Abstract
Fine mapping followed by candidate gene analysis of erd - a canine hereditary retinal degeneration characterized by aberrant photoreceptor development - established that the disease cosegregates with a SINE insertion in exon 4 of the canine STK38L/NDR2 gene. The mutation removes exon 4 from STK38L transcripts and is predicted to remove much of the N-terminus from the translated protein, including binding sites for S100B and Mob Proteins, part of the protein kinase domain, and a Thr-75 residue critical for autophosphorylation. Although known to have roles in neuronal cell function, the STK38L pathway has not previously been implicated in normal or abnormal photoreceptor development. Loss of STK38L function in erd provides novel potential insights into the role of the STK38L pathway in neuronal and photoreceptor cell function, and suggests that genes in this pathway need to be considered as candidate genes for hereditary retinal degenerations.
Keywords: Retinal degeneration, Leber Congenital Amaurosis, STK38L, animal model
1. INTRODUCTION
Early retinal degeneration (erd) is a canine early onset autosomal recessive disease [1] corresponding to Leber Congenital Amaurosis in humans. It is characterized by aberrant functional and structural development of rod photoreceptor inner and outer segments, and of both rod and cone synapses, with subsequent retinal degeneration [1]. The locus was previously mapped to a 30 cM interval on canine chromosome 27 (CFA27), corresponding to a part of human chromosome 12p [2].
In this report, the candidate region was first reduced by fine-scale mapping to a 4.6 cM zero recombination interval on CFA27, a 1.6 Mb region in the canine genome sequence assembly (CanFam2). Positional candidate gene analysis then identified a tRNA-like short interspersed repeat (SINE) insertional mutation in exon 4 of STK38L that cosegregates with erd.
The gene STK38L, also called NDR2, codes for serine/threonine kinase 38-like protein -- a protein kinase in the nuclear Dbf2-related (NDR) family involved in neuronal cell division and morphology [3, 4, 5]. STK38L kinase is activated by S100B (a Ca++-binding protein) and Mob (Mps one binder), enabling STK38L to autophosphorylate at Thr-75, Ser-282, and Thr-444 [5, 6]. STK38L and its partners have been implicated in neuronal cytoskeletal development, neurite outgrowth and synaptic remodeling. Despite this, much remains to be determined concerning their exact role and mechanisms of action. A particular gap in this understanding is their role in the retina. Until now, neither STK38L nor its partners, has been implicated in any hereditary retinal degeneration, or as having critical roles in photoreceptor development. The erd mutant dog therefore offers potential insights into the role of this pathway in both normal and abnormal photoreceptor and neuronal development.
2. MATERIAL AND METHODS
2.1. Animals
The erd strain of dogs, maintained at the Retinal Disease Studies Facility (RDSF) in Kennett Square, PA, derives from 2 erd-affected Norwegian elkhound dogs, highly inbred siblings, that were outbred to Beagles and other dogs, and their progeny used to generate pedigrees segregating erd. Selected dogs from this strain were evaluated by DNA and RNA analysis. Blood samples were also collected from privately owned dogs of several breeds, and DNA extracted as previously described [7]. All procedures involving animals were undertaken according to IACUC approved protocols at Cornell University and the University of Pennsylvania, and in adherence to the ARVO Resolution for the Use of Animals in Ophthalmic and Vision Research.
2.2. Gene-associated markers for linkage mapping
When fine mapping of erd began, the canine genome assembly was not available. The erd locus had previously been mapped to the CFA27 interval flanked by genes PFKM, LALBA, PTHLH, and IAPP [2], homologously located on HSA12p. From comparison of the canine and human genomes [8], 7 genes (BICD1, PTHLH, ITPR2, SSPN, SHARP1, LRMP, and SIAT8A) were selected from HSA12p between LALBA and IAPP, for fine mapping erd.
The RPC181 canine 8.1-fold BAC library from Roswell Park Cancer Institute (http:www.chori.org/bacpac/mcanine81.htm) was screened to identify BAC clones specific for SIAT8A, BICD1, and SHARP1 as previously described [9]. Gene-associated polymorphic markers were developed from identified BAC clones (Supplementary Table 1A) as previously described [9].
