Abstract
The molecular mechanisms involved in the derivation of induced pluripotent stem cells (iPSCs) from differentiated cells are poorly understood. Here we report that caspases 3 and 8, two proteases associated with apoptotic cell death, play critical roles in induction of iPSCs from human fibroblasts. Activation of caspases 3 and 8 occurs soon after transduction of iPSC-inducing transcription factors. Oct-4, a key iPSC transcription factor, is responsible for the activation. Inhibition of caspase 3 or 8 in human fibroblast cells partially or completely prevents the induction of iPSCs, respectively. Furthermore, retinoblastoma susceptibility (Rb) protein appears to be one of the factors that act downstream of the caspases. We propose that caspases are key facilitators of nuclear reprogramming in iPSC induction.
Keywords: iPSC, epigenetic reprogramming mechanism, caspase 3, caspase 8, retinoblastoma, Oct-4
Introduction
The ability to derive induced pluripotent stem cells (iPSCs) from various differentiated cells has ushered in a new era in stem cell research. Since the initial success in producing iPSCs from murine fibroblasts (Takahashi and Yamanaka, 2006), a significant number of advances have been made. Validation of the process was provided by many independent studies (Blelloch et al., 2007; Hanna et al., 2007; Meissner et al., 2007; Okita et al., 2007; Wernig et al., 2007). The authenticity of the embryonic stem cell like-properties of iPSCs came from studies showing the ability of murine iPSCs to participate in formation of various tissues when injected into murine embryos at the blastocyst stage (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). Another advance came from studies showing that it is possible to derive iPSCs without the use of the myc oncogene (Huangfu et al., 2008; Kim et al., 2008; Nakagawa et al., 2008), one of the original 4 transcriptional factors used by the Yamanaka group. This success is important because it significantly reduces the risk of carcinogenesis from the iPSC induction procedure.
A major milestone in the field was the successful derivation of iPSCs from human fibroblasts (Lowry et al., 2008; Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007), which generated great excitement because of its implications in human regenerative medicine.
Despite the tremendous progress made in the iPSC field, one key question remains unresolved: what is the molecular mechanism(s) involved in the iPSC derivation process? Many laboratories have made heroic efforts to try to identify the molecular factors involved in inducing or maintaining pluripotency. Examples include numerous gene profiling and epigenetic mapping studies (Azuara et al., 2006; Brambrink et al., 2008; Efroni et al., 2008; Jiang et al., 2008; Meissner et al., 2008; Mikkelsen et al., 2008; Mikkelsen et al., 2007; Pan et al., 2007; Sridharan et al., 2009; Zhao et al., 2007) conducted in either iPSCs or embryonic stem cells (ESCs). In addition, recent reports of the important roles of tumor suppressor genes p53, p16, and p19ARF shed some light on the iPSC induction process (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). Another interesting advance came from a study that showed that activation-induced cytidine deaminase (AID) may be required for epigenetic reprogramming towards pluripotency in human cells (Bhutani et al., 2010). However, the key molecular pathways/processes through which iPSCs are derived from differentiated cells remain unknown.
In this study, we report a surprising finding—that caspases 3 and 8, two proteases previously associated with regulation of apoptotic cell death, play key roles in mediating the reprogramming of human fibroblasts into iPSCs. Both caspases are activated by Oct-4, the most important transcription factor in iPSC induction. The functional importance of the activation of the caspases was demonstrated by the fact that blocking caspase 8 led to a complete shutdown of the iPSC reprogramming process. Furthermore, we provide evidence that the retinoblastoma susceptibility (Rb) protein is a critical target of caspases 3 and 8 during iPSC nuclear reprogramming from human fibroblast cells.
Results
Activation of caspases 3 and 8 during the iPSC induction process
We were interested in caspase activities in the iPSC induction process because it had been known that some caspases play critical roles in cellular differentiation. For examples, caspases are known to be involved in the differentiation of hematopoietic stem cells (Kang et al., 2004), osteoclasts (Szymczyk et al., 2006), and embryonic stem cells (Dejosez et al., 2008; Fujita et al., 2008). We reasoned that because of their important roles in differentiation, caspases might also play some role in the reversal of the process: iPSC induction, which is essentially a de-differentiation process.
In order to monitor caspase activities in the iPSC induction process, we developed two non-invasive caspase reporters to monitor caspase 3 and 8 activities non-invasively. These two caspases were chosen because of previous reports of their involvement in differentiation (Dejosez et al., 2008; Fujita et al., 2008; Kang et al., 2004; Szymczyk et al., 2006). Our caspase reporters consist of a firefly luciferase-GFP fusion protein (LucGFP) linked to a polyubiquitin domain. In between the two moieties, a caspase cleavage site (either -IETD- for caspase 8 or –DEVD- for caspase 3) is inserted (Figure 1A, see supplementary Figure S1 for data validating the functionality and specificity of the reporters). The assumption is that when caspases are inactive, the reporter proteins will be recognized by proteasomes and degraded immediately because the polyubiquitin domain serves as tag for protein destruction by proteasomes. However, when caspase 3 or 8 is active, the polyubiquitin domain will be cleaved off the reporter protein, leading to enhanced GFP and luciferase signals because these reporter proteins are now not subject to direct proteasome recognition and degradation.
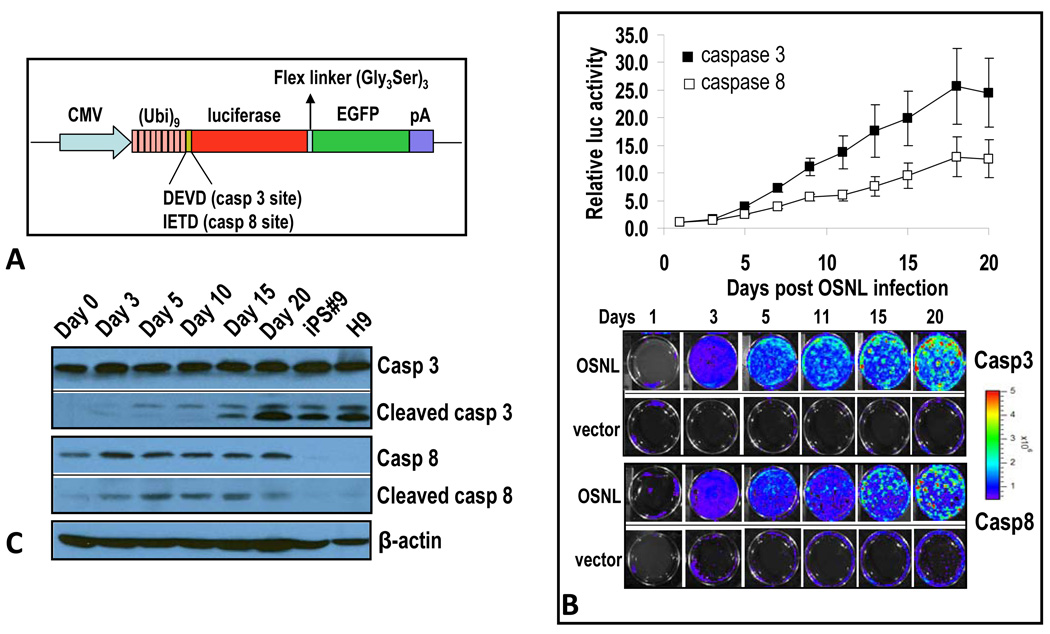
Figure 1. Caspases 3 and 8 are activated in the iPSC induction process.
A) A schematic diagram of caspase reporters based on the proteasome system. Our reporter consists of a fusion reporter domain where a firefly luciferase gene is fused with the enhanced green fluorescence protein (GFP) gene. A polyubiquitin domain is linked to the reporter with caspase 3 (DEVD) and caspase 8 (IETD) cleavages sites. In some cases, the CMV promoter was changed to that of EF1α.
B) Caspase 3 and 8 activation induced by iPSC-inducing transcription factor cocktail. IMR90 cells stably transduced with caspase 3 or caspase 8 reporters were infected with lentiviral vectors encoding OSNL. Reporter activities were then followed through bioluminescence imaging. Top panel, quantitative data showing time-dependent increases in caspase 3 and 8 activities in OSNL-transduced IMR90 cells. Lower panel, representative images of bioluminescence in OSNL-transduced cells. Error bars represent standard error of the mean (SEM, n=3).
C) Western blot analysis of caspase 3 and 8 in IMR90 cells transduced with OSNL. Cells transduced with OSNL were analyzed for total and activated caspase 3 and 8 activities at different times after lentiviral infection. H9 is a commonly used ESC line and iPS#9 is an IMR90-derived iPSC clone. β-actin was used as a protein loading control.
