols to analyze the axonal transport of mitochondria in Drosophila neuronal axons at the late larval stage (third instar) and will describe a variation on the method of Pilling et al (2006) in which we selectively label mitochondria in one peptidergic motor neuron per segmental nerve.
2. PROTOCOLS TO IMAGE AXONAL TRANSPORT OF MITOCHONDRIA IN RAT HIPPOCAMPAL NEURONS
2.1 Embryonic Neuronal Dissection
Day 18 rat embryos were taken out from a euthanatized pregnant rat and decapitated. Hippocampal tissues were dissected and collected and washed with chilled Ca2+, Mg2+-free HBSS buffer (Hanks' Balanced Salt Solution, Invitrogen Catalog No. 14170-112). The hippocampal tissues were incubated with 15 ml Digestion Solution (stock: 30 ml HBSS, 75 μl 50 mM EDTA pH8.0, 1ml 15 mg/ml Papain) at 37 °C for 30 minutes, and harvested in a tabletop centrifuge at 1300 rpm at room temperature for 5 minutes. Digestion solution was then replaced with 5 ml Digestion Inhibition Solution (stock: 27 ml HBSS, 3ml Solution A, 30 μl 1% DNase (Sigma, MO)). Solution A stock: 100 ml HBAA, 1 g BSA, 1 g Trypsin Inhibitor (Sigma). The tissue was triturated 10 times with pipettes and added to 10 ml Digestion Inhibition Solution and 5 ml Solution A, before centrifugation at 1300 rpm for 10 minutes. The pellet of dissociated hippocampal neurons was resuspended with Neurobasal medium (Invitrogen, Carlsbad, CA).
2.2 Primary Neuronal Culture
Coverslips (12 mm diameter, #1.5) were ethanol sterilized and autoclaved, and coated with filter-sterilized poly-ornithine (1mg/50ml, Sigma) and laminin (170 μg/50ml, Invitrogen) in 24-well plates the night before plating. The next day, coated coverslips were washed twice with sterile water and once with Neurobasal medium (Invitrogen). Dissociated neurons were then plated onto the coated coverslips at 50-100,000/well in 24-well plates in Neurobasal supplemented with B27 (Invitrogen) and 100 units/ml penicillin and 100μg/ml streptomycin for one hour at 37 °C 5% CO2 which allows most healthy neurons settle down and attach to the coverslips, and the medium was replaced with fresh medium to remove debris and dead cells. Neuronal cultures were maintained at 37 °C in 5% CO2 for 8-11 days prior to transfection.
2.3 Constructs
The following constructs were used: pLPS-RFP-mito (Colwill et al., 2006), (No. 11702, Addgene, Cambridge, MA); pEYFP-Synaptophysin, pEYFP-PSD95 (Chang et al., 2006).
2.4 Transient Transfection and Dye Application
For transient transfection, each well of cultured hippocampal neurons was washed twice with pre-warmed 500 μl plain Neurobasal (without B27 and antibiotics). 1 μg of pLPS-RFP-mito DNA and 200 ng of synaptophysin-YFP (or PSD95-YFP) DNA were then added together with 2.5 μl Lipofectamine 2000 (Invitrogen) in 100 μl plain Neurobasal. The DNA-lipofectamine 2000 mixture was removed two hours later, and neurons were washed three times with plain Neurobasal before being returned to the conditioned medium (the medium that had been used to culture the neurons before transfection). Transfected neurons were imaged 2-4 days later. If a fluorescent dye was used rather than the RFP-mito, Rhodamine 123 (Invitrogen) was applied to the cultures at 10μg/ml for 10 minutes at 37 °C 5% CO2 and the cultures were washed with fresh Neurobasal three times and imaged.
2.5 Image Acquisition
A coverslip with live neurons was placed into a 35 mm round dish and maintained in CO2-independent medium (Hibernate E, Brainbits, IL, recipe online: www.brainbitsllc.com) on a 37°C heated stage for less than an hour. Images were collected on a Zeiss LSM 510 confocal microscope (LSM 510 META/NLO; Carl Zeiss MicroImaging, Inc., Thornwood, NY) with LSM software 3.2 (Carl Zeiss MicroImaging, Inc.) using a 63×/N.A.0.90 water IR-Achroplan objective with excitation at 488 and 543nm. Lasers were used at 10% to minimize damage, and pinholes were opened maximally to allow the entire thickness of the axon to be imaged. Images were captured every 2 seconds. Images were processed with Photoshop 10.0 (Adobe, San Jose, CA) using only linear adjustments of contrast and color, and Illustrator 13.0 (Adobe). The sensitivity of detecting small movements is potentially limited by two factors: the pixel size and drift of the image. Drift can be determined by measuring mitochondrial movement in fixed tissue and, assuming that this is minimal, the threshold for movement detection will be 1 pixel displacement per time interval between sweeps (typically 2 seconds).
