Abstract
An all-atom multiscale computational modeling approach, Molecular Dynamics/Order Parameter eXtrapolation (MD/OPX), has recently been developed for simulating large bionanosystems. It accelerates MD simulations and addresses rapid atomistic fluctuations and slowly-varying nanoscale dynamics of bionanosystems simultaneously. With modules added to account for water molecules and ions, MD/OPX is applied to simulate the swelling of cowpea chlorotic mottle virus (CCMV) capsid solvated in a host medium in this study. Simulation results show that the N-terminal arms of capsid proteins undergo large deviations from the initial configurations with their length extended quickly during the early stage of capsid swelling. The capsid swelling is a symmetry-breaking process involving local initiation and front propagation. The capsid swelling rate is ~0.25 nm/ns (npn) during early stage of the simulation and propagation of the structural transition across the capsid is roughly 0.6npn. The system conditions that affect swelling of the capsid are analyzed. Prospects for creating a phase diagram for CCMV capsid swelling and using predictions to guide experiments are discussed.
Keywords: molecular dynamics/order parameter extrapolation (MD/OPX), all-atom multiscale, cowpea chlorotic mottle virus (CCMV)
I. Introduction
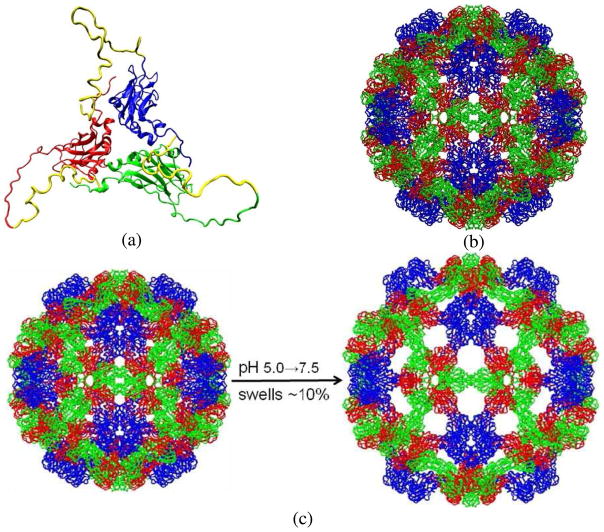
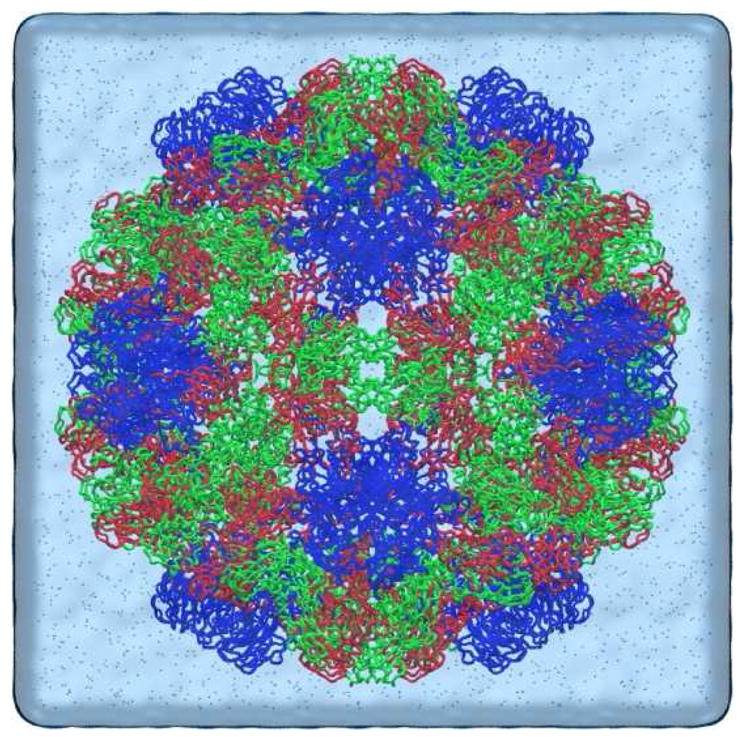
CCMV is a member of the bromovirus group of the Bromoviridae family. Its genome consists of four positive-sense single-stranded RNA molecules, two of which are encapsulated separately in two virions and the remaining two are packaged together into a third type of particle. The crystal structure of wild-type CCMV was solved at 3.2Å resolution by X-ray crystallography[1]. Its capsid is comprised of 180 chemically identical protein subunits that form a 286Å-diameter icosahedral shell displaying a T=3 quasi-symmetry. Each protein subunit is composed of 190 amino acids taking three quasi-equivalent positions on the capsid surface. As such, one asymmetric unit (i.e., protomer) of the capsid includes three subunits (A, B, and C). In the X-ray crystal structure, there are 41 N-terminal residues in subunit A and 26 residues in both B and C that are not visible. Fig. 1a shows the three quasi-threefold related subunits with the missing N-terminal residues added and optimized [2]. The icosahedral capsid can also be divided into 12 pentamers and 20 hexamers with 5 A subunits in each pentamer, and 3 B and 3 C subunits in each hexamer (Fig. 1b).
Fig. 1.
(a) X-ray crystal structure of CCMV protomer with the missing N-terminal residues added and optimized. Three quasi-threefold related protein subunits are colored in blue (A), red (B) and green (C) respectively, and the N-terminal arms in yellow; (b) native CCMV capsid organized in 12 pentamers and 20 hexamers with 5 blue A subunits in each pentamer and 3 red B and 3 green C subunits in each hexamer; and (c) Native CCMV swells ~10% as pH is increased from 5.0 to 7.5 in the absence of divalent cations.
Native CCMV is well known to undergo a reversible swelling transition (~10% increase in capsid diameter) when pH is increased from 5.0 to 7.0 or above in the absence of divalent cations and at low ionic strength (I=0.2M) (Fig. 1c) [1, 3–5]. In the expansion scheme proposed by Liu et al.[6], its swollen state can be generated by taking the pentamers and hexamers through rigid-body transformations from their native configurations as the following: translating pentamers by 24Å radially outwards and rotating them counter-clockwise by 9° around their 5-fold axes; translating hexamers by 21Å radially outwards and rotating them counter-clockwise by 8° around their 3-fold axes. Many other viruses undergo similar structural transitions (STs) in response to changes in their microenvironment, such as Nudaurelia capensis ω virus (NωV) [7–8], HK97 bacteriophage [9] and poliovirus [10–11]. Because the purified RNA and coat proteins of CCMV can reassemble in vitro to produce infectious virions [12], and the empty CCMV capsid assembled from its protein subunits displays many behaviors similar to those of the complete virus [4, 13], it is an excellent system for studying protein-protein and protein-RNA interactions, which provide important information on the STs, self-assembly and disassembly of viruses and other biological nanostructures.
Theoretical approaches including (1) computational molecular dynamics (MD) [14–17], (2) single-scale coarse-grained models[18–19], (3) symmetry-constrained models[15], (4) normal mode analysis (NMA) [20–22], and (5) Poisson-Boltzmann (PB) solvers [2, 23–24] have been applied to simulate viral properties and dynamics, but they have suffered from one or more limitations as reviewed earlier [23, 25–26]. In the following, we focus on the studies on viruses, notably CCMV.
MD has been applied to study RNA release from CCMV by constructing a hypothetical short single-stranded RNA, 5′-R(PGpGpApCpUpUpCpGpGpUpCpC)-3′[17]. In the MD simulation, only one asymmetric unit (i.e., a protomer) of CCMV capsid was included. Results indicate that the RNA fragment loses its secondary structure and moves into the channel along the three quasi-threefold axis of CCMV capsid by free diffusion. However, the conclusions are not based on simulations of whole-capsid, full-length RNA behaviors.
Coarse-grained models were used to simulate a set of viral capsids [19], though they have not been directly applied to CCMV. In the simulations, shape-based clusters of atoms were lumped into structureless beads for a reduced description of the viral capsids to allow more efficient sampling and a larger timestep than that used in MD. While the simulations provide insights into the stability of studied viral capsids, these models require parameter recalibration before simulation starts. They lack viral atomistic details needed to address local atomic interactions (e.g., the binding of divalent cations to capsid proteins and small-molecule drug/virus interactions) and miss the inter-scale feedback whereby nanoscale structural variables affect the statistical distribution of atomic-scale fluctuations which, in turn, mediate the entropic and free energy effects that drive the dynamics of the nanoscale features. This feedback loop is central to a complete understanding of the dynamics of viruses as well as other bionanosystems [27–28].
NMA was used to explore low-frequency, large-scale structural changes of CCMV capsid during its swelling [20]. It provided candidate structures of intermediate and swollen CCMV, and a putative pathway for the swelling. In the study, icosahedral symmetry was used to facilitate normal mode calculations via group theory and various methods including elastic network models and the use of block rotation/translation were applied to reduce the number of degrees of freedom. However, an icosahedral virus does not necessarily maintain the symmetry during its STs [14, 28–29]. It is likely that initiation of instability is local, starting with the motion or deformation of a single structural unit, and then propagates across the virus. Based on single-well (harmonic) potential approximations, NMA cannot capture the local, highly nonlinear, diffusive (friction-dominated) character of viral STs. Nor can it be readily used to study viral interactions with drugs or cell surface receptors for similar reasons.
Electrostatics of CCMV capsid were previously modeled by solving the PB equation [24]. The electrostatic interaction between RNA and CCMV capsid was simulated with a coarse-grained RNA model and a Monte Carlo approach [2]. In the model, each RNA nucleotide segment was treated as a sphere with a charge of −0.25e and no connectivity was enforced between the spheres. Results showed that there is a very strong interaction between RNA segments and the highly positive N-terminal residues of the capsid proteins. RNA segments were predicted to form a shell close to the capsid with the highest densities associated under protein dimers. These high-density regions are connected to each other in a continuous net of triangles. Medium density of RNA was found under protein pentamers. These studies revealed interesting features, but they do not capture the structural constraints imposed by RNA connectivity. Furthermore, they have the limitations of coarse-grained models and do not provide a general, parameter calibration-free approach to whole-virus modeling. Also it is assumed that the electrical field is fixed (i.e., is imposed by the capsid), rather than being co-evolved with the RNA (and possible capsid ST) during a simulation.