Please also see Figure S1 and Figure S2 for additional related data.
We transduced the caspase reporters into human IMR90 fibroblasts via recombinant lentivirus vectors. After puromycin selection to ensure stable integration of the reporters, we initiated the iPSC reprogramming process by infecting caspase reporter-transduced IMR90 cells with recombinant lentivirus vectors carrying 4 transcription factors (Oct-4, Sox2, Nanog, and Lin28, or OSNL in abbreviation) that are known to induce iPSC formation (Yu et al., 2007). We chose to conduct the studies with the OSNL protocol to avoid the use of myc, which was part of the original Yamanaka OSKM protocol (Takahashi and Yamanaka, 2006), because it was shown not to be essential for iPSC induction (Huangfu et al., 2008; Kim et al., 2008; Nakagawa et al., 2008). In addition, it has known abilities to induce caspase activation (Evan et al., 1994; Evan et al., 1992; Hueber et al., 1997), which might confound our results.
Interestingly, increased caspase 3 and 8 reporter activities were observed starting at day 3, continued to rise, and remained elevated during the rest of the observation period (Figure 1B). The specificity of the reporter was confirmed in supplementary Figure S1D, which shows that expression of dominant negative caspase 3&8 genes in OSNL transduced IMR90 cells blocked caspase 3&8 reporter activation, respectively. In addition, these data show the dependency of caspase 3 activation on caspase 8 but not vice versa. Furthermore, our results indicated that reporter activation was not due to a decrease in proteasome function as a control reporter with no caspase cleavage site showed a small decrease in signals instead of an increase after OSNL transduction (supplementary Figure S1E).
In iPSCs that eventually emerge from the OSNL-transduced cells, most (over 60%) of the colonies were positive for the caspase 3 reporter (supplementary Figure S2A) when observed through GFP fluorescence(supplementary Figure S2B).On the other hand, caspase 8 reporter activities were observed in less than 20% of iPSC colonies.
To confirm the above imaging results, western blot analyses of caspases 3 and 8 were carried out in IMR90 cells transduced with OSNL (Figure 1C). Our results indicated that while the levels of intact caspase 3 remained roughly the same in OSNL-transduced cells over time, there were progressive increases in activated caspase 3 with time (as represented by cleaved caspase 3 levels), indicating a steady increase in total caspase 3 levels. The level of activated caspase 3 remained robust in iPSCs. Interestingly, it was also quite high in H9 human embryonic stem cells (ESCs), similar to previous observations made in murine embryonic stem cells (Fujita et al., 2008). There were significant increases in both total and activated caspase 8 levels in OSNL-transduced fibroblast cells (Figure 1C). The levels of both remained significant high during the entire course of observation. However, in contrast to caspase 3, there were lower amounts of total and activated caspase 8 in iPSCs that emerged from OSNL-transduced IMR90 cells (Figure 1C). Similarly, H9 human embryonic stem cells displayed low levels of total or activated caspase 8 (Figure 1C). We also carried out experiments to compare caspase 3 levels in iPSCs and iPSC-derived embryoid body (EB) cells because previously it was reported that caspase 3 levels increased during mouse ESC differentiation (Fujita et al., 2008). Our results indicated that in EB cells derived from human iPSCs, total and activated caspase 3 levels held steady (Figure S2C). In contrast, EB cells from mouse iPS cells had even higher levels of total and activated caspase 3 than mouse ESCs (Figure S2C), consistent with previous observations (Fujita et al., 2008).
Quantitative PCR analysis showe dincreased caspase 3 and 8 mRNA levels after OSNL transduction in IMR90 cells (supplementary Figure S2D), consistent with western blot analysis results. In both IMR90-derived iPSCs and human H9 ESCs, caspase 3 mRNA levels are at about the same level as in IMR90 cells. However, caspase 8 mRNA levels in H9 human ESCs or iPSCs dropped precipitously to about 1/10 of the level in IMR90 cells (supplementary Figure S2E), again consistent with western blot analysis.
Oct-4 expression alone is sufficient to induce caspase 3 and 8 activation
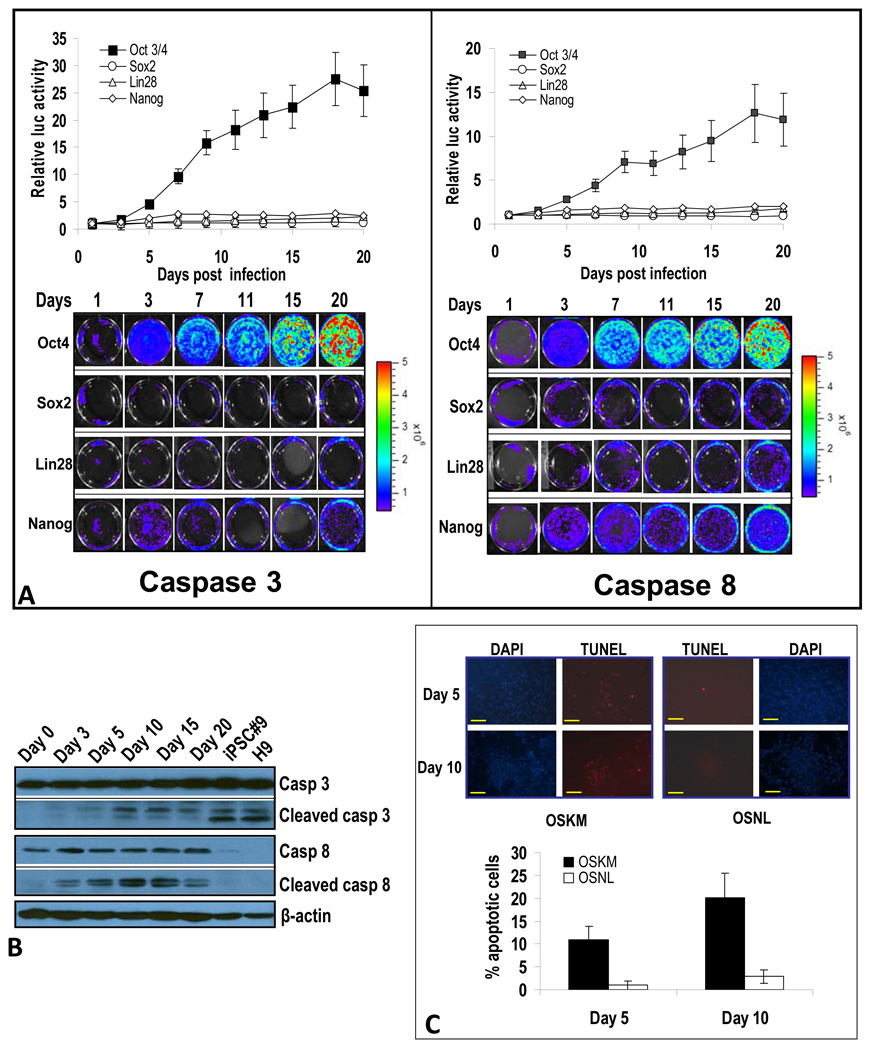
We next attempted to determine which of the four iPSC-inducing factors (e.g., OSNL) is responsible for caspase 3 and 8 activation. Our results (Figure 2A) with caspase reporters indicate that Oct-4, which is consistently the most important transcription factor required in different iPSC induction protocols (Maherali and Hochedlinger, 2008) and is able to induce iPSCs alone under certain circumstances (Kim et al., 2009a; Kim et al., 2009b), was responsible for caspase 3 and 8 activation. Among the other three factors, Nanog had a relatively small effect, and Sox 2 and Lin28 had minimal effects on caspase 3 and 8 activation (Figure 2A). Therefore, we conclude that Oct-4 is the factor primarily responsible for caspase 3 and 8 activation during OSNL-induced iPSC formation.
Figure 2. Oct-4 induced activation of caspases 3 and 8 and its effect on fibroblast cell viability.
A) Oct-4 is responsible for activation of caspases 3 and 8 in OSNL-transduced IMR90 cells. Reporter-transduced IMR90 cells were infected with lentiviral vectors encoding individual transcription factors and their reporter activities were followed by bioluminescence imaging. Top panels show the quantitative results of imaging caspases 3 and 8. Lower panels show the bioluminescence imaging of IMR90 cells transduced with caspase 3 and 8 reporters. The error bars represent standard error of the mean (SEM, n=3).
B) Western blot analysis of caspase 3 and 8 activation in Oct-4-transduced IMR90 cells. IMR90 cells infected with Oct-4 encoding lentiviral vectors were examined for caspase 3 and 8 expression and activation at different times after Oct-4 transduction.