For ITPR2, PTHLH and SSPN, partial canine genomic sequences were obtained from the Institute for Genome Research (TIGR, E. Kirkness, pers. comm.) and used to develop gene-specific polymorphic markers (Supplementary Table 1A). A polymorphic (CA)n repeat was identified in the LRMP gene by amplifying and sequencing canine intron 1 using primers designed from human sequences.
When the canine genome assembly (CanFam2) became available the position of all markers was identified in CanFam2 using UCSC Bioinformatics tools (electronic PCR, Blat; <http://genome.ucsc.edu/cgi-bin/hgGateway>).
2.3. Genotyping and linkage analyses
Pedigrees segregating erd were genotyped for informative markers (Supplementary Table 1). Markers were amplified from 50 ng of genomic DNA as per: 96°C for 2 minutes; 30 cycles of 96°C (20 seconds), 58°C (20 seconds), and 72°C (20 seconds); and a final extension at 72°C for 5 minutes. PCR products were analyzed either in 12% polyacrylamide gel or on an ABI310 Genetic Analyzer (Applied Biosystems, Foster City, CA). For the latter method, forward primers had a 5′-end fluorescent label 6Fam, Hex, or Tet (GIBCO® Invitrogen, Carlsbad, CA).
Marker genotypes were checked for Mendelian segregation and analyzed to establish linkage and map order using MultiMap [10] as previously described [11]. A sex-averaged framework map was constructed with LOD 3.0 support starting with the most informative markers (BICD1 and SIAT8A). Further markers were added with LOD 2.0 support (Figure 1). Haplotype analysis of erd pedigrees was performed manually following “the minimal recombination number” rule.
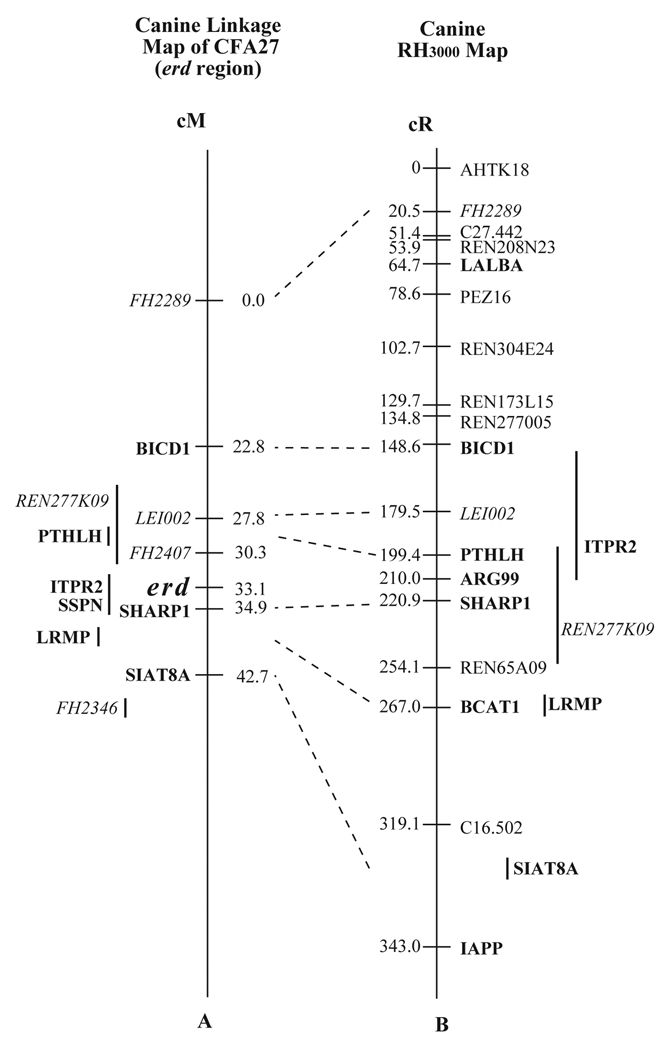
Figure 1. Fine mapping of the erd interval on canine chromosome 27.
A. Meiotic linkage map of the erd interval on canine chromosome 27 (CFA27) established by multipoint linkage analysis. The erd locus lies in a 4.6 cM zero recombination interval between microsatellite FH2407 and a gene specific polymorphism in SHARP1. The gene specific polymorphism in PTHLH co-segregates with FH2407.