Therefore, while above approaches reveal interesting features of CCMV stability and dynamics, they do not meet the challenge of simulating the whole-virus dynamics. Viruses like CCMV are normally composed of millions of atoms and their STs involve both atomistic fluctuations and nanoscale processes (e.g., large-scale translation and rotation of protein capsomeres) spanning across multiple time and length scales. It is natural to adopt a multiscale approach to study their dynamics. Multiscale analysis has been of interest since work on Brownian motion by Einstein [30]. It was shown that the wandering of a nanoparticle arises due to the interplay of short-scale random collisions and large-scale, slow motions of the whole nanoparticle created by the separation in the magnitude of the mass of an atom versus that of the nanoparticle. In earlier studies, Fokker-Plank (FP) and Smoluchowski equations were derived from the Liouville equation for nanoparticles without internal atomistic structure [31–38]. The development of projection operators [39] is not directly applicable because it requires integration over atomistic configurations and thereby makes it difficult to capture internal structure and involves memory kernels that are not readily evaluated using standard MD. We have advanced the multiscale approach to develop an all-atom multiscale analysis (AMA) theory for dynamical nanosystems [25] by (1) accounting for atomic-scale fluctuations of the system internal structure; (2) introducing general, automatically-generated collective modes and order parameters (OPs); (3) constructing ensembles of atomic configurations constrained to fixed values of the slowly varying OPs, as needed to construct thermal-average forces and friction/diffusion coefficients for their stochastic equations; (4) applying perturbation expansion to the solution of N-atom Liouville equation in terms of a smallness parameter (length or time scale ratio of one atom to the entire system), and (5) deriving stochastic equations (FP or Smoluchowski) for the probability density of coarse-grained system OPs. The AMA approach captures the cross-talk between the OPs and atomistic variables rigorously [25, 40–42], and provides the conceptual framework for our whole-virus simulation approach.
Based on concepts from AMA theory of nanosystem dynamics, we recently developed a multiscale MD/OP eXtrapolation (MD/OPX) approach for simulating viruses and other large bionanosystems[26, 28]. In the present implementation, it is optimized for non-enveloped viruses or the core of genomic complex of enveloped viruses, such dengue virus [43–44]. OPs constructed with orthogonal polynomials of atomic coordinates of a reference configuration or atomic displacements between two know configurations are introduced to capture the system slowly-varying nanoscale features. Replica short MD runs with random atomic velocity initializations are implemented to estimate the ensemble-average rate of change in OPs, which is then used to extrapolate the system over a time period that is much longer than the 10−14-second timescale of fast atomic vibrations and collisions. Since the timescale for the evolution of OPs is much larger than that of atomic vibrations and collisions, the OP extrapolation time can be many orders of magnitude greater than the MD simulation timestep. The resulting MD/OPX algorithm addresses rapid atomistic fluctuations and slowly-varying coherent dynamics of bionanosystems simultaneously and accelerates their MD simulations. Its underlying all-atom description of bionanosystems enables the use of a universal inter-atomic force field, avoiding recalibration with each new application as needed for coarse-grained models. The equation free multiscale analysis (EFM) approach developed by Kevrekidis et al.[45–46] shares much of the flavor of MD/OPX, i.e., short bursts of MD simulations can be used to extrapolate coarse variables (i.e., OPs here) over large time intervals and thus project the system over long time. It has been applied to an alanine dipeptide for molecular dynamics study by using the dipeptide dihedral angle (N-Cα-C-N) as its coarse variable[46]. However, the dipeptide is a very small system and thus its dihedral angle does not appear to be slowly-varying compared with our automatically constructed OPs for large bionanosystems.
In our earlier study[28], MD/OPX was used to simulate the swollen state of CCMV capsid in vacuum (i.e., gas phase). The swollen CCMV capsid shrinks significantly during 200ns simulation and it was found to be an energy-driven, symmetry-breaking process that starts locally and then propagates across the capsid. It involves large-scale cooperative motions of protein pentamers and hexamers in the capsid. In this study, MD/OPX is applied to the swelling of CCMV capsid solvated in a host medium at pH 7.0 and ionic strength I=0.2M. We first identify key elements of a model needed to simulate the swelling of native CCMV capsid through short-time MD simulation. Then MD/OPX is applied to capture the long-time dynamics of CCMV capsid swelling with the mechanisms of the capsid swelling and its dependence on system conditions discussed, and conclusions are drawn.
II. Computational Methods
CCMV capsid structure and MD Simulation protocols
The X-ray crystal structure of CCMV protomer (i.e., asymmetric unit) was downloaded from online Protein Data Bank (PDB ID: 1CWP). The missing N-terminal residues of protein subunits were added and further optimized with residues in the crystal structure fixed by using NAMD. Therefore, they do not introduce unphysical stress to the capsid (refer to the scheme provided in Zhang et al. [2]). The native CCMV capsid was generated by reorienting 60 copies of the protomer according to the icosahedral symmetry. The capsid was then solvated in water cube with different water boundaries by using VMD[47] (see Table 1). Protonation states of the amino acid residues were set according to specific pH, e.g., 5.0 or 7.0. As amino acids usually have one protonation state over a range of pH values, a simulation model prepared at pH 7.0 actually works over a pH range, e.g., 7.0 – 8.0. K+ and Cl− were added randomly to neutralize the solvated system achieving an ionic strength of 0.2M.
Table 1.
Simulations showing CCMV capsid swelling is strongly dependent on system conditions.
| Simulation | pHa | [Mg2+] (mol/L) | Water boundaryb | Ensemblec | Natomsd | Performance(days/ns)e | Time | Increase in capsid diameter (Å)f | Increase in capsid radii (Å)g | |
|---|---|---|---|---|---|---|---|---|---|---|
| outer | average | |||||||||
| sim1 | 7.0 | 0 | 7Å | NPT | 2,411,918 | 2.60 | 3ns | 10.23 | 5.74 | 1.35 |
| sim2 | 7.0 | 0 | 15Å | NPT | 2,845,915 | 2.95 | 3ns | 10.45 | 7.61 | 1.58 |
| sim3 | 7.0 | 0 | 15Å | NVT | 2,845,915 | 2.36 | 5ns | 26.08 | 18.05 | 10.48 |
| sim4 | 5.0 | 0 | 15Å | NPT | 2,846,275 | 2.95 | 3ns | 8.54 | 6.20 | 1.94 |
| sim5 | 5.0 | 0.05 | 15Å | NPT | 2,841,943 | 2.95 | 3ns | 6.86 | 4.17 | 1.31 |
pH of the system is set such that amino acid residues in the capsid are protonated/deprotonated corresponding to their pKa,
water boundary is the minimum distance between atoms on the capsid surface and faces of the water box;
NPT means constant pressure (1atm) and temperature (298.15K) ensemble and NVT is constant volume (unchanged from dimensions of the initial system prepared with VMD) and temperature (298.15K);
there are 522,000 atoms in CCMV capsid and the total number of atoms in the systems ranges from ~2.41 to ~2.85 million depending on the thickness of water boundary and salt concentration used for solvation;
performance here is for running simulations with 256 processors on Indiana University Big Red cluster;
the capsid diameter is calculated as the maximum distance between two Cα atoms in the capsid; and
the capsid outer and average radius are calculated as the average and maximum distance from heavy atoms in the capsid backbone to their center of mass respectively.
With the above resulting system and its structural topology, NAMD[48] was applied to run molecular dynamics simulation by using the CHARMM22 force field[49–50] for capsid proteins and TIP3P model for water molecules[51]. The simulation proceeded with an initial energy-minimization of the entire system for 10,000 steps using the conjugate gradient algorithm until the potential energies level off. Following the energy-minimization, the system was gradually heated to 298.15K and equilibrated for 20ps, after which the system temperature and pressure (in the case of pressure control) stay stable around specified values. Product MD trajectory was then obtained for investigating the capsid structural changes.
Periodic boundary conditions were applied to remove surface effects of the system. Either a constant volume/temperature (NVT) or a constant pressure/temperature (NPT) ensemble can be used for running MD. The temperature set in the simulations in this study was 298.15K and the pressure was 1atm for the NPT simulation. The volume of the system was determined by the lattice vectors specified for its unit cell. The lattice vectors were initially calculated according to the dimensions of the resulting system prepared with VMD. They were kept constant for NVT simulation, while scaled isotropically in the NPT simulation by applying the Nose-Hoover Langevin piston pressure control. The integration timestep used in our MD simulation was 1fs and a multiple-time-stepping algorithm[48] was employed in the following manner: bonded interactions were computed for every timestep, short-range nonbonded interactions every two timesteps and long-range electrostatic interactions every four timesteps. A cutoff distance of 12Å was used for van der Waals and short-range electrostatic interactions and the long-range electrostatic interactions were computed with the particle-mesh Ewald summation method using a grid point density of 1/Å. SHAKE [52] constraints were applied on hydrogen-oxygen bonds in water molecules.
To investigate the controlling factors of CCMV capsid swelling, five MD simulations were performed on the native state of CCMV capsid that was solvated in a host medium under various conditions. The complete CCMV capsid with the missing N-terminal residues of protein subunits added has 522,000 atoms and the total number of atoms in the simulated systems ranges from ~2.41 to ~2.85 million depending on the thickness of water boundary and salt concentration used for solvation. Simulations were run on Indiana University Big Red cluster and the performance goes from 2.36 to 2.95 days/ns when using 256 processors. A summary of the characteristics of prepared systems and the simulations is listed in Table 1.