C) Cell death in OSNL- and OSKM-transduced cells. IMR90 cells were infected with lentiviral vectors encoding OSNL or OSKM. At five or ten days post transduction, dying cells were quantified through TUNEL staining for DNA fragmentation, which is typical for cells at the end stage of apoptosis. The scale bars represent 200 µm.
Please also see Figure S3 for additional related data.
One important question is what kind of changes were induced by Oct-4 in the fibroblast cells. We observed significant morphological changes after Oct-4 transduction into IMR90 cells. These changes include smaller cell sizes, increased cellular densities, and faster growth rates (supplementary Figure S3A, left panels). When these cells were stained for stem cell markers, robust Oct-4 expression was observed, as expected. However, only sporadic expression of other stem cell markers was observed (supplementary Figure S3A, right panels). Despite clustered, colony type cellular growth, no iPS-like cells were observed over an extended period (10 weeks) of continuous culturing. These results indicated that forced Oct-4 expression induced significant morphological changes in IMR90 cells towards a more pluripotent state. However, its expression is not quite enough to push the cells into a full iPSC phenotype.
Western blot analysis confirmed that activities of both caspase 3 and caspase 8 were induced by Oct-4 transduction. The patterns of caspase 3 and 8 activation induced by Oct-4 paralleled those induced by OSNL (Figure 2B), thereby confirming the primary role of Oct-4 in mediating OSNL-induced caspase activation.
Additional western blot analysis confirmed induction of caspase 3 and 8 activation in IMR90 cells transduced with the original “Yamanaka Factors” (OSKM, or Oct-4+Sox2+Klf4+Myc) (Takahashi et al., 2007) (Figure S3B). A noteworthy difference is that OSKM appeared to induce caspase 3 and 8 activation at higher levels at day 10 (comparing Figures 1C, 2B, and S3B). Enhanced induction of caspase 3 and 8 activation is consistent with the extensive body of literature showing that myc, one of the four original Yamanaka factors, is able to induce caspase activation and cellular apoptosis (Evan et al., 1994; Evan et al., 1992; Hueber et al., 1997).
It is not known whether observed caspase activation in OSNL- or OSKM-transduced cells causes apoptosis. Because myc is known for its ability to induce apoptosis in human fibroblast cells and was expressed at significantly higher levels in OSKM transduced cells than in OSNL transduced cells (Figure S3C), we used the TUNEL assay to detect DNA fragmentation, which occurs at the end stage of apoptosis. Our results showed a significant amount of apoptosis (10–20% as determined by positive TUNEL signals) in OSKM-transduced IMR90 cells (Figure 2C). In comparison, only 1%-3% of OSNL-transduced cells were TUNEL positive (Figure 2C). This is consistent with previous findings that myc expression promotes fibroblast cells to undergo apoptosis (Evan et al., 1994; Evan et al., 1992; Hueber et al., 1997).
Critical roles of caspase 3 and 8 activation in the iPSC induction process
In order to determine whether the observed caspase 3 and 8 activation had any functional relevance in iPSC induction, we attempted to down-regulate caspase activities in OSNL-transduced cells. In our initial experiment, we stably transduced CrmA (through lentiviral vectors), a cow pox virus protein known to suppress caspase 1 and 8 activities (Gagliardini et al., 1994; Ray et al., 1992), into IMR90 cells. After infecting these cells with an OSNL-encoding lentiviral vector cocktail, we found a significant (i.e., around 80%) suppression in the frequency of iPSC induction (Figure 3A). This was a very surprising result because logically one would expect that inhibiting caspase activation would lead to increased iPSC generation because of reduced cell death from stress induced by OSNL or OSKM transduction, which can induce apoptosis in transduced fibroblast cells (Figure 2C).
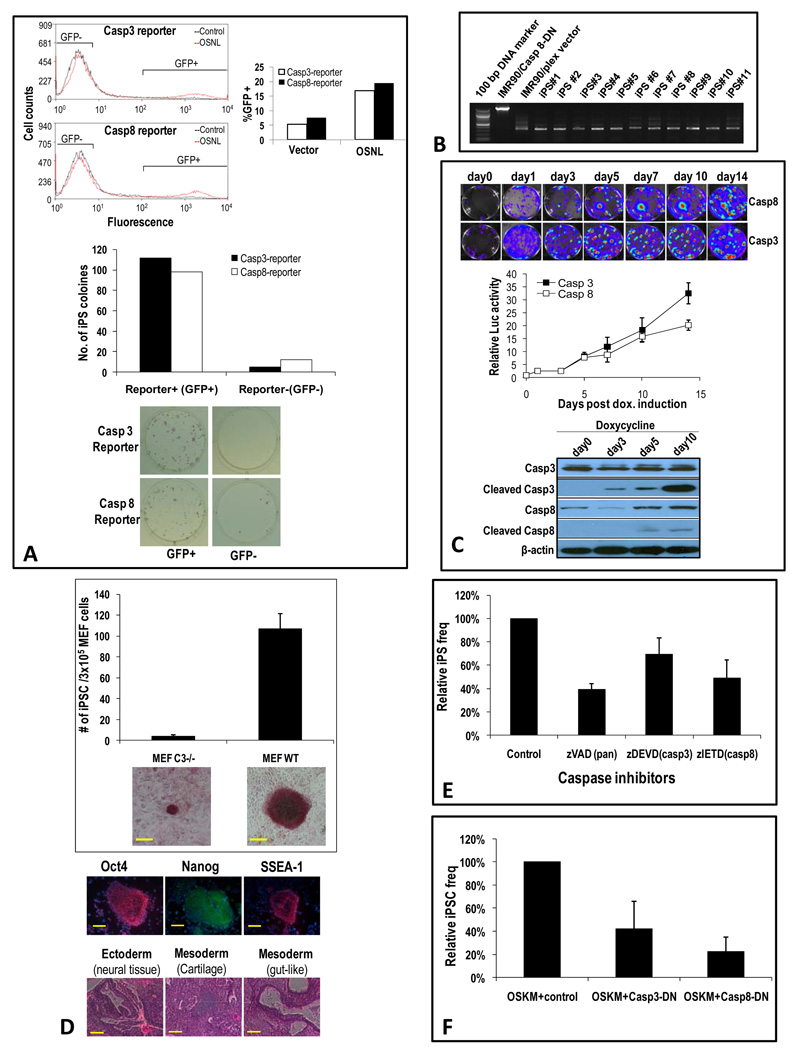
Figure 3. Caspases 3 and 8 are essential for iPSC induction.
A) CrmA-mediated suppression of iPSC formation in OSNL-transduced IMR90 cells. Top panel, relative frequencies of iPSC cellular formation. Error bars represent SEM (n=4). The inset shows the western blot of HA-tagged CrmA expression. Lower left panels, examples of alkaline phosphatase-stained iPSC-containing Petri dishes. Lower middle and right panels, examples of iPSC colonies with AP staining (lower middle) or no AP staining (lower right panels). The yellow scale bars represent 200 µm while the red scale bars represent 500 µm.
B) Caspase-targeted shRNA-mediated suppression of iPSCs. IMR90 cells were transduced with OSNL together with lentiviral vectors encoding shRNA minigenes targeted to caspase 1, caspase 8, or caspase 3, and the frequency of iPSC formation was quantified. Error bars represent SEM (n=4). Western blots showing knockdown of endogenous caspases are also shown.
C) C-FLIP-mediated attenuation of iPSC formation. IMR90 cells were infected with lentiviral vectors encoding the short form of c-FLIP in addition to the OSNL factors. Error bars represent SEM (n=5). The inset shows the western blot of exogenous HA-tagged cFLIP.
D) Complete blockade of iPSC formation by use of a dominant-negative caspase 8. IMR90 cells were stably transduced with DN-caspase 8 or DN-caspase 3. They were then transduced with lentiviral vectors encoding OSNL. Top panel, relative frequencies of iPSC formation. Error bars represent SEM (n=5). Western blot analysis of HA-tagged DN-caspase 3 and DN-caspase 8 expression are also shown. Lower panel, representative images of iPSC formation.
Please see Figure S4 for additional related data.