B. Radiation hybrid map of the corresponding interval. Marker order is in agreement with the linkage map (A) for markers mapped both ways. The erd interval corresponds to the region between PTHLH and SHARP1, a distance of 1.5 cR3000.
2.4. CFA27 Radiation Hybrid Mapping
To refine marker order on CFA27 and saturate the map with gene-specific markers, an RH Map of CFA27 was built using a commercially available 3000 Rad canine radiation hybrid panel (RH3000) (Research Genetics, Huntsville, Al). The 22 markers selected for RH mapping, comprised 3 microsatellites linked meiotically to erd (FH2289, LEI002, REN277K09), 12 markers selected from the CFA27 RH Map 5000 cR [8], and 7 gene-specific markers (BICD1, ITPR2, ARG99, SHARP1, BCAT1, LRMP, SIAT8A) derived from sequence of gene-specific canine BAC clones or the 3′UTR and intron sequences of the canine genes (Figure 1; Supplementary Table 1A). Markers were amplified and an RH map constructed as described previously [9]. When the canine genome assembly CanFam2 became available, the position of all markers was identified using UCSC Bioinformatics tools (electronic PCR, Blat; <http://genome.ucsc.edu/cgi-bin/hgGateway>).
2.5. Candidate gene selection
Within the final LD region, 15 Refseq genes were identified. Initial candidate gene screening was limited to genes expressed in retina or eye (either mRNA or ESTs deposited in database) and mapped to HSA12p12.1-11.22, which exhibits conserved synteny with CFA27. The most proximal gene (KLHDC5) and the most distal two genes (SSPN and ITPR2) were also excluded from initial screening because of their proximity to the ends of the interval. As none of the remaining 7 genes had previously been implicated in an hereditary retinal degeneration, or otherwise identified as critical for photoreceptor development, all were considered equally viable candidates (Supplementary Tables 1B and 1Ci).
2.6. PCR amplification and sequencing
Primers were designed to amplify coding sequences or genomic fragments within genes under standardized amplification conditions (Tm between 56°C and 63°C). Products were sequenced using the Applied Biosystems Automated 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Sequences were then analyzed and compared using Sequencher® 4.2.2 Software (Gene Codes Corporation, Ann Arbor, MI).
2.7. Population screening for the 4 bp deletion in intron 3
PCR screening of the 4-base deletion was done using three primers: two gene-specific primers flanking the 4 bp polymorphism (Supplementary Table 1Cii, primer pair 7) and a fluorescent-labeled FAM-M13(−21) universal primer: 5′-FAM-TGTAAAACGACGGCCAGT-3′. Unlabeled M13(−21) sequence (18 bp) was fused to the 5′ end of the forward primer. The reaction used: 2 pmol FAM-M13(−21) forward primer; 4 pmol reverse primer; and 1 pmol M13-tailed forward primer, made to a final volume of 11 µl with master mix (Invitrogen standard PCR reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 unit of Taq polymerase and 20 ng of DNA). PCR cycles included an initial denaturing step of 96°C (2 min), 25 cycles of 95°C (30 s), 56°C (45 s), 72°C (30 s), then 10 cycles of 95°C (30s), 53°C (45s), 72°C (30s) with a final extension step of 72°C for 10 minutes. Subsequently, 1 µl of the PCR product was mixed with 0.2 µl of size marker Liz-500 (Applied Biosystems, Foster City, CA) and 9.8 µl of formamide and ran on the ABI 3730×l DNA Analyzer. Fragment size was analyzed using GeneMapper v3.0 (Applied Biosystems, Foster City, CA).
2.8. Population screening, exon 4 SINE insertion
To screen canine populations for this SINE insertion, primers flanking it were designed (Supplementary Table 1Cii, primer pair number 5). Reactions used GoTaq green master mix (Promega, Madison, WI), an initial denaturing step of 94°C (10 min), 30 cycles of 94°C (1 min), 58°C (1 min), 72°C (1.5 min), and a final extension step of 5 minutes at 72°C. Products were visualized on 1.8% agarose gel: a normal dog showed one 625 bp band (homozygous for the normal allele), an affected dog showed one 854 bp band (homozygous for the insertional mutant allele), and a carrier dog had 2 bands representing both alleles, normal and mutant.