MD/OPX for simulating solvated systems
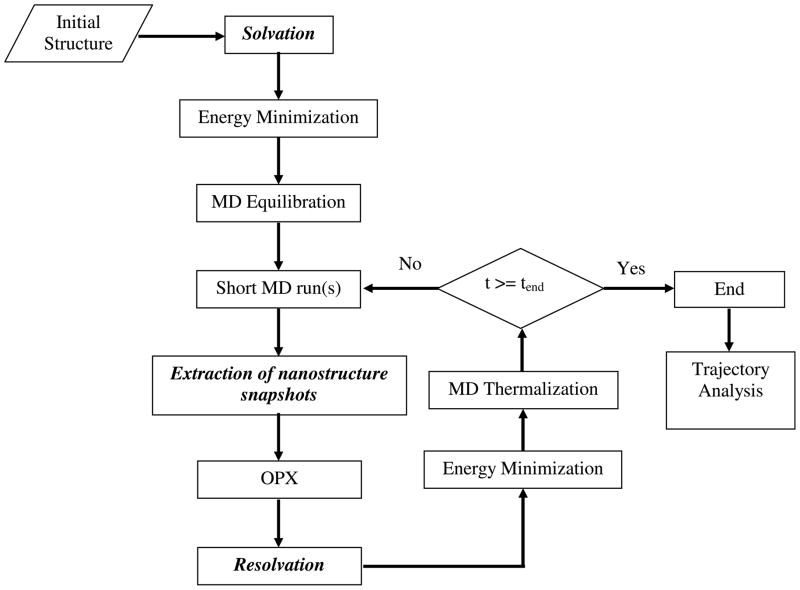
While earlier MD/OPX studies were focused on simulating nanostructures in vacuum (i.e., gas phase) [26, 28], new modules have been developed to account for water molecules and ions for simulating nanostructures solvated in a host medium. A flowchart of MD/OPX with new modules for “solvation”, “Extraction of nanostructure snapshots” and “resolvation” integrated is shown in Fig. 2. MD/OPX is implemented by using NAMD[48] as the MD platform. VMD is used for “solvation” and “resolvation” of simulated nanostructures and trajectory analysis.
Fig. 2.
Schematic flowchart of MD/OPX for simulation of nanostructures solvated in a host medium with new modules for “solvation”, “extraction of nanostructure snapshots” and “resolvation” integrated.
MD/OPX starts with an initial atomic structure that can be determined from X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy and cryo-electron microscopy. For system preparation, the structure is solvated in water and neutralized under certain pH and ionic strength (see example procedure described above for CCMV capsid). The resulting system along with its structural topology is then taken as input to NAMD for initial energy minimization and equilibration, after which the simulation is performed in MD/OPX cycles until it reaches the end of specified time. In each MD/OPX cycle, short MD run(s) are used to generate the time evolution of the system atomic configuration. With several snapshots chosen from the system trajectory, nanostructure is extracted from the system without water molecules and ions for calculating its structural OPs (see Appendix) and therefore estimating their rates of changes, i.e., time derivatives, which are then used to extrapolate OPs over long time. The atomic configuration of nanostructure is then reconstructed from the newly obtained OPs and resolvated in the host medium as explained below. Energy minimization and MD thermalization are applied to the resulting system before running short MD for another cycle.
In the “resolvation” module, the OPX resulting nanostructure is first put back to the host medium by using the final configuration of the preceding short MD run, which keeps the same number of water molecules and ions (i.e., same system structural topology) as needed for continuing the simulation in NAMD. Water molecules and ions that are within a certain distance of the nanostructure (denoted ds) are removed and then redistributed as follows since many of them may overlap with the nanostructure or be embedded in it. First, a water box that is large enough to solvate the nanostructure with at least a water boundary of ds (3403 Å3 for simulation of CCMV capsid swelling) is prepared and equilibrated under 298.15K and 1atm by running MD for 100ps. With the nanostructure placed in the center of the equilibrated water box, the coordinates of water molecules, which are selected as within ds and not overlapping with the nanostructure, are retrieved to replace those of the above removed water molecules and ions. Note that the number of the retrieved water molecules may not be exactly the same as that of the needed for redistribution due to the water density difference between the two systems. Thus ds is adjusted iteratively by small increments for selecting water molecules in the equilibrated water box until they are enough for the redistribution of the removed water molecules and ions. The ions are moved to positions that are nearest to their original ones and the overlapping water molecules are removed. This not only generates the final configuration that has no overlapping between the nanostructure and host particles, but also maintains local density and pressure of the host medium for the nanostructure across the OPX timestep without introducing unphysical stress.
MD/OPX simulation of CCMV capsid swelling
MD/OPX simulation of CCMV capsid solvated in a water cube with 15Å water boundary continued with the resulting structure of its MD simulation (sim2 in Table 1) at 1ns as described. It proceeded with cycles of short MD run and projection of the atomic structure by extrapolating its structural OPs. In each cycle, one 500fs MD run was obtained by running NAMD and 5 snapshots were chosen from the trajectory, with which 27 capsid OPs were constructed by using Legendre polynomials of atomic coordinates over order (0, 1, 2) in X, Y, and Z-directions (see Appendix). The capsid OPs were extrapolated over long time intervals by using least-squares fitting. The newly obtained OPs were taken to reconstruct atomic configuration of the capsid for resolvation in the water cube as described above followed by a 5000-step energy minimization, 6-stepwise graduate heating to 298.15K, and 5ps MD thermalization. Atomic accelerations of the energy-minimized and MD-thermalized configurations were computed as indicators to determine whether the structure is mature for the next MD/OPX cycle (i.e., no unphysical large forces on the atoms and atomic accelerations need to be smaller than a tolerance) and the timestep for extrapolation of OPs is adjusted adaptively to ensure the simulation is stable. As explained above, resolvation in MD/OPX is implemented in such a way that local density and pressure of the host medium for the capsid structure is maintained across the OPX timestep, and thus there is no significant configurational change in the capsid and host medium interface during maturation (i.e., energy minimization, graduate heating and thermalization) of the resolvated system. The 5ps MD run is used to thermalize the system, rather than structurally equilibrate it. It is short allowing large acceleration of MD/OPX over direct MD.
By repeating such cycles, native CCMV capsid (denoted Ncap) was simulated for 3ns. Analysis of the MD/OPX trajectory showed that the capsid swells in the water cube (see Results and Discussion). At 1.5ns, the capsid expands to a limit such that it starts to interact with the replicas of its neighboring cells. Therefore, the capsid was extracted from the trajectory snapshot and solvated in a larger water cube (25Å water boundary) for further swelling simulation. This capsid intermediate (denoted Icap) was simulated with MD/OPX by following the same procedure as used for Ncap. MD/OPX trajectory of Icap lasting 17.04ns was used for investigating swelling of the capsid.
The OP timestep obtained from adaptive MD/OPX cycles of simulating Ncap and Icap was found to undergo periodic oscillations and its average was determined to be 3.5ps. With this, the ratio of average OP timestep to the time interval of the short MD run is 70. In the case of simulating Icap solvated in a water cube with 25Å water boundary, there are 3,614,545 atoms in the system. The overall speedup of MD/OPX over MD is 2.14 times faster when using 128 processors on Indiana University Big Red cluster and 1.58 with 512 processors, and the resulting performance is 2.50 days/ns and 0.91 days/ns respectively. The reason for downgrading of the speedup when using more processors is that extraction of trajectory snapshots and resolvation of the capsid executed by calling VMD on only one node (4 processors) take up larger portion of the MD/OPX running time while parallel MD is accelerated by running on more processors.
In the above MD/OPX simulation, 5000 steps of energy minimization were applied to the entire system as a maturation process of the OPX resulting configuration to remove its unphysical structures, e.g., overlapping atoms and stretched bonds/angles. Instead, energy minimization can be applied to the OPX resulting configuration of the capsid only before resolvation. This is not implemented in present MD/OPX because the latest NAMD [48] has problems with embedding a NAMD run (for energy minimization of the nanostructure only) inside NAMD for the top level MD/OPX simulation. However, when this is resolved, the speedup of MD/OPX over MD can be improved to be 4.40 by using 128 processors or 2.67 with 512 processors.
Analysis of CCMV Capsid: Volume, SASA, and Water Density
To measure the volume of CCMV capsid (i.e., protein shell), the capsid snapshot extracted from simulation trajectories is placed at the center of the MD-equilibrated homogenous water box that has been used for “resolvation” in MD/OPX. The average density of water in the box is computed as 975.51kg/m3. By selecting water molecules within 1.4Å of the capsid, the volume of the capsid is considered as the total volume of selected water molecules, which can be calculated by summing up their masses and dividing it by the water density. The volume of the capsid cavity is determined as the total volume of water molecules that are not within 1.4Å of the capsid and for which the radial distance to the capsid COM is smaller than the capsid average radius. Once the volume of the capsid and that of its cavity are determined, the volume for the space outside of the capsid is calculated by subtracting them from the total volume of the water box. As these volume calculations are performed by immersing the capsid in a water box, they are applicable to nanostructures of any shape without symmetry constraints and not affected by the symmetry-breaking process of CCMV capsid swelling described below. With this, the average density of water inside the capsid cavity and outside of the capsid can be calculated respectively and their time evolution can be studied along the simulation trajectories. For calculating the capsid solvent-accessible surface area (SASA), the radius of protein atoms in the capsid is extended by 1.4Å to determine whether they are exposed to the solvent.
III. Results and Discussion
Insights into CCMV capsid swelling through Short-Time MD Simulation
To identify key elements needed to carry out long-time MD/OPX simulation of CCMV capsid swelling, short-time MD simulation was first run on native CCMV capsid that is solvated in a host medium at pH 7.0 and ionic strength I=0.2M. Native CCMV capsid was generated by reorienting 60 copies of its protomer (i.e., asymmetric unit) according to the icosahedral symmetry and solvated in the center of a water cube with 7Å boundary in each direction and neutralized with K+ and Cl− to I=0.2M by using VMD. A schematic representation of the resulting system is shown in Fig. 3a. Then the system along with its structural topology file were taken as input for NAMD[48] simulation by using the CHARMM22 force field[49–50] for capsid proteins and TIP3P model for water molecules[51] (see Computational Methods). The simulation proceeded with initial energy-minimization, thermalization with graduate heating and MD equilibration under constant temperature (298.15K) and pressure (1atm) control (i.e., NPT ensemble). The system was equilibrated after 20ps when the system temperature and pressure fluctuate around target values. Product MD run lasting 3ns was obtained to investigate structural changes of the capsid to identify essential features needed to model swelling of the capsid.
Fig. 3.
Schematic representations of CCMV capsid solvated in a water cube used for our simulations with the water box created using VMD “volmap” plugin and ions represented in CPK.