Because CrmA inhibits both caspase 1 and caspase 8, we subsequently used more specific approaches to pinpoint the exact caspase that was involved. First, we adopted an siRNA-based knockdown approach. To do this, we used lentiviral vectors encoding either caspase 1- or caspase 8-targeted shRNA to infect IMR90 cells. After selecting for cells with stable shRNA knockdown (as verified by western blot analysis), they were infected with the OSNL lentiviral vector cocktail. Our results indicate that expression of shRNA targeted to caspase 8 suppressed about 55% of iPSC formation (Figure 3B). In contrast, shRNA targeted to caspase 1 had no significant effect on iPSC induction frequency. These results indicated that the main inhibitory effect of CrmA on iPSC formation is mediated through caspase 8. Similarly, the importance of caspase 3,which can be activated by caspase 8, was demonstrated by targeted knockdown of its expression through shRNA. A significant reduction (around 50%) in iPSC induction was observed in caspase 3 knockdown IMR90 cells (Figure 3B).
The importance of caspase 8 was further demonstrated through transduction of the short version of the c-FLIP gene, which is a known cellular inhibitor of caspase 8 (Scaffidi et al., 1999; Yeh et al., 2000). Stable transduction of c-FLIP into IMR90 cells significantly attenuated the frequency of iPSC induction (about 50% reduction) (Figure 3C), again confirming the importance of caspase 8 in iPSC induction.
To provide definitive proof that the cleavage activities of caspases 3 and 8 are necessary for iPSC formation, we constructed lentiviral vectors encoding dominant-negative forms of caspases 3 and 8. The dominant negative form of the human caspase 8 gene (casp8-DN) encoding a protein virtually identical to the wild type except for a single amino acid change (i.e. C360A) (Stennicke and Salvesen, 1997), which disables its proteolytic function, was cloned into a recombinant lentiviral vector. Similarly, the dominant negative form of caspase 3 (casp3-DN) (Stennicke and Salvesen, 1997), which contains a single mutation (C163A) that disables its proteolytic function, was also cloned into a lentiviral vector. When IMR90 cells stably transduced with a lentiviral vector encoding casp8-DN were infected with the OSNL lentiviral cocktail, the iPSC induction process was completely shut down (Figure 3D, see Figure S4A for an amplified photo of the alkaline phosphatase stained, iPSC-containing Petri dishes). No iPSC clones were observed after 25 days of culture, compared with around 550 iPSC clones/5×105 cells for wild type IMR90 cells. The dramatic inhibitory effect achieved through dominant negative caspase 8 expression indicated that the cleavage activity of caspase 8 was essential for iPSC induction.
Similar to caspase 8, we found that stable transduction of a dominant negative caspase 3 could significantly reduce the frequency of iPSC induction (about 80%, Figure 3D). However, the induction process, despite being significantly attenuated by casp3-DN, was not completely shut down as in the case of casp 8-DN, indicating that there is some redundancy for the function of caspase 3, but not for that of caspase 8, in the iPSC derivation process.
The importance of caspase 8 was also confirmed in experiments conducted with the original Yamanaka protocol (Takahashi et al., 2007; Yamanaka and Takahashi, 2006). Transduction of casp 8-DN together with OSKM suppressed iPSC induction almost completely (over 95% suppression, see supplementary Figure S4B), indicating the general importance of caspase 8 activation in different human iPSC induction protocols.
Experiments were also conduced to examine the importance of caspase inhibitors at various stages of iPSC induction. Our results (supplementary Figure S4C) indicate that addition of caspase inhibitors can influence iPSC induction at all three stages (early, middle, and late) after OSNL transduction with early and late additions slightly more effective than middle stage addition.
A further experiment that supports positive roles for caspase 3 and 8 activation in iPSC formation came from following the fate of FACS sorted, caspase reporter-positive or -negative cells. IMR90 cells with caspase 3 or caspase 8 reporters (Figure 1A) were transduced with OSNL and FACS sorted 3 days later for cells that were either positive (as determined by GFP expression) or negative for caspase 3 or caspase 8 reporter activities. It is estimated that around 20% of OSNL-transduced cells were positive for caspase 3 or caspase 8. Around 9×104 of those GFP+ or GFP- cells (Figure 4A, top panel) were then plated on feeder cells and observed for iPSC formation. We found that caspase reporter-positive cells were 6 times more likely to form iPSCs than caspase reporter-negative cells (Figure 4A, middle and lower panels), strongly suggesting a positive relationship between caspase activation and iPSC formation.
Figure 4. The effect of caspase 3 and 8 activation on iPSC formation in OSNL-transduced IMR-90 cells.
A) Positive correlation of iPSC formation frequency with activated caspases. Top panel, FACS profile of IMR90 cells stably transduced with caspase 3 and 8 reporters and subsequently infected with lentiviral vectors encoding OSNL. Shown are FACS profiles at 3 days after OSNL transduction. Reporter positive and negative cells (as indicted by brackets in the FACS profiles) were then sorted and plated onto feeder cells at 9×104 cells/well in 6-well plates. Middle panel, frequencies of iPSC colonies from caspase+ and caspase- cells. Lower panel, alkaline phosphatase staining of iPSC colonies after 3 weeks of culturing.
B) Cell-intrinsic requirements for caspase 8 activation in the iPSC induction process. IMR90 cells transduced with either a control lentiviral vector (pLEX) or the same lentiviral vector encoding a dominant negative caspase 8 (casp8-DN) were mixed at a ratio of 1:9, infected with OSNL, and then plated onto feeder cells. Twenty days later, eleven randomly chosen iPSC colonies from the OSNL-transduced cells were picked and analyzed by PCR for the presence of either the control vector or casp8-DN.
C) Activation of caspases 3&8 in iPSC induction from murine fibroblast cells. Murine tail tip fibroblast transduced doxycycline-inducible OSKM were transduced with caspase 3 or 8 reporters. Reporter activation was then observed at different times after doxycycline addition. Top panel: representative images of reporter activities. Middle panel: quantitative plot of reporter activities. Error bars represent SEM (n=3). Lower panel: western blot analysis of total and activated caspases 3&8 in doxycycline treated cells.
D) iPSC colony formation from wild type and caspase 3−/− MEF cells. MEF cells were transduced with OSKM and evaluated for iPSC colony formation through AP staining 20 days later. Right panel, quantitative data showing average iPS colony numbers from triplicate plates. Error bars represent SEM (n=3). Representative AP staining of iPSC colonies from wild type and casp3−/− MEF cells were also shown. Scale bars represent 200 µm. Top right panels, verification of the stemness of putative iPS colonies through immunofluorescence staining of typical ESC markers. Lower right panel: formation of three germ layers in terotomas formed in nude mice from select wild type MEF cells.
E) Relative frequency of iPSC induction from MEF cells in the presence of chemical caspase inhibitors. The concentrations of the inhibitors used are at 5 µM. For control a structurally similar but non-inhibitive compound was used. Error bars represent SEM (n=3).
F) Relative frequency of iPSC Induction from MEF cells in the presence of dominant-negative caspase 3&8 genes. Error bars represent SEM (n=3).
One key question regarding the role of caspase 8 is whether the effects we observed were cell-autonomous or non-autonomous. It is theoretically possible that inhibition of caspase 8 attenuated some unknown, secreted paracrine factors that promote the formation of iPSCs from OSNL- or OSKM-transduced cells. To examine this possibility, we carried out experiments with IMR90 cells transduced with either casp8-DN or a control vector. The cells were mixed at the casp8-DN:control ratio of 9:1, and transduced with OSNL. The transduced cells were subsequently observed for iPSC colony formation. If the effects of caspase 8 were totally paracrine (or non-cell autonomous), we would expect that the ratio of iPSC colonies that originate from the two cell populations would equal to the ratio of the initial cellular mix, which is 9:1. If iPSC colonies were found mostly originate from control vector-transduced cells despite the fact they only accounted for 10% of the initial cell mix, this would indicate that the effect of casp8-DN expression is cell-autonomous. When iPSC colonies that emerged from the mixed cell population were selected, expanded, and analyzed through PCR, all 11 selected clones were proven to contain only the control vector sequences (Figure 4B). The probability for this to occur purely by chance is 10−11 (because the expected frequency for each of the 11 iPSC clones to originate from control vector transduced cells is 10−1 if the effect of caspase 8 suppression is non-cell autonomous) if we assume a non-cell autonomous mechanism. Therefore, our results strongly suggest that the effects of caspase 8 activation on iPSC induction are cell-autonomous.
Additional experiments also confirmed the importance of caspase activation in induction of iPSCs from mouse cells. In order to observe the activation of caspases 3&8 in murine iPSC induction process, we used a previously published secondary iPSC reprogramming system using mouse fibroblast cells derived from transgenic mice with doxycycline inducible OSKM genes already embedded within their genome (Stadtfeld et al., 2010). These cells were transduced with our caspase reporters (Figure 1A) and observed for caspase 3&8 induction. Our results indicate that both caspase 3&8 are strongly activated, as shown in Figure 4C. As a matter of fact, more than 50% of individual colonies from doxycycline induced cells showed strong activation of caspase 3&8 (top and middle panels) and many of these caspase activated colonies transformed into iPSC-like colonies after day 7. Western blot analysis confirmed the activation of both caspase 3&8 (Figure 4C, lower panel).