2.9. Tissue collection, RNA extraction, cDNA preparation, Northern analysis
Tissue samples were collected as previously described [7]. Total RNA was extracted from frozen normal, carrier and affected tissues at 4.7, 8.7 and 14.1 weeks old using TRIZOL (Invitrogen, Carlsbad, CA) following the manufacturer's protocol, with 1µl of RNase inhibitor (SUPERase-In, Ambion, Austin, TX) added to prevent RNA degradation. RNA was also extracted from a normal 8.7 weeks old dog from kidney, large intestine, small intestine, stomach, cerebellum, occipital cerebral cortex, frontal cerebral cortex, amygdala, hippocampus, heart, skeletal muscle and testis. Testis RNA from an 8.3 weeks affected dog was also extracted. Five to ten micrograms of total RNA in 2µl volume was used to prepare cDNA using Thermoscript RT-PCR kit (Invitrogen, Carlsbad, CA). Extension of cDNA sequence by 5′ and 3′ RACE was obtained through the SMART™RACE cDNA amplification kit (Clontech, Mountain View, CA) following the manufacturer’s protocol (Supplementary Table 1Cii, primers 1 and 2). RNA membranes were prepared as previously described [7]. A probe for northern analysis was generated from normal retinal cDNA using gene-specific primers from the last exon of the gene, exon 14, which shows the weakest homology to STK38 (NDR1) another member of the STK gene family (Supplementary Table 1Cii, primer pair 8). Probe was prepared, labeled, and hybridized to membrane as previously described [7]. Blots were exposed to X-ray film (Kodak, Rochester, NY) at −70°C for 5 days with two intensifying screens (Kodak, Rochester, NY). The same membranes were subsequently hybridized to a canine beta-actin probe as a control for RNA loading, and exposed over night at room temperature.
2.10. In-silico analysis
The predicted canine STK38L sequence was aligned to human, murine, chicken and zebrafish STK38L proteins using MegAlign software (DNASTAR, Lasergene, Madison, WI). Protein structure and prediction of domains used the SWISS-PROT (http://us.expasy.org/sprot) and NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi) databases.
3. RESULTS
3.1. Fine scale mapping of the erd interval
Genotypes were obtained from 17 erd-segregating pedigrees, including 141 animals in informative generations, for 5 previously published canine microsatellite markers [8] and gene-specific microsatellite and SNP markers identified in seven genes: BICD1, PTHLH, ITPR2, SSPN, SHARP1, LRMP, SIAT8A (see material and methods, section 2.2, and Figure 1). Multipoint analysis placed the erd locus and six markers at unique map positions at LOD 2 support, and the most likely location of other markers was assigned (Figure 1). Two-point linkage analysis confirmed the tightest linkage between erd and markers SHARP1 (theta = 0.014; lod = 36.075) and FH2407 (theta = 0.028; lod = 25.355). From two-point linkage and haplotype analyses, markers PTHLH, FH2407, LEI002, and REN277K09 form a cluster with no internal recombinations observed, but sharing one recombination with erd. Similarly, one recombination was detected between erd and SHARP1. The position of the erd locus, its final absolute linkage disequilibrium (LD) interval, was thus refined to the 4.6 cM interval flanked by genes PTHLH and SHARP1, corresponding to the 1.6 Mb CanFam2 region of CFA27 between nucleotides 22,803,789 and 24,476,056.
The order of markers on CFA27 established by RH mapping was consistent with those by linkage mapping (Figure 1) and in CanFam2 (Supplementary Table 1). Gene order in this RH3000 Map was also highly conserved compared to homologous regions of the human GB4 map and genome sequence assembly.