While the capsid average radius (i.e., the average distance between atoms in the capsid backbone and their COM) might better account for the distribution of capsid atoms, the capsid size is normally described by its diameter in experiments [1, 8, 53]. Besides, N-terminal arms of viral capsid proteins are known to be highly dynamic fluctuating in the capsid interior [1, 54], the capsid minimum radius (i.e., the minimum distance between atoms in the capsid backbone and their COM) is not suitable for measuring the capsid size. Therefore, we use capsid diameter calculated as the maximum distance between two Cα atoms in the capsid along with the maximum radius of the capsid backbone (i.e., the maximum distance from atoms in the capsid backbone to their COM) to characterize CCMV capsid size and thus describe swelling of the capsid in the following.
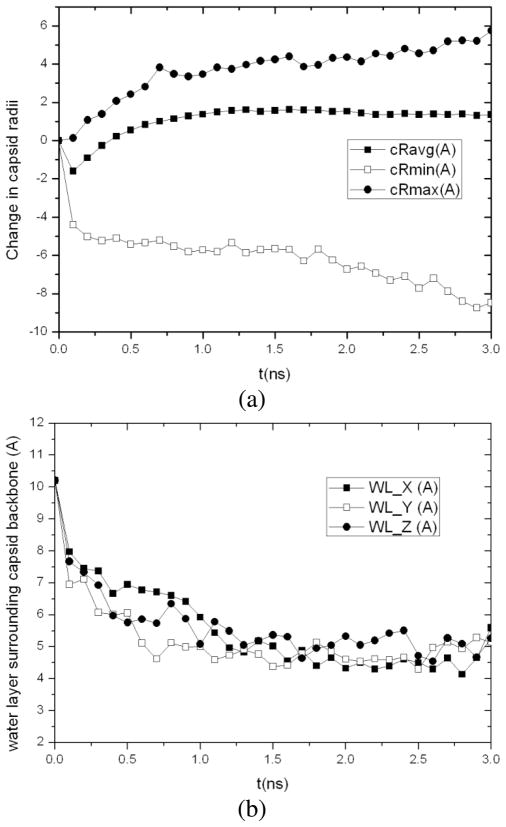
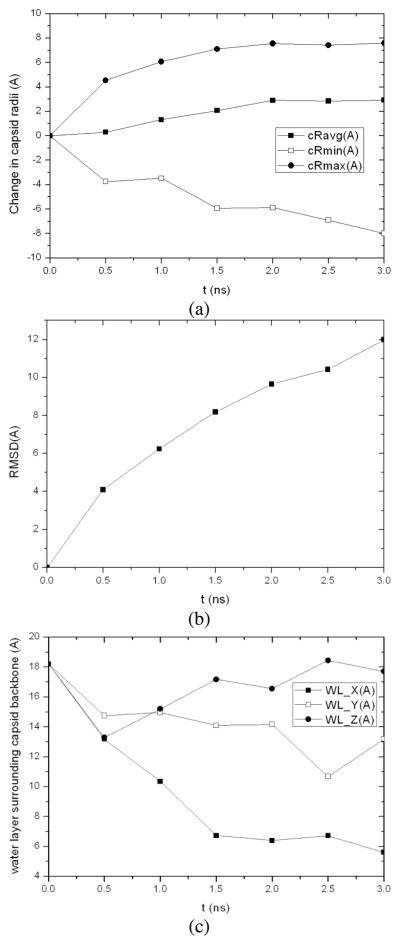
Analysis of the MD trajectory shows that the capsid size stays roughly constant during initial energy minimization, thermalization, and NPT equilibration with the outer radius of capsid backbone increased by only 0.12Å, although the average and inter radius of capsid backbone is deceased by 1.60Å and 4.40Å, respectively. The capsid starts to swell after equilibration. For the resulting structure at 3ns, its outer radius is 5.74Å greater than the native CCMV capsid and 1.35Å greater for the average radius. The capsid diameter is increased by 10.23Å. It was also found to be 13.99Å thicker than the native state with its maximum radius (i.e., the maximum distance of capsid backbone atoms to the capsid COM) increased from 142.49Å to 148.23Å and the minimum radius decreased from 82.32Å to 73.85Å (Fig. 4a). This indicates that the capsid proteins undergo fluctuations during capsid swelling. The thickness of water layer surrounding the capsid backbone in X, Y and Z directions, which are calculated by averaging the minimum distance between non-hydrogen atoms in the capsid backbone (excluding those in protein side chains) and the faces of water box on two sides of each direction, were found to decrease as the capsid swells (Fig. 4b). As a result, the distance between the capsid surface and that of its replicas in neighboring periodic cells decreases. When it reaches the cutoff distance of non-bonded atomic interactions (usually 12Å for short-range van der Waals and electrostatic interactions in MD simulation), the capsid starts to interact with the replicas of its neighboring cells, which slows down swelling of the capsid.
Fig. 4.
3ns MD simulation of native CCMV capsid solvated in a water cube with an initial 7Å water boundary: (a) changes in the average, minimum and maximum radii of the capsid backbone from the starting configuration and (b) averaged thickness of water layer surrounding the capsid backbone in X, Y and Z directions.
The above MD simulation shows that native CCMV capsid swells in 3ns when placed in a host medium at pH 7.0. However, the simulation takes 2.60 days to finish 1ns by using 256 processors on Indiana University Big Red cluster. And as the capsid swells, the water box needs to be expanded to appropriately simulate swelling of the capsid (see discussions below). With the system size largely increased, the performance of MD simulation is downgraded and it becomes even more time-consuming to simulate the swelling of CCMV capsid. Since viral STs involve processes taking place across multiple time and length scales, in the following we apply multiscale MD/OPX to CCMV capsid swelling, which accelerates MD for long-time simulation and addresses the atomistic fluctuations and nanosystem coherent motions simultaneously.
MD/OPX Simulation of CCMV Capsid Swelling
Since earlier MD/OPX studies [26, 28], new modules have been added to account for water molecules and ions and thus achieve simulating bionanosystems in a host medium. Here MD/OPX is used to simulate swelling of CCMV capsid in a host medium. As native CCMV capsid is expected to expand by ~24Å to reach the size of its swollen state[1], simulation of its swelling is divided into the following two phases for efficiency: (1) increase the water boundary from 7Å used in the above MD simulation to 15Å for solvating native CCMV capsid (Ncap) and run MD/OPX on the resulting system until the capsid swells to the size limit of the water cube, and (2) extract the intermediate capsid structure (Icap) obtained from (1) and solvate it in a larger water cube for further simulation of capsid swelling.
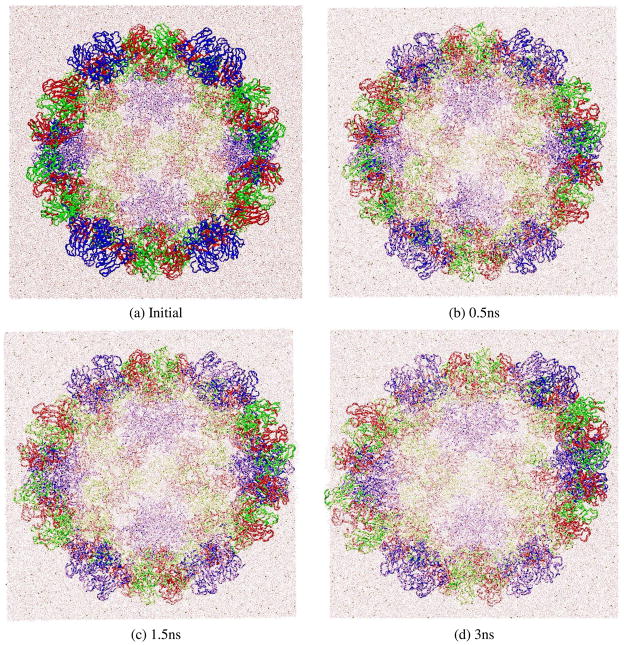
For the first phase of the MD/OPX simulation, we started with native CCMV capsid (Ncap) solvated in a water cube of 15Å water boundary. MD/OPX trajectory lasting 3ns was obtained to study the structural changes of the capsid (see Computational Methods). Fig. 5 shows trajectory snapshots of the capsid in water cube at: (a) the starting configuration, (b) 0.5ns, (c) 1.5ns, and (d) 3ns. The capsid was seen to expand in the MD/OPX trajectory. Quantitative analysis of the MD/OPX trajectory shows the capsid swells with the outer radius of its backbone increased by 7.58Ǻ in 3ns and 2.92Ǻ for the average radius (Fig. 6a). The capsid swells fast up to 2ns at a rate of ~0.22npn in the outer radius and the swelling slows down thereafter. The predicted capsid swelling rate and the time course of capsid radii are in good agreement with the direct MD simulation (sim2 in Table 1 as described below). RMSD of atomic positions between capsid snapshots along the trajectory and its starting structure increases from 0Ǻ to 12Ǻ in 3ns (Fig. 6b). Calculations show the thickness of water layer surrounding the capsid backbone decreases as a result of capsid swelling. It reaches 6Ǻ (half of the cutoff distance for atomic non-bonded interactions) at 1.5ns (Fig. 6c), within which the capsid starts to interact with the replicas of its neighboring cells. Therefore, it is necessary to extract the capsid structure from the trajectory snapshot at 1.5ns and solvate it in a larger water cube for further swelling simulation.
Fig. 5.
MD/OPX trajectory snapshots showing the expansion of native CCMV capsid (Ncap) in water cube with an initial 15Å water boundary: (a) the starting configuration, (b) 0.5ns, (c) 1.5ns and (d) 3ns.
Fig. 6.
Analysis of 3ns MD/OPX trajectory of native CCMV capsid (Ncap): (a) changes in the average, minimum and maximum radii of the capsid backbone from its starting configuration, (b) RMSD of atomic positions between snapshots of capsid backbone along the trajectory and the starting configuration, and (c) averaged thickness of water boundary surrounding the capsid backbone in X, Y and Z directions.