The functional importance of caspase activation was further demonstrated by use of mouse embryonic fibroblast cells derived from caspase 3 knockout mice. Our data (Figure 4D, top panel) indicate that caspase 3 deficiency in MEF cells significantly attenuated the number of iPSC colonies formed after OSKM transduction. Furthermore, even the few iPSC-like colonies that emerged from casp3−/− MEF cells were significantly smaller in size when compared with those from wild type cells. More importantly, they failed to expand when picked and plated with new feeder and ES medium. Not a single colony grew in three independent attempts (5 colonies were picked in each experiment). These data indicated that caspase 3 deficiency had a strong effect in preventing the full epigenetic reprogramming of MEF cells into iPS cells. In comparison, all iPSC-like colonies derived from wild type MEF cells expanded in secondary cultures successfully, indicating successful self renewal. In addition to strong AP staining (Figure 4D, middle panels), they stained positive for key ES markers (lower panels) and could form all three germ layers when injected into nude mice (Figure 4D, lower panels).
Further results supporting a role for caspases 3&8 come from experiments involving the use of chemical inhibitors of caspases (either general caspase inhibitors or specific caspase 3 or 8 inhibitors), which significantly inhibited iPSC formation from OSKM-transduced MEF cells (Figure 4E). In addition, the transduction of dominant negative caspase 3 or 8 genes significantly attenuated OSKM-induced iPSC formation from MEF cells (Figure 4F). These results suggest significant roles for caspases 3 and 8 in OSKM-mediated iPSC formation in murine fibroblast cells, consistent with our results from human iPSC induction experiments.
Exogenous expression of caspase 8 stimulates induction of iPSCs
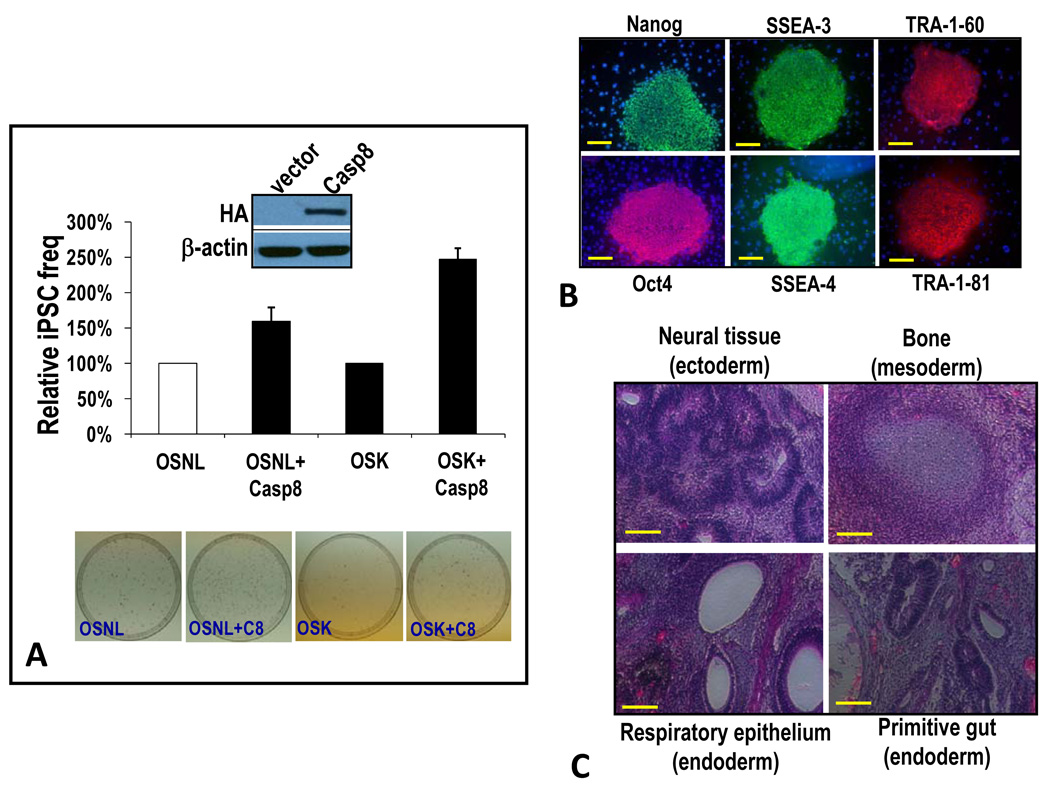
The experiments described so far clearly indicate the importance of caspase 8 activation in iPSC induction by the transcription factor cocktails. We next attempted to determine the effect of exogenous caspase 8 expression on the iPSC induction process. When we used IMR90 cells stably transduced with an exogenous caspase 8 gene, the frequency of iPSC induction with the OSNL protocol showed a 50% increase when compared with control (Figure 5A). Similarly, a 150% boost in iPSC induction was observed in IMR90-casp8 cells when we used the three factor Yamanaka protocol (OSK) (Figure 5A). These results again confirmed a key role for caspase 8 in the iPSC induction process.
Figure 5. Over-expression of caspase 8 enhances the frequency of iPSC formation.
A) Increased frequency of iPSC formation as a result of exogenous caspase 8 expression. IMR90 cells were infected with lentiviral vectors encoding OSNL or OSK together with a lentiviral vector encoding caspase 8. The frequency of iPSC formation in the transduced cells was then evaluated and normalized against control OSNL- or OSK-transduced cells, respectively. Error bars represent SEM (n=3). Western blots show HA-tagged exogenous caspase 8 expression.
B) Expression of key ESC markers in putative iPSCs. Putative iPSC clones were subcultured and examined for various ESC marker expressions through immunofluorescence staining. Scale bars represent 200 µm.
C) Teratoma formation from putative iPSCs. Putative iPSC clones from OSNL+casp8-transduced IMR90 cells were injected subcutaneously into scid mice. Teratoma formation was examined 9 weeks later. Mice with teratoma were sacrificed and the tumors excised and sectioned for H&E staining. Shown are representative images with features of selected differentiated tissues. Scale bars represent 200 µm.
Please see Figure S5 for additional related data.
Further experiments were conducted to verify the “stemness” of putative iPSCs induced through the OSNL+casp8 protocol. iPSC clones induced by use of this protocol expressed various markers of ESCs (Figure 5B). In addition, they formed three germ layers when injected into SCID mice (Figure 5C). Examination of the methylation status of the endogenous Oct-4 gene promoter of putative iPSCs further confirmed their close resemblance to ESCs (supplementary Figure S5A), as did quantification of the mRNA levels of key ESC-specific genes (supplementary Figure S5B). Further experiments also demonstrated that these cells could form embryoid bodies that express markers for a variety of differentiated cell types (Figure S5C), consistent with properties of iPSCs.
Retinoblastoma susceptibility protein is a key downstream target of caspases 3 and 8 in the iPSC induction process
We next tried to identify the mechanism through which caspase 8 facilitates iPSC induction. Because caspase 8 cleavage activities are clearly necessary in facilitating the iPS process, we reasoned that one or more of the cleavage substrates of caspases might be involved in the iPSC induction process. We further reasoned that caspase 8 may function through deactivation of factors involved in maintaining the differentiated state of fibroblasts. We eventually focused our attention on the retinoblastoma susceptibility (Rb) gene, which is a tumor suppressor gene that regulates cell cycle progression. It also has a key role in promoting and maintaining cellular differentiation (Dannenberg et al., 2000; Jori et al., 2007; White et al., 2005). Previous studies have indicated that activated caspase 8 can cause cleavage and deactivation of the Rb protein through the activation of caspase 3 (An and Dou, 1996; Dou et al., 1997; Janicke et al., 1996; Tan et al., 1997). Inactivation of Rb through viral protein E1A (Ferrari et al., 2008) or genetic deletion has been shown to facilitate cellular reprogramming to a stem cell-like state (Liu et al., 2009).