3.2. Evaluation of Positional Candidate genes
Of the 15 Refseq genes in the final LD interval for erd (Figure 2A), the coding regions of 7 were amplified by RT-PCR and sequenced. MRPS35 (accession # GU811152), PPFIBP1 (GU811154; exons 1 – 7 only), MED21 (GU811151), TM7SF3 (HM013658), FGFR1OP2 (GU811150), and C12orf11 (GU811149), showed no size differences between products from affected and normal samples, and no significant sequence differences between cases and controls. Several polymorphisms were identified that were not associated with erd disease status (see accession deposits for details). Significant differences in size and sequence were detected for STK38L (GU811153).
Figure 2. Schematic illustration of the erd interval on canine chromosome 27.
A. The 1.6 Mb zero recombination interval is drawn to scale and includes 15 Refseq genes between PTHLH and SHARP1. Their position on the UCSC CanFam2 dog assembly is indicated by nucleotide numbers, and their annotation by black arrows.
B. Canine STK38L comprises 14 exons. Filled boxes represent coding regions and empty boxes the 5′ and 3′ untranslated regions. The sizes of the exons and introns are marked above and below the boxes, respectively. Primer pairs 1 and 2, represented by black arrows, were used to amplify the complete coding region from normal and affected dogs, while primer pair 5 was used to characterize the genomic region flanking exon 4.
C. Structure of the inserted SINE. The nucleotide sequence of the tRNA-like SINE is depicted in bold upper case letters. Exon 4 sequence observed in non-affected dogs is indicated in unbold upper case letters. The 15 nucleotide sequence representing the direct duplication at the insertion site is boxed. Introns 3 and 4 are indicated in lower case letters. Underlined is the polymorphic 4 nucleotide sequence (agta) site deleted in erd-affected dogs and in a small number of normal dogs from other breeds.
3.3. Mutation identification in STK38L
3.3.1 Canine STK38L mRNA
BLAT alignment of the human STK38L mRNA (NM015000.3) -- comprising 14 exons, 13 of which code for a 464 amino acid protein -- against the CanFam2 genomic sequence, identified 14 predicted canine exons, with the first methionine located in the second exon (Figure 2B), a structure shared with its human and murine orthologs. The predicted canine STK38L/NDR2 gene and mRNA share about 78% identity with the predicted canine STK38 paralogous gene (also known as NDR1, on CFA12). Two primer pairs (Supplementary Table 1Ci, primers 1–2) specific for STK38L successfully amplified the complete coding region in 2 overlapping fragments from retinal cDNA of a 10.4 weeks old normal dog (accession # GU811153, Supplementary Figure 1A1). By 5′ RACE, (Supplementary Table 1Cii, primer 1) the cDNA sequence was extended 48 bases upstream to the first ATG located in exon 2. This result is similar to that in the homologous mouse sequence AY223819 [4]. Because the 5′ end of the sequence is open, further upstream extension was considered possible. However, in-silico analysis of the 5′ end of canine STK38L identified several transcription elements and signals that support the assumption that this is the end of the gene: three SP1 elements (gggcgg) are located upstream to the first ATG at positions −110 bp, −111 bp and −288 bp in exon 1. One CRE element is found in position −228, and a TATA box is present in position −987 bp.
3′ RACE using primer 2 in Supplement Table 1Cii resulted in two products: one extended 265 nt beyond the stop codon in exon 14, and included a polyadenylation signal 28 bases preceding the poly A tail. The second extended 921 nt after the stop and included a polyadenylation signal 25 bases preceding the poly A tail (Supplementary Figure 1A1).
3.3.2 Evaluation of STK38L in erd-affected dogs
Comparison of RT-PCR products from erd-affected, carrier and normal retina (9.6, 9.1 and 6.9 weeks old, respectively) showed no sizes differences when amplifying from exon 6 to 14 (Supplementary Table 1Ci, primer pair 2). However, when amplifying from exon 1 to 7 (Supplementary Table 1Ci, primer pair 1), the product size varied with erd genotype. The affected dog yielded an amplimer 123 nt shorter than that of the homozygous normal dog, and the heterozygote yielded both amplimers (Figure 3A). Sequence analysis revealed that exon 4 was absent from the affected mRNA. No other differences were observed between affected and normal dogs (Supplementary Figure 1B1).
Figure 3. A SINE insertion in STK38L cosegregates with disease in erd-affected dogs.