2.6 Image Analysis
To analyze the characteristics of movement of individual mitochondria, Metamorph software (MDC, CA) was used to generate kymographs from the time-lapse live-imaging movies. All the LSM pictures from one time-lapse file can be imported into Metamorph as an image stack. A region of the axon of a desired length (typically 70 to 150μm) can be selected from the image stack and analyzed. In each kymograph, the x axis represents the position along the axon and the y axis represents time. Vertical lines indicate stationary mitochondria with no displacement during the time elapsed and diagonal lines represent moving mitochondria and their direction. Their velocity is reflected in the slope of the lines. From kymographs, one can also determine additional dynamic properties of movement, such as whether a mitochondrion is in constant motion, stops and starts again, or changes its directions. For a comprehensive description of the dynamics of mitochondria, it is advisable to calculate the number of mitochondria that are stationary, moving anterograde, or moving retrograde, the velocity of each mitochondrion that is in motion, for each direction of movement, and the percentage of time the mitochondria spend moving in each direction in the sample. The duration of continuous movements and the frequency of pauses or reversals of direction can give information about the processivity of mitochondrial movement. The length of each mitochondrion can also be determined. An example of such analysis can be found in Wang and Schwarz (2009).
2.7 Expected Results
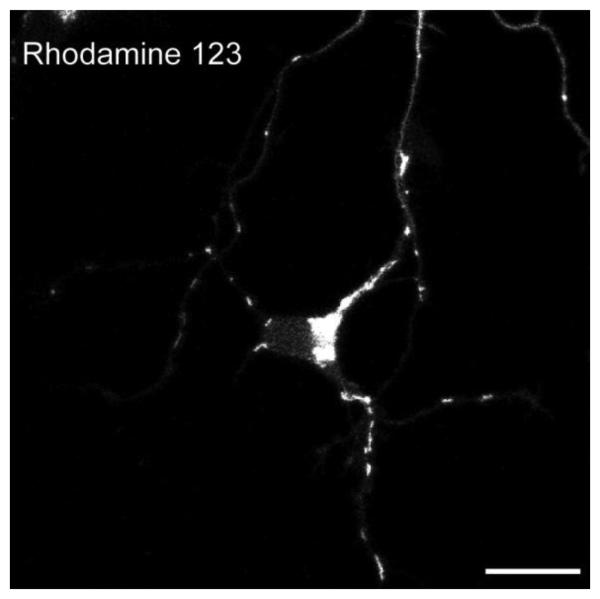
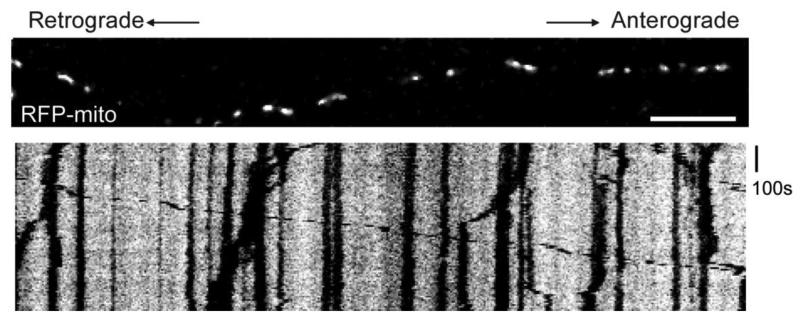
Neuronal mitochondria can be labeled by both mitochondrial dyes (such as MitoTrackers, Rhodamine 123 etc) and fluorescent protein-tagged mitochondrial protein (such as RFP-mito used here). We found that MitoTracker dyes diminish mitochondrial motility and alter their morphology, probably by affecting mitochondrial membrane potential (Buckman et al., 2001), and therefore avoid MitoTracker for live-imaging of neuronal mitochondria. Rhodamine 123 is another widely-used mitochondrial dye (Morris and Hollenbeck, 1995). In cultured hippocampal neurons, Rhodamine 123 labeled mitochondria have normal morphology (Figure 1) and are highly motility (data not shown). Therefore, Rhodamine 123 might be a suitable choice for cultured neurons with uniform properties, such as neurons from knock-out animals. However, as mentioned above, by labeling all the mitochondria in the dish, the dye can make it difficult to discern which mitochondria are in a single identified neurite. The neurons therefore need to be plated at a very low density in order to distinguish individual axons and hence to know the orientation of movement. Tagging mitochondria by transfection with a construct encoding a mitochondrially-targeted fluorescent protein (RFP-mito) can be a better choice than mitochondrial dyes for this reason, but also is preferable because it can be co-transfected with multiple proteins of interest into the neurons (also see Wang and Schwarz, 2009). Because neurons that have taken up the RFP-mito plasmid are very likely to have