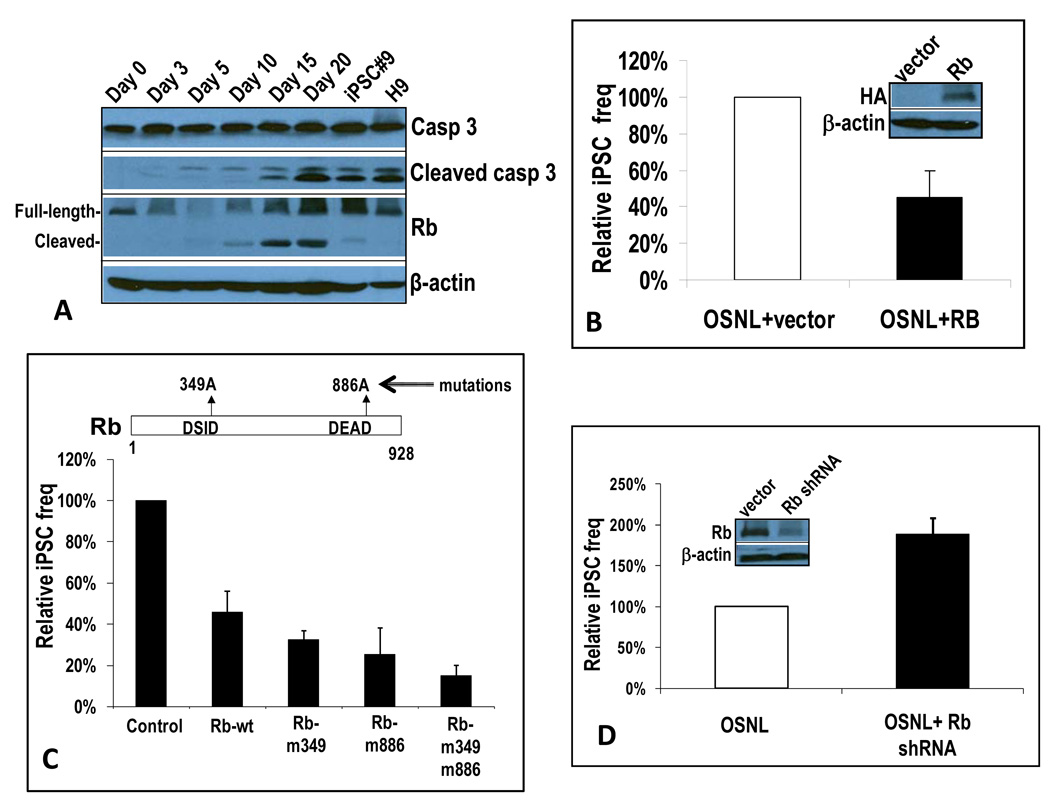
We initiated our investigation into the role of the Rb protein by probing its status during the iPSC induction process. Western blot analyses (Figure 6A) indicated that Rb protein was intact before the transduction of OSNL. However, a reduction in the full-length protein and an increase in cleaved Rb protein were obvious by day 3. By day 5 almost all full-length Rb protein was gone. Interestingly, starting at day 10, increases in both total and caspase-cleaved Rb fragments were observed. Furthermore, in iPSCs as well as in H9 hESCs, full-length Rb levels were similar to parental IMR90 fibroblast cells, indicating that Rb attenuation during iPSC induction was transient. The pattern of Rb cleavage during iPSC induction was almost identical to the patterns of caspase 3 and caspase 8 (Figure 1C). In particular, activated caspase 3 levels correlated very well with those of cleaved Rb, suggesting a very close relationship between the two. Rb proteins were mostly present at full-length in both iPSCs and ESCs at very robust levels, although weak, cleaved bands were clearly visible. Transduction of Oct-4 alone induced a similar Rb cleavage pattern (supplementary Figure S6A), consistent with the primary role of Oct-4 in activating caspases 3 and 8. A further examination showed that Rb proteins are mostly phosphorylated in iPSCs (supplementary Figure S6B), consistent with previous findings that Rb was inactive in ESCs (Burdon et al., 2002; Savatier et al., 1994).
Figure 6. Rb protein as a key downstream target of caspases 3 and 8 in the iPSC induction process.
A) Western blot analysis of Rb expression during the iPSC induction process. IMR90 cells tranaduced with control vector, casp3-DN, or casp8-DN were infected with OSNL or Oct-4. At different time points they were lysed and examined for Rb and other cell cycle regulator/tumor suppressor protein expression through western blot analysis.
B) Rb-mediated suppression of iPSC induction in OSNL-transduced IMR90 cells. A lentiviral vector encoding a full-length Rb gene was co-transduced with OSNL into IMR90 cells. The induction of iPSCs was then examined and quantified. The error bar represents SEM (n=4). Western blots show exogenous Rb expression.
C) The effect of mutations in caspase 3 cleavage sites on the ability of Rb to suppress ONSL induced iPSC formation in IMR90 cells. The error bars represent SEM(n=3).
D) shRNA-mediated knockdown of Rb led to increased iPSC induction in OSNL-transduced IMR90 cells. A lentiviral vector encoding an shRNA minigene targeted to Rb was transduced into IMR90 cells. Cells with stable shRNA expression were then transduced with OSNL and observed for iPSC formation. Relative frequency is shown. The error bar represents SEM (n=4). Western blots show reduced endogenous Rb expression after shRNA transduction .
Please see Figure S6 for additional related data.
We next determined if Rb is functionally important for the iPSC induction process. We cloned the human Rb gene into a lentiviral expression vector and then stably transduced Rb into IMR90 cells. As shown in Figure 6B, over-expression of the Rb gene significantly reduced the frequency (about 50%) of OSNL-induced iPSC formation. The importance of caspase-mediated cleavage of Rb was further confirmed when Rb proteins with their caspase 3 cleavage sites mutated were evaluated for their ability to suppress iPSC formation in OSNL-transduced IMR90 cells (Figure 6C). Indeed we observed that caspase 3-resistant mutants could inhibit iPSC induction more effectively than wild type Rb. These results therefore clearly established that caspase-mediated Rb inactivation is an important step in iPSC induction.
The importance of deactivating Rb was further demonstrated through shRNA-mediated Rb knockdown. When we stably transduced an shRNA targeted to Rb into IMR90 cells, the frequency of OSNL-induced iPSC formation was almost doubled (Figure 6D), again consistent with Rb as an important barrier for iPSC induction.
Comparing Rb with p53 and p21 cell cycle regulating proteins in the iPSC induction process
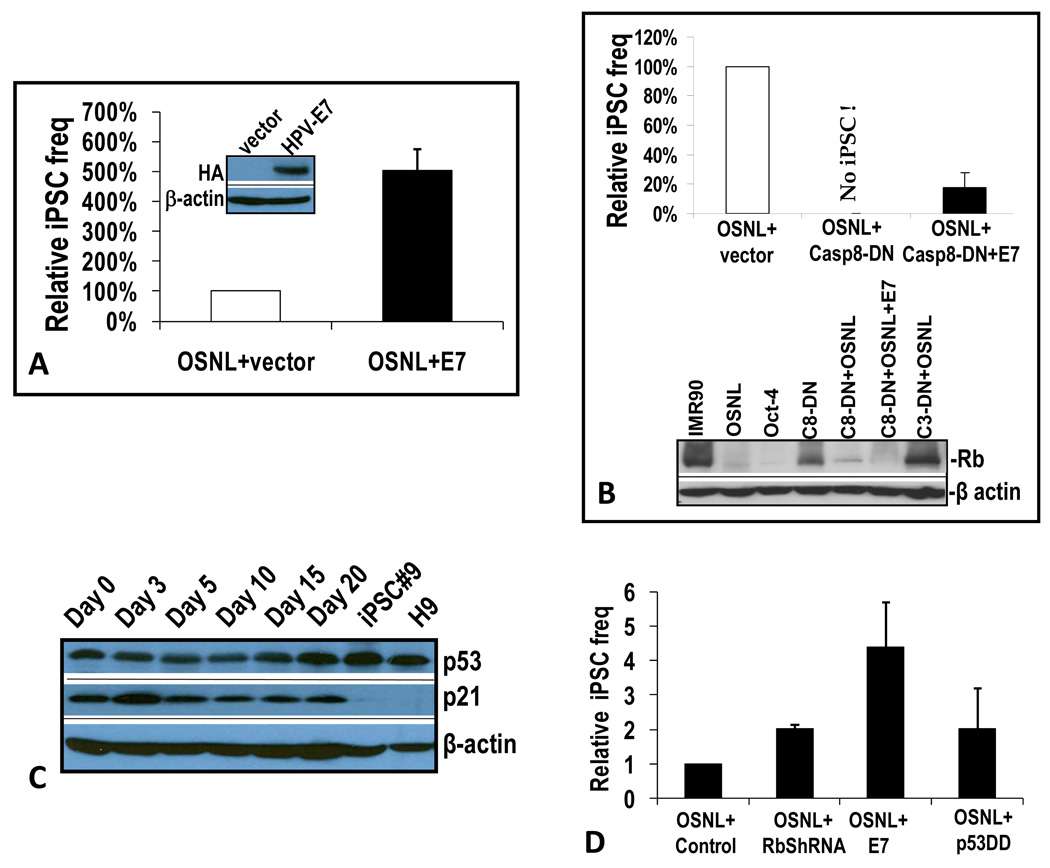
In order to further examine the importance of caspase-mediated Rb cleavage in iPSC induction, we decided to examine the effect of human papillomavirus type 16 E7 (HPV16 E7) protein on the iPSC induction process. HPV16 E7 has been shown to effectively inhibit Rb function (Barbosa et al., 1990; Dyson et al., 1989; Gage et al., 1990; White et al., 1994). Co-transduction of E7 with OSNL increased the frequency of iPSC formation almost 5-fold (Figure 7A). Furthermore, E7 was able to partially alleviate the complete blockade of iPSC induction caused by casp8-DN (Figure 7B, top panel), strongly suggesting that caspase 8-mediated reprogramming proceeds at least partially through deactivating Rb. Consistent with these results, shRNA-mediated Rb knockdown was able to rescue the defect in iPSC induction partially in caspase deficient (casp3−/−) MEF cells mediated by OSKM (supplementary Figure S7A).