A. Gel electrophoresis of the complete coding sequence of canine STK38L, amplified in two overlapping fragments (5'UTR-exon 7; exon 6 - 3'UTR) from normal (N), carrier (C), and affected (A) dogs. Primers located on the 5′ UTR and exon 7 amplify a 661 bp amplicon from a normal dog, a 538 bp amplicon from an affected dog, and both bands from a heterozygous dog. No size differences among normal, affected and heterozygous products were observed for the amplification product including exons 6 to 14 (exon 6-3′ UTR).
B. Segregation of the SINE insertion in a 3-generation erd-informative pedigree. The larger band (854 bp) represents the affected amplicon which includes the SINE insertion; the 625 bp band represents the normal amplicon. All affected dogs (1, 2, 4, 6, 8, 10) yield only the 854 bp band; the homozygous normal grandsire (12) yields only the 625 bp band; and the heterozygous dogs (3, 5, 7, 9, 11) yield both bands (and fainter heterodimer bands that run slowly). There is complete cosegregation between the SINE insertion and the disease.
To determine why exon 4 was absent, genomic donor-acceptor sites in introns 3 and 4 were sequenced from erd-affected, carrier and normal dogs (Supplementary Table 1Cii, primer pairs 3 to 6). Primer pair 5 amplified the expected 625 bp long product from the normal dog but an 854 bp product from affecteds, and both products from carriers. Sequence analysis showed a 4 nt deletion in intron 3 (−68 to −72) and insertion of a tRNA-like short interspersed repeat (SINE) in exon 4 including a 15 nt duplication (Figure 2C). The 854 bp amplicon cosegregated with erd (Figure 3B). Sequence analysis of 8 affected and 3 carrier colony dogs from generations closer to the proband showed concurrence of the SINE insertion and the 4 nt deletion, suggesting that neither mutation arose within the colony.
3.3.3. Population Screening for the 4 nt deletion and the SINE insertion in STK38L
To establish the disease relevance of the 4 nt deletion in intron 3 and the SINE insertion in exon 4, we first screened a set of 82 purebred dogs from 16 different breeds that did not include Norwegian elkhounds for the 4 nt deletion. Amplified genomic DNA flanking the 4 nt deletion site established that 74 of these dogs were homozygous for the wild type allele (no deletion, 279 bp product), 6 were heterozygous for the deletion (yielding a 275 bp and a 279 bp product, 3 American Eskimos, 2 Border Collies and one American cocker spaniel) and 2 dogs were homozygous for the 4 nt deletion (one American cocker spaniel and one Border collie, Supplementary Table 2A). The latter two dogs were not affected with erd or any other form of retinal degeneration, and their amplicons were sequenced and confirmed to have the same 4 nt deletion. All 8 dogs with the 4 nt deletion were screened for the SINE insertion by amplification of genomic DNA flanking the insertion. No SINE insertion in exon 4 was found in these 8 dogs. These results suggest that the 4 nt deletion in intron 3 represents the minor allele of a polymorphism that does not cause erd, and that the erd mutation occurred on a chromosome bearing this minor allele.
A further set of control dogs (141 dogs from 24 breeds) were screened for the SINE insertion and all were homozygous wild type (Supplementary Table 2B).
Seven Norwegian elkhound dogs segregating a retinal degeneration clinically distingushable from erd were also tested for both the SINE insertion and the 4 nt deletion; all were homozygous wildtype by both tests, confirming that this breed has more than one form of inherited retinal degeneration segregating.
3.4. RNA expression analysis
Northern analysis using an STK38L specific 3′ end probe (see material and methods, section 2.9) identified 2 transcripts (4.4 Kb and 5.1 Kb) in retina from normal and carrier dogs at ages ranging from 4 to 14 weeks old (Figure 4A). In the 4.3 weeks old affected retina (Figure 4A, lane 3) a different pattern was observed, composed of 4 “shadow-like” bands, but this pattern was not clearly seen at other time points in the affected retinas.
Figure 4. Northern analysis of canine STK38L expression in retina and testis.