taken up the additional plasmids, one is selectively imaging from those neurons that are expressing the transgene of interests. Figure 2 shows an axon transfected with RFP-mito and synaptophysin-YFP. Mitochondrial movement labeled by RFP-mito was recorded and transformed to a kymograph. The majority of mitochondria are stationary. About 30% of mitochondria are moving during the interval shown (also see Wang and Schwarz, 2009). This method to analyze axonal transport of mitochondria in cultured neuronal system may be used to study the impact of overexpressed proteins, neurotrophic factors, toxins, stress conditions or similar manipulations on the axonal transport and morphology of mitochondria. These approaches will not only illuminate the basic mechanisms of mitochondrial motion and the regulation of those dynamics, but will also provide a better understanding of the significance of mitochondrial motility in normal and pathological cellular processes.
Figure 1. Rhodamine 123 Staining of Mitochondria in Rat Hippocampal Neurons.
A neuronal cell body with a few neurites, stained with Rhodamine 123 as in text. Neurons were dissected and dissociated from embryonic rat hippocampus, and cultured in vitro for 4 days. Scale bar, 10 μm.
Figure 2. RFP-mito Movement in Cultured Rat Hippocampal Neurons.
A cultured hippocampal neuron axon, transfected with RFP-mito and synaptophysin-YFP, with the axonal terminal to the right and cell body to the left. Neurons were dissociated and cultured as in text. The upper panel is the first frame of the time-lapse movie of RFP-mito and the lower panel is the kymograph (see text for details) generated from the movie showing the movements of RFP-mito during the time lapse. Vertical or diagonal black lines represent mitochondrial movements in the kymograph. Scale bar, 10 μm.
3. PROTOCOLS TO IMAGE AXONAL TRANSPORT OF MITOCHONDRIA IN DROSOPHILA LARVAL NEURONS
3.1 Fly Stocks and Culture
The following fly stocks were used: CCAP-GAL4 (Park et al., 2003), D42-GAL4 (Pilling et al., 2006), and UAS-mito-GFP (Pilling et al., 2006). CCAP-GAL4 drives protein expression in a particular class of neurosecretary neurons that make the neuropeptide CCAP (Crustacean Cardio-Active Peptide). This is a very sparse population of cells, thought to be just one per hemisegment of the VNC, whose cell bodies are laterally located in the VNC and which send an axon out in the segmental nerve to end in peptidergic boutons located on a single body wall muscle (Muscle 12) in each hemisegment (Park et al., 2003; Vomel and Wegener, 2007). Thus, if this line is used to drive expression of a fluorescent marker, only the axon of this CCAP neuron will be visible in the segmental nerve. D42-GAL4 drives protein expression chiefly in motor neurons (Yeh et al., 1995) and not at all in sensory neurons. Thus, if this line is used to drive expression of a fluorescent marker, all the efferent axons, but none of the sensory afferents, will be visible in the segmental nerve. Mito-GFP is a GFP-tagged mitochondrial targeting sequence (Pilling et al., 2006). Placed, behind a UAS promoter, it will be expressed in whatever population of neurons is expressing GAL4.