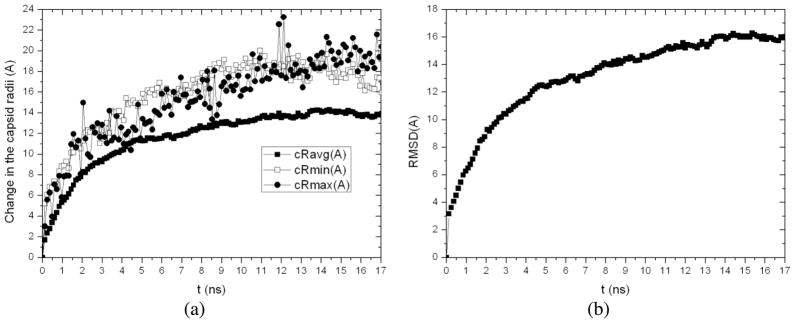
With the intermediate capsid structure (Icap) extracted from the above MD/OPX trajectory at 1.5ns, we solvated the capsid in a larger water cube (25Å water boundary) and then perform 17.04ns MD/OPX simulation. Analysis of the output MD/OPX trajectory shows the capsid swells by 20.41Ǻ and 15.95Ǻ with small fluctuations in its backbone maximum and minimum radii respectively and 13.93Ǻ in the capsid average radius during 17.04ns (Fig. 7a). RMSD of atomic positions between capsid snapshots along the trajectory and its starting structure (Icap) increases from 0Ǻ to 16.05Ǻ in 17.04ns (Fig. 7b). The capsid was found to swell fast during early stage of the simulation with quick increase in both the capsid average radius and RMSD. Notably, the capsid swells by ~12.94Ǻ in its outer radius during 5.16ns, i.e., ~0.25npn. The swelling slows down as the capsid changes towards its near-equilibrium state, i.e. swollen form of the capsid.
Fig. 7.
17.04ns MD/OPX simulation of the swelling of capsid intermediate (Icap): (a) time course of the change in the capsid average, minimum and maximum radii and (b) RMSD of atomic positions between snapshots of capsid backbone along the trajectory and the initial structure.
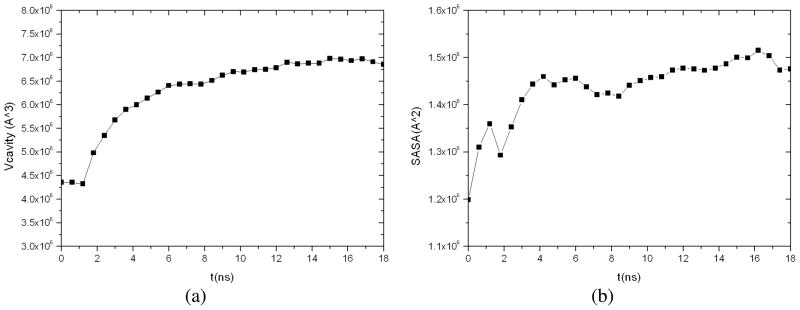
Combining the above two MD/OPX trajectories into one, we obtained 18.54ns dynamics of CCMV capsid swelling from its native state. Total increase in the capsid outer radius is 26.49Ǻ and 14.93Ǻ in the average radius. Analysis of the entire trajectory (Fig. 8) showed that the capsid cavity volume increases from 4.36×106 Ǻ3 to 6.86×106 Ǻ3 by 57% during the course of swelling. The capsid solvent-accessible surface area (SASA) was found to increase from 1.20×106 Ǻ2 to 1.47×106 Ǻ2 in 18ns with moderate fluctuations. These indicate that regions of capsid proteins buried in the capsid interior in its native state are exposed to the solvent during capsid swelling, which also leads to increase in capsid volume.
Fig. 8.
Analysis of CCMV capsid along the entire 18.54ns MD/OPX trajectory: (a) the volume of capsid cavity, and (b) capsid SASA.
CCMV Capsid Swelling Mechanisms
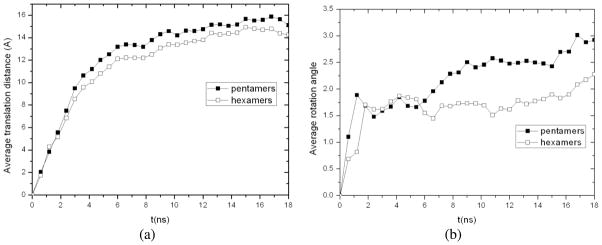
In our earlier study [28], cooperative motions of protein capsomeres have been described for the swollen form of CCMV capsid simulated in vacuum (i.e., gas phase). To characterize the capsomere cooperative motions during swelling of CCMV capsid in a host medium, we calculated the average COM translation distances and rotation angles of 12 pentamers and 20 hexamers along the trajectory obtained from the above MD/OPX simulation. As shown in Fig. 9, pentamers translate radially outwards by 15.10Ǻ on average in 18.54ns and 14.24Ǻ for hexamers; they also undergo counter-clockwise rotation by 2.92° and 2.28° respectively with fluctuations along the trajectory. Though the 18.54ns MD/OPX simulation has not completed captured the entire swelling process of CCMV capsid, these predictions agree with experimental data on the expansion of CCMV capsid in water from its native to the swollen state through rigid-body transformations of the capsomers [1, 6]. Therefore, the swelling of CCMV capsid in a host medium involves large-scale translation and rotation of the pentamers and hexamers, which was also found in the shrinkage of swollen CCMV capsid in vacuum studied earlier [28]. The capsomeres undergo cooperative motions through strongly-coupled allosteric interactions during viral capsid STs.
Fig. 9.
Cooperative motions of pentamers and hexamers during 18.54ns CCMV capsid swelling: time courses of (a) their average COM translation distances and (b) their average rotation angles calculated through structure fitting to the initial configurations in native CCMV capsid.
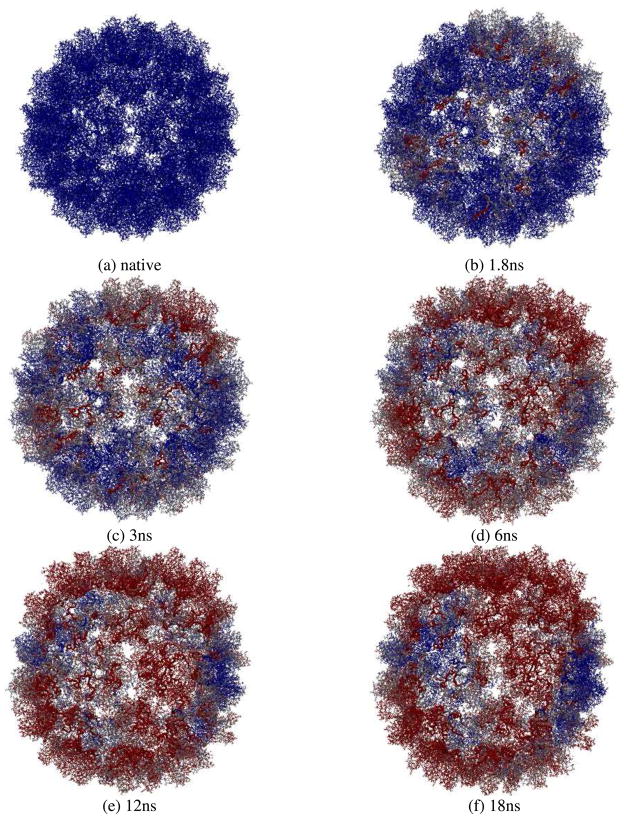
We now explore the swelling mechanisms of CCMV capsid by observing the atomistic motions of the capsid. Fig. 10 shows the interior view of the capsid trajectory snapshots at (a) initial native state, (b) 1.8ns, (c) 3ns, (d) 6ns, (e) 12ns, and (f) 18ns. Atoms in the capsid are represented by their displacements from the original positions with a blue-white-red color scale (i.e., 0 Å for blue, 24 Å for red, and white as the midpoint) to investigate conformational changes in the capsid. N-terminal arms of the capsid proteins are highlighted as ribbons and they are colored according to the displacements of atoms as well. It is seen that atomic displacements are not constrained to the icosahedral symmetry, i.e., atoms in the capsid do not change their color simultaneously in a symmetry-preserving manner. Especially, the N-terminal arms change their color from blue to red during the early stage of capsid swelling (see trajectory snapshots at 1.8ns and 3ns, and movie in the Supporting Material).
Fig. 10.
Interior view of the capsid swelling trajectory snapshots with atoms colored by their displacements from the original positions in a BWR color scale (0Å for blue, 24Å for red, and white as the midpoint): (a) initial native state with all atoms in blue, (b) 1.8ns, (c) 3ns, (d) 6ns, (e) 12ns, and (f) 18ns. N-terminal arms of the capsid proteins are highlighted as ribbons and they are colored according to the displacements of atoms as well. They change their color from blue to red during early stage of the simulation and their fast fluctuations precede and induce global expansion of the capsid. Swelling of CCMV capsid is a symmetry-breaking process involving local initiation and front propagation.
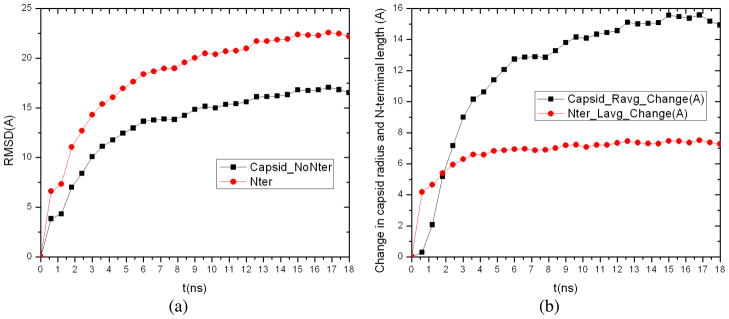
Quantitative calculations show that the atomic RMSD between trajectory snapshots and the initial PDB structure for Cα atoms in the N-terminal arms of capsid proteins (the first 26 residues in each protein subunit) displays much greater increase than that for the rest of capsid protein backbone with their difference being 2.78Å at 0.6ns, increased to 5.32Å at 8.4ns, and becoming nearly constant thereafter (Fig. 11a). This indicates that the N-terminal arms undergo larger deviations from their initial configurations during capsid swelling, especially its early stage. The change in N-terminal arm length (i.e., the distance between the Cα atom of the first and 26th residues) as averaged over 180 capsid proteins is found to increase quickly by 4.18Å at 0.6ns, reach 6.82Å at 4.8ns, and remain constant afterwards (Fig. 11b). In contrast, change in the capsid average radius increases much more slowly during early stage of simulation, and keeps increasing near the end. This shows that the N-terminal arms extend their length much more quickly than global expansion of the capsid.