A. STK38L expression in normal [N], carrier [C] and affected [A] canine retinas at 3 time points [lanes 1–3 ~4.5 weeks, lanes 4–6 ~8.5 weeks; lanes 7–9 ~14 weeks]; and in normal [N] and affected [A] testis at ~ 8.5 weeks old [lanes 10–11]. Expression of β-actin is shown for evaluation of loading among lanes. Two STK38L transcripts (~4.4 Kb and ~5.1 Kb) are observed in normal retina and testis. The ~4.5 week old affected retina (Lane 3) demonstrates a different pattern with 4 “shadow-like” bands, but this pattern was not observed in affected retina at later ages (Lanes 6 and 9).
B. STK38L expression profile in Kidney (K, lane 1), Large intestine (LI, lane 2), small intestine (SI, lane 3), stomach (St, lane 4), cerebellum (CBL, lane 5), occipital cerebral cortex (OCC, lane 6), frontal cerebral cortex (FCC, lane 7), amygdala (A, lane 8), hippocampus (Hip, lane 9), heart (H, lane 10), and skeletal muscle (SM, lane 11) from an 8.7 weeks old normal dog. Expression of β-actin is shown for evaluation of loading among lanes. STK38L is expressed strongly in heart (Lane 10), at much lower levels in kidney, large intestine, small intestine, stomach and brain (Lanes 1–9) and is not detected in skeletal muscle (Lane 11).
Both transcripts were present in testis from a normal and affected dog (Figure 4A, lanes 10–11, and normal heart (Figure 4B, lane 10). Very low expression levels were observed in kidney, large intestine, small intestine, stomach and brain, and none was detected in skeletal muscle (Figure 4B). The 2 transcripts differed by about 600–700 bp, which corresponds to the 655 bp difference between the 2 polyadenylation products identified by 3′ RACE. Thus the 2 bands are presumed to represent polyadenylation variants, and they appear to be co-expressed at similar levels.
3.5. In-silico analysis
The putative canine STK38L protein, like the human, is 464 amino acids long (Supplementary Figure 1A2, Supplementary Figure 2). In the N terminus an S100B binding region is identifiable by similarity (swissprot in-silico analysis). This region extends from amino acid 63 to 88. A protein kinase domain extends from amino acid 90 to 383. Three post-translational modification residues are in position 75 (threonine), 282 (serine) and 442 (threonine).
Comparison of the putative canine STK38L sequence to that of 4 other species (human, mouse, chicken and zebrafish) revealed the strong conservation among these species by the high percentage of amino acid identity. Closest to the canine sequence was human (99.1%) followed by mouse (97.4%), zebrafish (90.3%) and chicken (89.8%) (Figure 5A, Supplementary Figure 2). In erd-affected dogs, 41 amino acids are missing from the putative protein. These 41 amino acids include the complete S100B binding region (26 amino acids, that includes threonine 75), and part of the protein kinase domain (15 amino acids, Figure 5B).
Figure 5. Conservation analysis of STK38L.
A. Pairwise percent nucleotide identities (upper right cells) and divergence (lower left cells) calculated for 5 species (D = dog, H = human, M = mouse, Z = zebrafish, C = chicken) for the STK38L complete coding sequence. The human and canine sequences are the most similar to each other, being 99.1% identical. Sequence distances were calculated using MegAlign 6.1 software (Lasergene, DNAstar, Inc.).
B. Multiple sequence alignment of the 5′ end (first 120 codons) of the canine STK38L putative protein sequence (labeled D) with that of human (H), mouse (M), zebrafish (Z) and chicken (C). The consensus sequence for the 5 species is listed above each set (1..60; 61…120) of aligned sequences. Differences among species are bold and boxed. In the consensus sequence for codons 61–120 (i.e. the lower panel), the 41 amino acids (KKL…FGE) missing from the putative protein in erd-affected dogs is boxed and shaded. Within this region, the S100B domain (KKL…GLD) is indicated in bold. The autophosphorylation residue at position 75 is labeled with an asterisk and boxed. The protein kinase domain (FES…) starts at position 90. See Supplementary Figure 2 for the corresponding analysis of the complete protein. Alignment was done using MegAlign 6.1 software (Lasergene, DNAstar, Inc.).