Flies were cultured in a 12 hour light-dark cycle at 25 °C on standard soft medium seeded with live yeast. UAS-mito-GFP was crossed to either UAS-CCAP-GAL4 or UAS-D42-GAL4 and the flies were placed in small fly cages with grape juice agar plates (see below) seeded with yeast as the only food source on which eggs could be laid. Grape juice plates were replaced every day after egg laying (AEL). The progeny of UAS-mito-GFP and CCAP-GAL4 will have mito-GFP expression in CCAP neurons, and the progeny of UAS-mito-GFP and D42-GAL4 will have mito-GFP expression in the broader pool of motor neurons.
3.2 Grape Juice Agar Plates
473 ml Welch's Grape Juice, 455 ml sterile water, and 29.3 g Granulated Agar were mixed in a 2L beaker, and heated in microwave for at least 8 minutes till all the agar was dissolved. It was stirred occasionally as it cooled down till 65 °C, and 9.9 ml 95% Ethanol and 9.5 ml Glacial Acetic Acid were added. Small tissue culture dishes (60 mm diameter) were filled with the mixed grape juice agar, and covered until the agar had hardened and prior to use.
3.3 Staging and Dissection of Live Larvae
Drosophila larvae reach the third instar stage approximately 4-5 days AEL and live about 48 hours before pupariation. Wandering third instar larvae were picked from grape juice agar plates and dissected promptly in fresh Schneider's medium (Invitrogen) with 5mM EGTA. Larvae were dissected as described (Hurd and Saxton, 1996) on a glass slide (25×75 mm, 1.0 mm thick, VWR International, PA) that had been coated with a thin layer of Sylgard (Dow Corning Corporation, MI) and sealed with a silicon imaging chamber (Grace Bio-labs, OR) to hold medium. Dissection pins (FST, CA) that had been cut to 2-3 mm to avoid touching the lens were used to immobilize the larvae. The larvae are cut open along the dorsal midline, the body wall muscles are pinned back, and the major gut structures and salivary glands are carefully removed. The dissection needs to be completed within 5 minutes and should expose an intact VNC, segmental nerves and muscles.
3.4 Image Acquisition
Live dissected larvae in the imaging chamber on the glass slide were maintained in fresh Schneider's medium with 5mM EGTA at 22 °C for less than an hour. Images were collected on a Nikon E800 epifluorescence upright microscope with SPOT software (Diagnostic Instruments, MI) using a Nikon NIR Apo 60×/N.A.1.0 water objective with excitation at 488 nm. Images were captured every 2 seconds for 200 frames. Images were processed with Photoshop 10.0 (Adobe, San Jose, CA) using only linear adjustments of contrast and color, and Illustrator 13.0 (Adobe).
3.5 Image Analysis
The original time-lapse pictures are saved as a tiff-stack by the SPOT software. This tiff-stack can be imported into Metamorph as described previously. Since the stage scale of the Nikon E800 microscope needs to be calibrated manually by the users, the actual displacements of these original tiff pictures will not be retained automatically when imported into Metamorph, but the pixel values of these pictures will still be displayed. Kymographs can be generated as in Section 2.6.
3.6 Expected Results
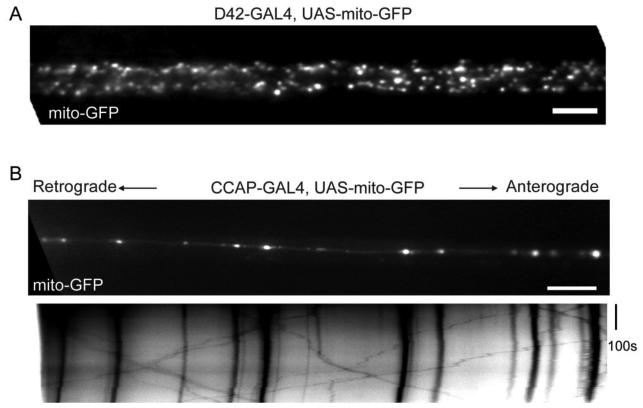
The in vivo imaging of axonal transport of mitochondria by mito-GFP in Drosophila larvae was first established by Piling et al. (2006) in motor neuron axons using D42-GAL4. Since mito-GFP is expressed in all motor neurons, segmental nerves contain numerous axons are intensely fluorescent with mito-GFP (Figure 3A and Pilling et al., 2006), to the extent that individual mitochondria can be difficult to resolve and difficult to track for any distance. To circumvent this problem, regional photobleaching was required (Pilling et al., 2006). After photobleaching a small area in an axonal bundle, only those mitochondria