Fig. 11.
(a) Atomic RMSD between trajectory snapshots and the initial PDB structure for Cα atoms in the N-terminal arms (i.e., the first 26 residues) of capsid proteins compared with that for the rest of capsid protein backbone, and (b) the change in N-terminal arm length as averaged over 180 capsid proteins compared with the change in capsid average radius along the simulation.
Similar to the shrinkage of swollen CCMV capsid in vacuum simulated earlier[28], the ST of CCMV capsid during its swelling involves “local initiation and front propagation” (see movie in Supporting Material). The nucleation takes about 6ns to occur as seen by the color change of two entire capsomers from blue to red in the trajectory snapshot (i.e., the upper two shown in Fig. 10d). This is 14ns shorter than the nucleation time needed for the shrinkage of swollen CCMV capsid in vacuum. One reason is that N-terminal arms of capsid proteins were added in the present simulation of CCMV capsid swelling in a host medium and their fast motions accelerate the nucleation of capsid ST. After nucleation, the capsid ST then propagates across the capsid indicated by spreading of the red areas. The propagation of cooperative motions from one capsomere to its neighbors takes place roughly on a timescale of 10ns (see snapshots Fig. 10(d) to (f) and the movie in Supporting Material). Thus the propagation speed can be calculated as ~0.6npn with the average capsomere distance of about 6nm, which is similar to the ST in swollen CCMV capsid during its shrinkage in vacuum. Therefore, STs of viral capsids start with local conformational changes that break icosahedral symmetry (e.g., N-terminal motions of capsid proteins), followed by global structural changes that involve large-scale cooperative translations and rotations of capsid structural units (e.g., capsomeres or protein subunits) through strong allosteric interactions.
Dependence of CCMV Capsid Swelling on System Conditions
The above simulations show that CCMV capsid swells over tens of nanoseconds. Though there have not been swelling experiments performed on empty CCMV capsids, such STs for the complete virus particles (i.e., capsids embedded with viral RNA) were observed to occur on timescales of milliseconds to minutes in experiments [55–57]. In the following, we present a list of simulations that indicate the swelling of CCMV capsid is strongly dependent on system conditions. Five different simulations starting from the capsid native state were given in Table 1. The capsid was solvated in a water cube with two different water boundaries, i.e., 7Å (sim1) and 15Å (sim2 to sim5). The water boundary here is the minimum distance between atoms on the capsid surface and the faces of water cube. Note that sim1 has been used to identify key elements for long-time simulation of the capsid selling as discussed above. There are 522,000 atoms in CCMV capsid and the total number of atoms in these systems ranges from ~2.41 to ~2.85 million depending on the thickness of water boundary and salt concentration used for solvation.
Periodic boundary conditions were applied for all MD simulations (see Computational Methods). Constant volume and temperature control (i.e., NVT ensemble) was used for sim3, and the others were performed under constant pressure and temperature control (i.e., NPT ensemble). The specified temperature was 298.15K and the pressure was 1atm for NPT simulations. With the above settings, NAMD was used to simulate the five systems to determine the controlling factors of CCMV capsid swelling. It was found that the unit cell for systems prepared with VMD “solvate” plugin by using its default parameters (notably 2.4Å for the minimum distance between water molecules and the solute) need to be compressed in order to reach 1atm pressure in NPT simulations (e.g., sim1 and sim2). Without pressure control, the systems would have a pressure much lower than 1atm in NVT simulation (e.g., sim3). The analysis of the output trajectories is as follows.
CCMV capsid was found to swell much faster and more dramatically in sim3 under NVT control (i.e., constant volume, low pressure) than in NPT simulations. While the capsid swells by 7.61Å in its outer radius and only 1.58Å in the average radius during 3ns NPT simulation in sim2, the capsid as simulated under NVT control swells with the outer and average radius increased by 15.33Å and 10.03Å in 3ns, and 18.05Å and 10.48Å respectively during 5ns in sim3. During the 5ns significant swelling, the capsid thickens by 16.91Å with the capsid SASA increased from 1.20*106 Å2 to 1.51*106 Å2; the volume of capsid cavity is increased by 45% from 4.09*106 Å3 to 5.95*106 Å3 (see calculation details in Computational Methods and Excel spreadsheet in Supporting Material). In the early stage of the simulation, water molecules flux from the capsid cavity into the capsid to solvate proteins and small regions of vacuum are formed inside the capsid. As the capsid continues swelling, water molecules flux from the capsid exterior into the capsid cavity. The average density of water inside the capsid cavity decreases significantly from 839.56kg/m3 to 670.07kg/m3 at the end of the 5ns simulation, while average density of water outside of the capsid stays roughly constant.
When pressure control is applied as in sim1 and sim2, the system is compressed to reach 1atm pressure and the swelling rate of CCMV capsid is decreased. The capsid cavity volume and its SASA are also found to increase accompanying the capsid swelling. The density of water in the capsid cavity decreases during early stage of the simulation due to swelling of the capsid and the flux of water molecules into the capsid to solvate proteins. It then increases as the capsid swelling slows down and the water molecules flux from the capsid exterior into the capsid cavity under the pressure control. In the case of sim2, the average density of water inside the capsid cavity decreases from 886.00kg/m3 to 841.39kg/m3 at 1ns and then increases to 866.82kg/m3 at 3ns.
To understand the different swelling behaviors of CCMV capsid found in the NPT and NVT simulations, we compare the system pressure profiles of sim2 and sim3. As no algorithm has been developed to analyze the radial pressure of a system like CCMV capsid here, we divided the simulation box into horizontal slices of ~10Å thickness in Z-direction and calculated the local pressure of each slice by using the pressure profile analysis module provided in NAMD [48] for the two simulations. Calculation results showed that the average pressure difference between slices that sandwich CCMV capsid at the bottom and the top (both from the capsid cavity surface to the capsid exterior) is 88atm at 0.1ns in sim3, which is ~13atm greater that in sim2. Though the pressure profile in NAMD is calculated by following from the algorithm proposed for lipid bilayers [58] and it could lead deviations for the CCMV capsid solvated in a water box here, these calculations suggest the pressure gradient across the capsid in sim3 is higher than that in the latter. As the capsid swells, the pressure of the system slice right below or above the capsid surface (i.e., pressure of water outside of the capsid) keeps increasing. It takes 0.6ns to decrease the pressure gradient across the capsid to zero in sim2, while 1ns (i.e., 0.4ns longer) in sim3. As a result, the capsid swells faster and more dramatically in NVT simulation sim3 than in NPT simulation sim2.
Complete wild-type CCMV virions swell radially by ~10% when the solution pH is raised to 7.0 at low ionic strength (I<0.2M) in the absence of divalent cations. The transition can be reversed when pH is lowered from 7.0 to 5.0[1]. The virus stays stable in its native compact form at pH 5.0. Though there has been no swelling experiment performed on empty CCMV capsids, here we adjust the system pH from 7.0 (sim1 and sim2) to 5.0 and investigate the pH dependence of CCMV capsid stability. As only the pKa of histidine side chain (6.04) lies in this pH range among all amino acids, the side chain of 360 histidines in CCMV capsid (i.e., 2 in each protein subunit) are protonated corresponding to the pH change. As a result, the net charge of CCMV capsid increases from +900e to +1260e. The capsid is solvated in a water cube and neutralized with 2282 K+ and 3542 Cl− for NPT-ensemble simulation. Trajectory analysis shows that size of the capsid characterized by its outer radius undergoes 1.41Å smaller increase from the intial X-ray crystal structure during 3ns at pH 5.0 (sim4) than that at pH 7.0 (sim2), though the capsid average radius displays slightly greater increase (0.36Å) at pH 5.0 than that obtained at pH 7.0 (see Figs. S2 and S3 in Supporting Material). The capsid diameter is found to increase from 284.69Å to 294.43Å in 2ns and level off thereafter with moderate fluctuations at pH 5.0. Increase in the capsid diameter is also 1.91Å smaller than that obtained from pH 7.0 simulation (see Fig. S1 in Supporting Material). This indicates empty CCMV capsid becomes less likely to swell (more stable) when pH is lowered from 7.0 to 5.0, which agrees with the experimental results on complete CCMV virions.
While empty CCMV capsid is shown to be more stable at pH 5.0 than that at pH 7.0 in the above analysis, the capsid still displays swelling in the early stage of the simulation. Divalent cations (e.g., Ca2+ and Mg2+) are known to bind CCMV capsid proteins, stabilize the capsid and eliminate swelling of the complete virus [1]. To examine their effects on the behavior of empty CCMV capsid, we add 0.05M MgCl2 to the system prepared in sim4 at pH 5.0 and run another 3ns MD simulation on the resulting system, i.e., at pH 5.0 and 0.05M MgCl2. Results show that the capsid is further stabilized with its diameter increased to only 291.07Å in 1ns and level off thereafter (Fig. S1). A number of Mg2+ ions are found to diffuse into the capsid (Fig. S4), 51 at 1ns and 60 (~9% of the total) at 3ns. Especially at the end of the simulation, 11 out of the 60 Mg2+ ions are bound to the divalent cation inter-protein binding sites, which are composed of residues Glu81, Gln85, Lys84 and Glu148 from a protein and Gln149* and Asp153* from its adjacent protein [1, 24]. A typical example is shown in Fig. S5a. Time courses of the distances between Mg2+ ion in the center and its ligand atoms show that the Mg2+ ion is diffused into the binding site from bulk water and then bound there (Fig. S5b). Such binding of divalent cations to the sites between capsid proteins stabilizes the capsid against swelling [1, 6, 53].