Figure 7. Comparing Rb and other cell cycle regulating proteins.
A) HPV16-E7-mediated increase in iPSC formation. Native IMR90 cells or IMR90 stably transduced with HPV16-E7 were transduced with OSNL and observed for iPSC formation. The error bar represents SEM (n=5). The western blot shows exogenous HA-tagged E7 expression.
B) Partial alleviation of casp8-DN mediated blockade of iPSC formation by HPV16-E7 in IMR90 cells. IMR90 cells stably transduced with casp8-DN were infected with OSNL or OSNL+E7 and observed for iPSC formation. The error bar represents SEM (n=4). The lower panel shows western blot analysis of the effects of dominant negative caspases and HPV16-E7 on Rb expression in OSNL-transduced IMR90 cells.
C) Western blot analysis of p53 and p21in OSNL-transduced IMR90 cells.
D) Effect of Rb knockdown by shRNA, HPV16 E7 transduction, and transduction of a dominant negative p53 on the frequencies of iPSC formation in OSNL-transduced IMR90 cells. Error bars represent SEM (n=3). Please see Figure S7 for additional related data.
Western blot analyses were carried out to examine the status of Rb after OSNL and/or E7 transduction (Figure 7B, lower panel). OSNL or Oct-4 transduction caused significant attenuation of Rb levels. Inhibition of caspase 8 activity partially blocked OSNL transduction-mediated Rb attenuation. On the other hand, a dominant negative caspase 3 significantly blocked Rb cleavage after OSNL transduction. These data indicated that caspase 3 activity was required for Rb cleavage. Furthermore, rescue of iPSC formation in casp8-DN-transduced cells correlated with almost complete degradation of Rb in IMR90 cells transduced with OSNL+casp8-DN+E7, consistent with published literature suggesting the ability of E7 to destabilize Rb (Jones et al., 1997).
Because recent reports indicated that the tumor suppressor gene p53 and its downstream gene p21 are involved in the iPSC formation process, we examined the status of these genes (Figure 7C). Our analysis did not reveal any caspase-mediated cleavage of p53 or p21 proteins. Furthermore, p53 protein levels did not significantly change during the course of iPSC induction or in iPSCs that eventually emerge. For p21, except a small transient increase at an early time (day 3), there was a small reduction of p21 protein levels at later times in OSNL-transduced IMR90 cells. In iPSCs and in H9 hESCs, p21 expression was completely suppressed, similar to previous reports (Hong et al., 2009; Utikal et al., 2009). In comparison, minimal changes in p53 and p21 levels were found in Oct-4-transduced IMR-90 cells (supplementary Figure S7B).
A further experiment was done to examine the relative importance of Rb and p53 in the iPSC induction process. Previous studies have indicated that suppression of p53 could enhance the formation of iPSCs by 3–100 fold (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). We carried out experiments to compare the effects of Rb down-regulation (through E7 expression or shRNA knockdown) versus p53 suppression in the iPSC induction process. Our results indicated a 2-fold increase in iPSC formation when a dominant negative p53 is co-expressed with OSNL, similar to Rb shRNA knockdown and lower than E7-mediated Rb suppression (Figure 7D), which caused a 5-fold increase in iPSC induction. When similar experiments were done in mouse MEF cells, the results were quite different: p53 knockdown mediated a 6-fold increase in iPSC formation while E7 transduction increased iPSC formation about 3-fold (supplementary Figure S7C). The exact mechanism for the reversed roles of p53 and Rb in human and mouse fibroblasts is not clear. However, when western blot analyses of p53 and p21 were carried out in mouse MEF cells, we observed a clear induction of p53 and p21 that were further strengthened by HPV16 E7 transduction (supplementary Figure S7D). Because previous studies have shown that Rb knockout results in activation of p19Arf, which can stimulate p53 and p21 expression through regulating MDM2 (Hsieh et al., 1999; Pomerantz et al., 1998; Zhang et al., 1998), we examined p19Arf status. Our results indicate that p19Arf was not expressed during any stage of iPSC induction from passage 1 or 2 mouse MEF cells. This result was consistent with recent publications that report significant expression of p19Arf only occur in MEF cells after passage 3(Li et al., 2009; Utikal et al., 2009). In addition, p19Arf was not expressed in the mouse iPSCs that eventually emerged. Similarly, in human iPSC induction from IMR90 cells (<passage 10), there was also an absence of p14Arf expression (supplementary Figure S7E) in cells transduced with OSNL or OSNL+shRb. Under those situations, knocking down Rb did not appear to promote p14Arf expression. Therefore, Rb and p53 pathways appear to play different roles in mouse and human iPSC induction, despite both being very important. The mechanisms for this difference are not clear at present.
Discussion
The ability to induce iPSC formation from differentiated human cells is a great leap forward for stem cell research and regenerative medicine. However, despite significant advances in the field, very little information is available on how the process of nuclear reprogramming is initiated and carried forward. Our discovery of the essential roles of caspase activation provides unexpected but significant insights into the nuclear reprogramming process.
Although our studies show that both caspase 3 and caspase 8 play very important roles in the iPSC induction process, there are some important differences between the two: 1) the expression levels of caspase 8 increased initially in OSNL-transduced IMR90 cells but decreased significantly in iPSCs (Figure 1C); 2) modest increases in total levels of caspase 3 in fibroblasts accompanied its activation in the iPSC induction process, with robust levels of total and activated caspase 3 in iPSCs; 3) inhibition of caspase 8 can completely block the iPSC induction process while inhibition of caspase 3 can only partially block the iPSC induction process. The reason for this difference is not clear. However, it is possible that caspase 8 activation is absolutely required because there is no redundancy, while caspase 3 activation has been shown to have some redundancy (e.g., caspase 7). From this perspective, it is interesting to note that caspase 8-deficient mice are embryonic lethal (Varfolomeev et al., 1998) while caspase 3-deficient mice can survive in some strains despite problems (Lakhani et al., 2006).
The involvement of two apoptotic caspases in the nuclear reprogramming process raises some important questions. For example, what are the additional targets of the caspases that are involved in the iPSC induction process? We have provided clear evidence that the retinoblastoma susceptibility protein (Rb) is a key target for caspases and that the cleavage and inactivation of Rb appeared to be critical for the iPSC induction process. However, it is eminently possible that other caspase cleavage targets may play important roles in this process.
How does caspase-mediated cleavage of Rb and other targets facilitate iPSC induction? Although we do not have a clear picture at this point, it is conceivable that Rb and other targets yet to be identified are part of the multiple layers of “epigenetic walls” that tightly guard the chromatin landscape to maintain a differentiated state (Hemberger et al., 2009). If this is the case, we can then view the roles of caspases as intracellular “wrecking crews” whose jobs in the iPSC induction process are to transiently dismantle that normally insurmountable barrier in differentiated cells (i.e., fibroblast cells). The critical importance of knocking down the epigenetic walls is underscored by our observation of the complete blockade of the iPSC induction process by dominant-negative caspase 8 expression.
In summary, our data provide strong evidence for the involvement of apoptotic caspases in the iPSC induction process. This surprising finding provides significant insights into the process of transcription factor-mediated iPSC induction.