that move into the bleached zone from either end will be visible in that zone. Although this is an excellent way to analyze moving mitochondria and allows multiple mitochondria moving in either the anterograde or retrograde directions to be analyzed, information about the pool of stationary mitochondria in the original photobleaching area is necessarily lost. Thus it is not possible to determine the percent of mitochondria that are motile vs stationary. In addition, a strong laser is required for photobleaching; in Piling et al., 2006, a 488-nm light at full intensity from the MRC600 confocal laser (60×objective, zoom factor 4) was used to bleach the segment. Therefore we have modified that technique by expressing mito-GFP in CCAP neurons which selectively innervate a single muscle (Muscle 12) per hemisegment (Park et al., 2003; Vomel and Wegener, 2007). The content of mito-GFP in axonal bundles is therefore greatly reduced and analysis without photobleaching is possible (compare Figure 3A and 3B). Mitochondria exhibit similar bidirectional movements as in cultured mammalian neurons (Figure 3B, Pilling et al., 2006; Wang and Schwarz, 2009). This method will permit this semi-intact in vivo system to be applied to study axonal transport of mitochondria in conjunction with Drosophila genetics. The axonal transport of mitochondria can therefore be imaged and analyzed in different mutant backgrounds, and the effect of pathological proteins, expressed as transgenes with UAS promoters, can be examined because the transgene will be expressed in the same subset of neurons that are marked with UAS-mito-GFP. An additional modification of this method has been made (Miller et al., 2005) in which imaging can be done in intact, living larvae, without the need for a dissection. This method was used for examining the transport of synaptic vesicles, but should be applicable to the examination of mitochondria as well. The method has the advantage of a true in vivo record of organelle motility, but it can be more difficult to keep a significant length of axon in the plane of focus of the microscope and requires the use of chloroform to immobilize the larvae.
Figure 3. Mito-GFP Expression and Movement in Drosophila Larval Neurons.
(A) A segmental axonal bundle of a third instar Drosophila larva with the VNC to the left, in which mito-GFP was expressed in all motor neuronal axons driven by D42-GAL4. (B) The upper panel shows the first frame of a time-lapse movie from the segmental axonal bundle of a third instar Drosophila larva, with the VNC to the left, in which mito-GFP was expressed in selected CCAP axons driven by CCAP-GAL4. The lower panel is the kymograph (see text for details) generated from the movie, showing the movements of individual mitochondria during the time lapse, in which vertical or diagonal black lines represent mitochondrial movements. Scale bars, 10 μm.
4. CONCLUSION
The advancements of modern microscopic techniques and neuronal culturing systems have greatly facilitated our ability to image the axonal transport of mitochondria in live neurons. Here we introduced two neuronal systems, one is in vitro cultured rat hippocampal neurons, and the other is in vivo Drosophila third instar larval neurons, to record live mitochondria in axons. Both systems have advantages and disadvantages, and both methods can be used together to establish mechanisms and regulatory systems underlying mitochondrial motility, and pathological influences that will figure in neurodegeneration.
ACKNOWLEDGEMENT
We thank Drs. A. M. Craig and T. Pawson for reagents; Drs. F. Sun, Z. Wills, M. Greenberg for assistance with hippocampal cultures; Dr. L. Bu and the Developmental Disorders Research Center Imaging Core and E. Pogoda for technical assistance. This work was supported by NIH RO1GM069808.