Though the MD simulations performed here are fairly short (due to the size of CCMV capsid) and longer trajectories are needed to conclude on the capsid stability and swelling dynamics, our results indicate that the swelling of CCMV capsid in a host medium is strongly dependent on system conditions (e.g., the pressure gradient across the capsid, pH, and the presence of divalent cations). We show that empty CCMV capsid becomes more stable as the solution pH is lowered from 7.0 to 5.0 and further stabilized when divalent cations are added to the system. This argues for the correctness of conditions under which our above long-time MD/OPX simulation was carried out.
In addition to factors discussed above, discrepancies between the timescale for capsid swelling obtained from our simulations and that for swelling of complete virus particles in experiments include the following. (1) The free energy landscape is populated by many minima. Thus along the pathway connecting the original to the final structure the system can reside for appreciable time at these minima. In the transition for one minimum to the next, many transition states and associated energy barriers are encountered. (2) The free energy landscape can be so populated with minima that the system is driven by a higher-order entropy, i.e., the system loses track of where it is going although it is driven by an overall energy gradient; this is a complex analog of a two well potential with a broad, shallow well and a narrow shallow one; in that case the system could prefer to be in the relatively high energy well. (3) The dynamics of viral capsid is likely highly frictional; thus despite a large free-energy gradient the kinetics can be slow. (4) Attractive capsid-RNA interactions will repress capsid swelling, which requires overcoming the free energy barriers.
IV. Conclusions
While STs of CCMV (e.g., pH-induced swelling) have been studied since 1975 [3–4, 53, 59–60], the exact transition mechanisms remain unclear [5]. With new modules added to account for water molecules and ions, MD/OPX is used to simulate the swelling of native CCMV capsid in a host medium. Insights into the stability of CCMV capsid were first obtained through short-time MD simulation. With essential features identified to allow the swelling of CCMV capsid, MD/OPX simulation was run on the capsid with its long-time dynamics captured. We validate the algorithm whereby the dynamics of capsid OPs is computed by using capsid atomic variables even though the capsid is immersed in a host medium. Furthermore, this algorithm is based on extrapolation of the capsid configuration followed by resolvation and re-thermalization, and the host fluid dynamics is accounted for indirectly since it interacts with the capsid as captured explicitly through the short MD run.
MD/OPX simulation results show that the N-terminal arms of capsid proteins undergo large deviations from the initial configurations with their length extended quickly during the early stage of capsid swelling. Similar to the shrinkage of swollen CCMV capsid in vacuum[28], swelling of native CCMV capsid in a host medium is also symmetry-breaking involving local initiation and front propagation. This suggests that it is not appropriate to use symmetry-constrained models to study the pathways and mechanisms of viral STs [15, 20].
The pH-induced swelling of CCMV has been suggested to be a first order phase transition at low salinity (I=0.2M) with hysteresis found in its titration curve [59]. However, divalent cations, such as Mg2+ and Ca2+, are known to bind CCMV capsid proteins at the quasi-threefold axes, and thus stabilize the capsid and the complete virus [1, 4]. By adding magnesium ions to the solution ([Mg2+]=0.01M), hysteresis in the titration curve of CCMV can be abolished [3, 59] and thus CCMV swelling becomes fully reversible. This indicates that virus swelling can become a second order phase transition in the presence of divalent cations. It was also found that the effect of a high concentration in potassium ions (e.g., 0.8M KCl) is identical to that of magnesium for eliminating hysteresis in the CCMV titration curve. Furthermore, as salinity is increased from 0.2M to 0.3M, the hysteresis loop size is reduced by a factor of 2. While the above controlling factors for the swelling of complete CCMV have been determined, those for the empty CCMV capsid swelling are still under investigation [53].
Our all-atom multiscale simulations can be used to create a phase diagram for CCMV capsid mapping out regions in the space of pH, salinity, concentration of divalent cations, and temperature wherein first and second order transitions take place. As capsid fluctuations are believed to play an important role in vial STs, identifying conditions where critical fluctuations are realized will be of great interest. The simulations will be used to suggest the experimental conditions wherein large fluctuations and the most dramatic STs occur. From our simulations, swelling of CCMV capsid was found to start locally and then spread across the capsid. However, one might suggest that viral STs occur simultaneously. If this is the case, there should be instantaneous communication of proteins across the viral capsid, e.g., as follows from Coulomb interactions. Therefore, decreasing salinity will increase the length scale of effective interactions, which is limited to the Debye length. In this way, we will determine theoretically if there is an observable transition between the nucleation/propagation and the global/simultaneous ST mechanism as salinity changes. This suggests that at low salinity the system should be highly cooperative and global so that critical fluctuations would be repressed; alternatively, the high salinity system is expected to act more like a two or three dimensional system and display critical behavior.
As found through our MD/OPX simulation, the N-terminal arms of CCMV capsid proteins are highly dynamic. They undergo large deviations from the initial configurations with their length extended quickly during the early stage of CCMV capsid swelling. Such motions are found to take place in other viral capsids. They are believed to play important roles in packaging viral genome during virus maturation or engineered nanomaterial synthesis by using viral capsids as molecular containers [54]. For example, the N-terminus of the poliovirus capsid protein VP1 that is entirely internal in native virion becomes externalized upon cell attachment, and the exposed N-terminus of VP1 was shown to be responsible for liposome binding [10, 61]. These conformational changes can be predicted from our simulations and used to guide experimental studies, e.g., chemical labeling[62]. If the protein terminal domains are protruding/retracting in a fluctuating manner, then the degree of labeling will increase when such fluctuations are large. Furthermore, the depth of penetration of the labeling should increase as critical fluctuation intensity increases.
While the water molecules and ions have been modeled explicitly in the simulation of CCMV capsid swelling in this study, an implicit solvent approach can be used instead. It will simplify MD/OPX making the workflow similar to the simulation of nanostructures in vacuum. With this, the performance obtained from our previous study on simulating the shrinkage of swollen CCMV capsid for 200ns is expected [28]. Given the capability of MD/OPX to simulate viral STs with their multiscale features captured, we suggest it will facilitate computer-aided design of antiviral drugs and vaccines, and nanocapsules for delivery of therapeutic agents to diseased tissues.
Supplementary Material
Acknowledgments
This project was supported in part by National Institutes of Health (NIH) through Stanford University via SimBios, National Science Foundation through the Collaborative Research in Chemistry Program, NIH National Institute of Biomedical Imaging and Bioengineering, and Indiana University College of Arts and Sciences and the METACyt project through the Center for Cell and Virus Theory. Y. Miao thanks D. Zhang for generously providing the complete CCMV protomer structure and J. Tillotson for help on running jobs on Indiana University Big Red supercomputer.
Appendix Construction of Order Parameters from Atomic Configurations
Consider a nanostructure embedded in a box (a cube for simplicity of presentation) of volume L3. Basis functions uk (x) (e.g., orthogonal polynomials or harmonic functions) labeled with integer index k are introduced such that
| (1) |
for Kronecker delta δkk′. Composite functions Uk ( s⃑) are defined such that
| (2) |
where s⃑ = (x, y, z) for box centered at s⃑ = 0⃑. By using Uk (s⃑), system collective modes can be constructed as with and the nanostructure s⃑ is considered to be deformation of a reference configuration s⃑0:
| (3) |
where σ⃑i is the residual due to finite truncation of k sum and Φ⃑k are system order parameters (OPs). Assume mi is the mass of atom i and . We determine Φ⃑k to minimize the mass-weighted root mean square residuals (RMSR), i.e., , which yields
| (4) |
The σ⃑i contribution is neglected in arriving at this definition of Φ⃑k as σ⃑i fluctuates with i and hence with space, while the basis functions that capture overall nanostructural features vary smoothly by design. When most of the space in the system is occupied with atoms, the i sum is essentially a Monte Carlo integration. The orthogonality of the normailized basis functions implies that Bqk ≈ δqk and (4) can be approximated as
| (5) |
Therefore, dimensionality reduction for nanostructures from the N-atom configuration to M OPs is achieved through
| (6) |
In the above formulation, atomic coordinates of a reference configuration are used to construct the nanostructure collective modes and their summation given by Eqn. (3) results in a new configuration that the system can evolve to. Instead atomic displacements calculated from two known configurations of the nanostructure can be used to construct its collective modes and their summations weighted by OPs would describe system coherent motions directly.