Experimental Methods
Cell strains and tissue culture
Early passage (passage 6) human fibroblast cells (IMR90) were obtained from the Coriell Institute for Medical Research (Camden, NJ). They were maintained in standard medium (MEM+Non-essential amino acid+10% bovine serum) and sub-cultured every 3–5 days as necessary. Passage P0 or P1 mouse embryonic fibroblast cells were derived from wild type and caspase 3−/− C57BL/6 mice following standard procedures. Doxycycline-inducible OKSM-transgenic mouse-derived tail tip fibroblast cells were provided by Drs. Matthias Stadtfeld and Konrad Hochedlinger of Harvard Medical School (Charlestown, MA).
Plasmids, molecular cloning, and lentivirus production
Please see details in the supplementary information section.
Bioluminescence observation of caspase reporter activation
To image caspase reporter activity through bioluminescence imaging, we used the IVIS200 instrument from Caliper Life Sciences (Hopkinton, MA). The manufacturer’s instructions were followed to image cell culture dishes containing fibroblast cells transduced with Oct-4 or OSNL. Before imaging, cell culture dishes were washed with PBS (phosphate-buffered -saline). Luciferin (purchased from Caliper Life Sciences, Hopkinton, MA) was then added to cells as a solution at 150 µg/mL. Ten minutes later, the cells were placed in the IVIS200 imaging chamber and imaged. Quantification of the luciferase activity was carried out by use of the Living Image 3.1 software supplied by the manufacturer.
Procedure for iPSC induction
For iPSC production from human cells, a protocol published by the Thomson lab at the University of Wisconsin was followed (Yu et al., 2007). Briefly, IMR90 cells at the exponential phase of growth were infected with a cocktail of lentiviral vector supernatants (either OSNL or OSKM) with the help of polybrene (at 5 µg/mL). Twenty-four hours after infection, infected fibroblast cells were trypsinized and replated onto SNL feeder cells at a density of 5×105 cells/10-cm Petri dish or 2×105/60 mm Petri dish. Twenty-four hours later, the medium was changed to hES medium supplemented with 4 ng/mL FGF. Medium was changed every 48 hours. The hES medium contains 80% DMEM/F12 medium (1:1)+20% knockout serum replacer. For iPSC induction from mouse fibroblast cells, a similar procedure was adopted except that the genes used were of murine origin. In addition, only OSKM was used for gene transduction. For all of our experiments involving calculation of iPSC frequency, we stop the human iPSC experiments at 20 days after gene transduction whereas the murine iPSC experiments were stopped at 14 days after gene transduction.
Alkaline phosphatase staining and enumeration of iPSC colonies
Alkaline phosphatase staining was carried out by use of a commercially available Alkaline Phosphatase staining kit (Millipore, Billerica, MA) according to the manufacturer’s protocols. Quantification of iPSC generation was carried out by counting the number of AP+ colonies.
Immunofluorescence staining
Information for various specific stem cell antibodies was obtained from different vendors (see supplementary information). Cells to be stained were first fixed in PBS with 4% paraformaldehyde. Primary antibodies were used at dilutions recommended by the manufacturer. Secondary antibodies labeled with either Alexa555or Alexa488 were obtained from Invitrogen (Carlsbad, CA). Cellular nuclei were stained with DAPI that was included in the Vectorshield mounting medium (Vector Laboratories, Burlingame, CA).
Teratoma formation
Putative iPSCs from 1 confluent single well of a 6-well plate were suspended in PBS and injected into the dorsal flank of individual SCID mice (NCI-Charles River, Wilmington, MA). Two months (about 9 weeks) later, the mice were sacrificed and the tumors excised, fixed in paraformaldehyde, sectioned, and stained in hematoxylin and eosin.
Western blot analysis
Standard protocols were used to carry out western blot analysis. Please see Supplementary Information section for more details on the antibodies.
PCR analysis of lentiviral vector or encoded casp8-DN sequences
To identify the presence of either the control vector (pLEX) or casp8-DN sequences in iPSC colonies, DNA from the colonies was extracted and subjected to PCR using the following PCR primers that flanked the multiple cloning sequence (MCS) of pLEX: 5’-CACCAAAATCAACGGGACTT-3’ (forward); 5’-ATATAGACAAACGCACACCGGCCT-3’ (reverse).
TUNEL assay
Cells undergoing apoptosis were visualized though the TUNEL assay, which identifies nuclear DNA fragmentation that is typical of end stage apoptotic cells. A commercially available kit for the TUNEL assay was obtained from Invitrogen (Carlsbad, CA). This kit labels fragmented nuclear DNA with Alexa594-labeled dUTP, which can be detected as red fluorescence under a fluorescence microscope. DNase I-treated cells were used as positive controls.
Supplementary Material
Acknowledgments
We thank the various investigators who have deposited many of the plasmids used in this study at Addgene (Cambridge, MA). These include the iPSC factors deposited by Drs. Thomson (U. of Wisconsin) and Yamanaka (Kyoto University) and the dominant/negative caspase plasmids deposited by Dr. Salvesen (Burnham Institute). We also want to thank Drs. Hochedlingder and Mahereli (Harvard Medical School) for their gift of the doxycycline inducible secondary reprogramming mouse fibroblast cells. This study was supported in part by grants CA131408 and CA136748 from the US National Cancer Institute (to C.-Y. Li), an NCI SPORE developmental grant in lung cancer research (to C.-Y. Li), and grant NNX09AH19G (to C.-Y. Li) from the NASA Space Radiation Biology Research Program. Qian Huang was supported by grant 2010CB529900 from the National Basic Research Project of China and Outstanding Young Scientist grants 30325043 & 30428015 from the China National Natural Science Foundation. None of the authors have any financial interest in the work that is presented in this study. We thank Melissa Stauffer, PhD, of Scientific Editing Solutions, for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information
The Supplementary Information section contains additional details of our experimental methods, primer sequences, shRNA sequences, and 7 supplementary figures.
References
- An B, Dou QP. Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as candidate. Cancer research. 1996;56:438–442. [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barbosa MS, Edmonds C, Fisher C, Schiller JT, Lowy DR, Vousden KH. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. The EMBO journal. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell stem cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell stem cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends in cell biology. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes & development. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou QP, An B, Antoku K, Johnson DE. Fas stimulation induces RB dephosphorylation and proteolysis that is blocked by inhibitors of the ICE protease family. Journal of cellular biochemistry. 1997;64:586–594. [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell stem cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G, Harrington E, Fanidi A, Land H, Amati B, Bennett M. Integrated control of cell proliferation and cell death by the c-myc oncogene. Philosophical transactions of the Royal Society of London. 1994;345:269–275. doi: 10.1098/rstb.1994.0105. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA, Thomson JA, Zwaka TP. Caspase activity mediates the differentiation of embryonic stem cells. Cell stem cell. 2008;2:595–601. doi: 10.1016/j.stem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JR, Meyers C, Wettstein FO. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. Journal of virology. 1990;64:723–730. doi: 10.1128/jvi.64.2.723-730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardini V, Fernandez PA, Lee RK, Drexler HC, Rotello RJ, Fishman MC, Yuan J. Prevention of vertebrate neuronal death by the crmA gene. Science. 1994;263:826–828. doi: 10.1126/science.8303301. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nature reviews. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O'Connor DJ, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Molecular cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nature biotechnology. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hueber AO, Zornig M, Lyon D, Suda T, Nagata S, Evan GI. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Walker PA, Lin XY, Porter AG. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. The EMBO journal. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nature cell biology. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Jones DL, Thompson DA, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- Jori FP, Galderisi U, Napolitano MA, Cipollaro M, Cascino A, Giordano A, Melone MA. RB and RB2/P130 genes cooperate with extrinsic signals to promote differentiation of rat neural stem cells. Molecular and cellular neurosciences. 2007;34:299–309. doi: 10.1016/j.mcn.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, Jurewicz A, Waisman A, Brenner O, Haffner R, Gustafsson E, et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol. 2004;173:2976–2984. doi: 10.4049/jimmunol.173.5.2976. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009a;461 doi: 10.1038/nature08436. 649-643. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009b;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J, et al. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell stem cell. 2009;4:336–347. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell stem cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature biotechnology. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell stem cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nature methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, −6, −7, and −8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- Szymczyk KH, Freeman TA, Adams CS, Srinivas V, Steinbeck MJ. Active caspase-3 is required for osteoclast differentiation. Journal of cellular physiology. 2006;209:836–844. doi: 10.1002/jcp.20770. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tan X, Martin SJ, Green DR, Wang JY. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes & development. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Molecular biology of the cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Takahashi K. [Induction of pluripotent stem cells from mouse fibroblast cultures] Tanpakushitsu kakusan koso. 2006;51:2346–2351. [PubMed] [Google Scholar]
- Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, et al. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell stem cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nature biotechnology. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.