REFERENCES
- Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 2007;27:422–30. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolino A, Bolis A, Previtali SC, Dina G, Bussini S, Dati G, Amadio S, Del Carro U, Mruk DD, Feltri ML, Cheng CY, Quattrini A, Wrabetz L. Disruption of Mtmr2 produces CMT4B1-like neuropathy with myelin outfolding and impaired spermatogenesis. J. Cell Biol. 2004;167:711–21. doi: 10.1083/jcb.200407010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buckman JF, Hernandez H, Kress GJ, Votyakova TV, Pal S, Reynolds IJ. MitoTracker labeling in primary neuronal and astrocytic cultures: influence of mitochondrial membrane potential and oxidants. J. Neurosci. Methods. 2001;104:165–76. doi: 10.1016/s0165-0270(00)00340-x. [DOI] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Mitochondrial movement and positioning in axons: the role of growth factor signaling. J. Exp. Biol. 2003;206:1985–1992. doi: 10.1242/jeb.00263. [DOI] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr. Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chang DT, Honick AS, Reynolds IJ. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J. Neurosci. 2006;26:7035–45. doi: 10.1523/JNEUROSCI.1012-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Owens GC, Crossin KL, Edelman DB. Serotonin stimulates mitochondrial transport in hippocampal neurons. Mol. Cell. Neurosci. 2007;36:472–483. doi: 10.1016/j.mcn.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Colwill K, Wells CD, Elder K, Goudreault M, Hersi K, Kulkarni S, Hardy WR, Pawson T, Morin GB. Modification of the Creator recombination system for proteomics applications--improved expression by addition of splice sites. BMC Biotechnol. 2006;6:13. doi: 10.1186/1472-6750-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Chapman AL, Tennant ME, Manser C, Tudor EL, Lau KF, Brownlees J, Ackerley S, Shaw PJ, McLoughlin DM, Shaw CE, Leigh PN, Miller CC, Grierson AJ. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum. Mol. Genet. 2007;16:2720–8. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreirinha F, Quattrini A, Pirozzi M, Valsecchi V, Dina G, Broccoli V, Auricchio A, Piemonte F, Tozzi G, Gaeta L, Casari G, Ballabio A, Rugarli EI. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 2004;113:231–42. doi: 10.1172/JCI20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J. Cell Sci. 2005;118:5411–9. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye AT, Davoren JE, Wipf P, Fink MP, Kagan VE. Targeting mitochondria. Acc. Chem. Res. 2008;41:87–97. doi: 10.1021/ar700135m. [DOI] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–85. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat. Protoc. 2006;1:2406–15. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- McDermott CJ, Grierson AJ, Wood JD, Bingley M, Wharton SB, Bushby KM, Shaw PJ. Hereditary spastic paraparesis: disrupted intracellular transport associated with spastin mutation. Ann. Neurol. 2003;54:748–59. doi: 10.1002/ana.10757. [DOI] [PubMed] [Google Scholar]
- Miller KE, DeProto J, Kaufmann N, Patel BN, Duckworth A, Van Vactor D. Direct observation demonstrates that liprin-α is required for trafficking of synaptic vesicles. Curr. Biol. 2005;15:684–689. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J. Cell Biol. 1995;131:1315–26. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overly CC, Rieff HI, Hollenbeck PJ. Organelle motility and metabolism in axons vs dendrites of cultured hippocampal neurons. J. Cell Sci. 1996;109 (Pt 5):971–80. doi: 10.1242/jcs.109.5.971. [DOI] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–56. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Pigino G, Morfini G, Pelsman A, Mattson MP, Brady ST, Busciglio J. Alzheimer's presenilin 1 mutations impair kinesin-based axonal transport. J. Neurosci. 2003;23:4499–508. doi: 10.1523/JNEUROSCI.23-11-04499.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–68. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Tiwari P, Xie Z, Zheng JQ. Acute impairment of mitochondrial trafficking by beta-amyloid peptides in hippocampal neurons. J. Neurosci. 2006;26:10480–7. doi: 10.1523/JNEUROSCI.3231-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Thies E, Mandelkow EM. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J. Neurosci. 2007;27:2896–907. doi: 10.1523/JNEUROSCI.4674-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol. 2004;24:8195–209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vomel M, Wegener C. Neurotransmitter-induced changes in the intracellular calcium concentration suggest a differential central modulation of CCAP neuron subsets in Drosophila. Dev. Neurobiol. 2007;67:792–808. doi: 10.1002/dneu.20392. [DOI] [PubMed] [Google Scholar]
- Wang X, Shaw WR, Tsang HT, Reid E, O'Kane CJ. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 2007;10:177–85. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+ regulation of kinesin-mediated mitochondrial motility. Cell. 2009 doi: 10.1016/j.cell.2008.11.046. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J, Smith SJ. Mitochondria and release at hippocampal synapses. Pflugers Arch. 2003;447:363–370. doi: 10.1007/s00424-003-1182-0. [DOI] [PubMed] [Google Scholar]
- Yeh E, Gustafson K, Boulianne GL. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc. Natl. Acad. Sci. USA. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]