Footnotes
Supporting Information Available
Animation of the MD/OPX simulation trajectory and the simulation data relevant to our discussions in Sect. III are provided. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Speir JA, et al. Structures of the Native and Swollen Forms of Cowpea Chlorotic Mottle Virus Determined by X-Ray Crystallography and Cryoelectron Microscopy. Structure. 1995;3(1):63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang DQ, et al. Electrostatic interaction between RNA and protein capsid in cowpea chlorotic mottle virus simulated by a coarse-grain RNA model and a Monte Carlo approach. Biopolymers. 2004;75(4):325–337. doi: 10.1002/bip.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adolph KW. Structural Transitions of Cowpea Chlorotic Mottle Virus. J gen Virology. 1975;(28):147–154. [Google Scholar]
- 4.Liepold LO, et al. Structural transitions in Cowpea chlorotic mottle virus (CCMV) Phys Biol. 2005;(2):S166–S172. doi: 10.1088/1478-3975/2/4/S11. [DOI] [PubMed] [Google Scholar]
- 5.Speir JA, et al. Enhanced Local Symmetry Interactions Globally Stabilize a Mutant Virus Capsid That Maintains Infectivity and Capsid Dynamics. Journal of Virology. 2006;80(7):3582–3981. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu HJ, et al. Pseudo-atomic models of swollen CCMV from cryo-electron microscopy data. Journal of Structural Biology. 2003;142(3):356–363. doi: 10.1016/s1047-8477(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 7.Canady MA, et al. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV) Journal of Molecular Biology. 2000;(299):573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- 8.Canady MA, Tsuruta H, Johnson JE. Analysis of Rapid, Large-scale Protein Quaternary Structural Changes: Time-resolved X-ray Solution Scattering of Nudaurelia capensis ω Virus (NωV) Maturation. Journal of Molecular Biology. 2001;(311):803–814. doi: 10.1006/jmbi.2001.4896. [DOI] [PubMed] [Google Scholar]
- 9.Wikoff WR, et al. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. Journal of Structural Biology. 2006:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Belnap DM, et al. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. Journal of Virology. 2000;74(3):1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogle JM. Poliovirus cell entry: Common structural themes in viral cell entry pathways. Annual Review of Microbiology. 2002;(56):677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bancroft JB, Hiebert E. Formation of an Infectious Nucleoprotein from Protein and Nucleic Acid Isolated from a Small Spherical Virus. Virology. 1967;32(2):354. doi: 10.1016/0042-6822(67)90284-x. [DOI] [PubMed] [Google Scholar]
- 13.Brumfield S, et al. Heterologous expression of the modified coat protein of Cowpea chlorotic mottle bromovirus results in the assembly of protein cages with altered architectures and function. Journal of General Virology. 2004;85:1049–1053. doi: 10.1099/vir.0.19688-0. [DOI] [PubMed] [Google Scholar]
- 14.Freddolino PL, et al. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14(3):437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Speelman B, Brooks BR, Post CB. Molecular Dynamics Simulations of Human Rhinovirus and an Antiviral Compound. Biophysical Journal. 2001;80:121–129. doi: 10.1016/S0006-3495(01)75999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phelps DK, Rossky PJ, Post CB. Influence of an antiviral compound on the temperature dependence of viral protein flexibility and packing: a molecular dynamics study. Journal of Molecular Biology. 1998;276(2):331–337. doi: 10.1006/jmbi.1997.1542. [DOI] [PubMed] [Google Scholar]
- 17.Isea R, Aponte C, Cipriani R. Can the RNA of the cowpea chlorotic mottle virus be released through a channel by means of free diffusion? A test in silico. Biophysical Chemistry. 2004;107(2):101–106. doi: 10.1016/S0301-4622(03)00193-5. [DOI] [PubMed] [Google Scholar]
- 18.Harries D, May S, Gelbart WM, Ben-Shaul A. Structure, Stability and Thermodynamics of Lanellar DNA-Lipid Complexes. Biophysical Journal. 1998;75:159. doi: 10.1016/S0006-3495(98)77503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arkhipov A, Freddolino PL, Schulten K. Stability and dynamics of virus capsids described by coarse-grained modeling. Structure. 2006;(14):1767–1777. doi: 10.1016/j.str.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Tama F, Brooks CL. The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle virus. Journal of Molecular Biology. 2002;318(3):733–747. doi: 10.1016/S0022-2836(02)00135-3. [DOI] [PubMed] [Google Scholar]
- 21.Tama F, Brooks CL. Diversity and identity of mechanical properties of icosahedral viral capsids studied with elastic network normal mode analysis. Journal of Molecular Biology. 2005;345(2):299–314. doi: 10.1016/j.jmb.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 22.van Vlijmen HWT, Karplus M. Normal mode calculations of icosahedral viruses with full dihedral flexibility by use of molecular symmetry. Journal of Molecular Biology. 2005;350(3):528–542. doi: 10.1016/j.jmb.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 23.Sayyed-Ahmad A, Miao Y, Ortoleva P. Poisson-Boltzmann theory of bionanosystems. Communications in Computational Physics. 2008;3(5):1100–1116. [Google Scholar]
- 24.Konecny R, et al. Electrostatic properties of cowpea chlorotic mottle virus and cucumber mosaic virus capsids. Biopolymers. 2006;82(2):106–120. doi: 10.1002/bip.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Y, Ortoleva P. All-atom multiscaling and new ensembles for dynamical nanoparticles. Journal of Chemical Physics. 2006;125(4):44901. doi: 10.1063/1.2218838. [DOI] [PubMed] [Google Scholar]
- 26.Miao Y, Ortoleva PJ. Molecular dynamics/order parameter extrapolation for bionanosystem simulations. J Comput Chem. 2008;30(3):423–437. doi: 10.1002/jcc.21071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pankavich S, et al. Stochastic dynamics of bionanosystems: Multiscale analysis and specialized ensembles. J Chem Phys. 2008;128(23):234908. doi: 10.1063/1.2931572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y, Ortoleva PJ. Viral structural transition mechanisms revealed by multiscale molecular dynamics/order parameter extrapolation simulation. Biopolymers. 2009;93(1):61–73. doi: 10.1002/bip.21299. [DOI] [PubMed] [Google Scholar]
- 29.Rader AJ, Vlad DH, Bahar I. Maturation dynamics of bacteriophage HK97 capsid. Structure. 2005;13(3):413–421. doi: 10.1016/j.str.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Annalen der Physik. 1905;(17):549–560. [Google Scholar]
- 31.Chandrasekhar S. Dynamical Friction.I.General Considerations: the Coefficient of Dynamical Friction. Astrophysical J. 1943;97:255–262. [Google Scholar]
- 32.Deutch JM, Oppenheim I. The Concept of Brownian Motion in Modern Statistical Mechanics. Faraday Discuss. Chem. Soc; London: 1987. pp. 1–20. [Google Scholar]
- 33.Deutch JM, et al. Light-Scattering from Systems with Chemical Oscillations and Dissipative Structures. Journal of Chemical Physics. 1972;57(10):4327. [Google Scholar]
- 34.Shea JE, Oppenheim I. Fokker-Planck equation and Langevin equation for one Brownian particle in a nonequilibrium bath. Journal of Physical Chemistry. 1996;100(49):19035–19042. [Google Scholar]
- 35.Shea JE, Oppenheim I. Fokker-Planck equation and non-linear hydrodynamic equations of a system of several Brownian particles in a non-equilibrium bath. Physica A. 1997;247(1–4):417–443. [Google Scholar]
- 36.Ortoleva PJ. Nanoparticle dynamics: a multiscale analysis of the Liouville equation. J Phys Chem B. 2005;109(45):21258–66. doi: 10.1021/jp051381b. [DOI] [PubMed] [Google Scholar]
- 37.Peters MH. Fokker-Planck equation, molecular friction, and molecular dynamics for Brownian particle transport near external solid surfaces. Journal of Statistical Physics. 1999;94(3–4):557–586. [Google Scholar]
- 38.Peters MH. Fokker-Planck equation and the grand molecular friction tensor for coupled rotational and translational motions of structured Brownian particles near structured surfaces. Journal of Chemical Physics. 1999;110(1):528–538. [Google Scholar]
- 39.Zwanzig R. Nonequilibrium Statistical Mechanics. Oxford University Press; New York: 2001. [Google Scholar]
- 40.Pankavich S, et al. J Chem Phys. 2008. Stochastic Dynamics of Bionanosystems: Multiscale Analysis and Specialized Ensembles; p. 029822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shreif Z, Ortoleva P. Curvilinear All-Atom Multiscale (CAM) Theory of Macromolecular Dynamics. J Stat Phys. 2008;(130):669–685. [Google Scholar]
- 42.Miao Y, Ortoleva PJ. Viral structural transitions: an all-atom multiscale theory. Journal of Chemical Physics. 2006;125(21):214901. doi: 10.1063/1.2400858. [DOI] [PubMed] [Google Scholar]
- 43.Kuhn RJ, Rossmann MG. Structure and assembly of icosahedral enveloped RNA viruses. Virus Structure and Assembly. 2005;64:263–284. doi: 10.1016/S0065-3527(05)64008-0. [DOI] [PubMed] [Google Scholar]
- 44.Shreif Z, et al. Enveloped viruses understood via multiscale simulation: Computer-aided vaccine design. Springer: Netherlands. 2008;15:363–380. [Google Scholar]
- 45.Kevrekidis IG, Gear CW, Hummer G. Equation-free: The computer-aided analysis of comptex multiscale systems. Aiche Journal. 2004;50(7):1346–1355. [Google Scholar]
- 46.Hummer G, I, Kevrekidis G. Coarse molecular dynamics of a peptide fragment: Free energy, kinetics, and long-time dynamics computations. Journal of Chemical Physics. 2003;118(23):10762–10773. [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Phillips JC, et al. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;(26):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackerell AD, et al. Self-Consistent Parameterization of Biomolecules for Molecular Modeling and Condensed Phase Simulations. Faseb Journal. 1992;6(1):A143–A143. [Google Scholar]
- 50.MacKerell AD, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen WL, et al. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 52.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23(3):327–341. [Google Scholar]
- 53.Lavelle L, Michel JP, Gingery M. The disassembly, reassembly and stability of CCMV protein capsids. Journal of Virological Methods. 2007;146(1–2):311–316. doi: 10.1016/j.jviromet.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 54.Douglas T, et al. Protein Engineering of a Viral Cage for Constrained Nanomaterials Synthesis. Advanced Material. 2002;14(6):415–418. [Google Scholar]
- 55.Taylor DJ, et al. Correlation of chemical reactivity of Nudaurelia capensis omega virus with a pH-induced conformational change. Chem Commun (Camb) 2003;(22):2770–1. doi: 10.1039/b310533d. [DOI] [PubMed] [Google Scholar]
- 56.Lee KK, et al. Evidence that a Local Refolding Event Triggers Maturation of HK97 Bacteriophage Capsid. Journal of Molecular Biology. 2004;340(3):419–433. doi: 10.1016/j.jmb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 58.Lindahl E, Edholm O. Spatial and energetic-entropic decomposition of surface tension in lipid bilayers from molecular dynamics simulations. Journal of Chemical Physics. 2000;113(9):3882–3893. [Google Scholar]
- 59.Jacrot B. Studies on the assembly of a spherical plant virus: II. The mechanism of protein aggregation and virus swelling. Journal of Molecular Biology. 1975;95(3):433–446. doi: 10.1016/0022-2836(75)90201-6. [DOI] [PubMed] [Google Scholar]
- 60.Klug WS, et al. Failure of viral shells. Physical Review Letters. 2006;97(22) doi: 10.1103/PhysRevLett.97.228101. [DOI] [PubMed] [Google Scholar]
- 61.Fricks CE, Hogle JM. Cell-Induced Conformational Change in Poliovirus - Externalization of the Amino Terminus of Vp1 Is Responsible for Liposome Binding. Journal of Virology. 1990;64(5):1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beardsley RL, Running WE, Reilly JP. Probing the struture of Caulobacter crescentus ribosome with chemical labeling and mass spectrometry. J Proteome Res. 2006;5:2935–2946. doi: 10.1021/pr060170w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.