4. DISCUSSION
In this study, fine mapping followed by positional candidate gene analysis established that erd is strongly associated with an STK38L/NDR2 allele characterized by a 4 nt deletion in intron 3 and a SINE insertion in exon 4. The deletion was also found as a minor allele (without the SINE insertion) in canine populations not segregating erd. The SINE insertion in exon 4 was only seen in erd affected or heterozygous dogs -- this mutation presumably took place originally on a CFA27 chromosome bearing the 4 nt deletion in intron 3. All erd-affected and -heterozygous dogs then inherited this allele identical by descent.
The SINE insertion identified in the STK38L gene belongs to the family of SINEC_Cf repeats that comprise a major subclass of canine-specific SINEs [13; 14]. SINEC_Cf are likely derived from tRNA and contain internal control elements for transcription by RNA polymerase III from a tRNA [15, 16]. Nearly half of all annotated genes in CanFam2 contain at least one SINEC_Cf. This element remains so active in the canine genome that of 87,000 young SINEC_Cf elements in CanFam2 nearly 8% are heterozygous [13]. Moreover, comparison of CanFam2 with the previous survey sequence of a standard poodle genome CanFamSS [17] estimate that 20,000 or more of these insertion sites are bimorphic, i.e. not fixed at homozygosity [13; 14]. SINE insertions in exonic regions or intron/exon boundaries have previously been implicated in two other canine diseases -- narcolepsy [18] and centronuclear myopathy [19] -- and in the merle coat color phenotype [20].
The erd SINE insertion results in the absence of exon 4 from the mature STK38L mRNA, presumably by splicing out during maturation of the mRNA, even though the insertion does not appear to introduce or remove any obvious splicing sites. Furthermore, the mutant transcript appears to be stable in both homozygous and heterozygous retinas, not being degraded by nonsense mediated decay or other mechanisms. The translated protein, however, would clearly be dysfunctional, lacking its S100B binding region, part of its Protein Kinase domain (15 amino acids) and the Thr-75 residue critical for autophosphorylation (Figure 5B, and Supplementary Figures 1B2 and 2). Both STK38L/NDR2 and its paralog NDR1 are regulated by binding of S100B, a Ca++-binding protein, to their S100B binding domains [3] and via binding to their N-termini by at least two different human Mob proteins [6, 21]. Binding of STK38L by S100B and or Mob proteins is proposed to induce a conformational change that enables STK38L to autophosphorylate at Thr-75, Ser-282, and Thr-444 [5, 6]. It is thus reasonable to propose a causative role for this STK38L mutation in erd.
Furthermore, the erd disease phenotype itself suggests a compatibility between the underlying disease mechanism and proposed roles for the STK38L/NDR2 pathway in neurons. In the erd-affected retina, photoreceptor development is not arrested, but aberrant [1]. Because photoreceptor differentiation occurs postnatally in dogs [22], unlike in humans, the abnormal development of the erd-affected retina becomes evident in dogs between 3 and 10 weeks old. The subsequent degeneration of the erd-affected retina is initially rapid, but becomes more gradual in dogs over 6 months old [1]. Morphologically, rod and cone inner- and outer-segments do develop, and their numbers, sizes and proportions on average are initially normal [1]. The primary morphological abnormality during this period of aberrant development is dyscoordination of the length and proportions of inner and outer segments among adjacent rods. This dyscoordination prevents the alignment that normally creates distinct inner and outer segment layers. This alignment starts at around 4 weeks postnatal in normal canine retina, when developing inner and outer segments first become closely packed together, and is essentially complete across the retina by 7–8 wks postnatal [1, 22]. In the same period in erd-affected retina, unusual and prominent villiform processes extend from the inner segments of rods and cones, and photoreceptor synapses fail to develop properly, morphologically and functionally. Subsequent to these abnormalities of development, at about the 10th week postnatal, rods and cones degenerate, rapidly at first and later more gradually [1]. The primary pathology in erd thus represents dysregulation of photoreceptor growth. Loss of STK38L function makes an appealing explanation for this abnormality.
This is the first time that the STK38L/NDR2 pathway has been implicated in photoreceptor development or disease. With hindsight, however, it is reasonable to expect such a role, given the importance of the polarized, highly regulated, circadian growth and renewal cycle in photoreceptors, and the proposed roles for STK38L in other, especially neuronal, tissues. STK38L and its interacting partners, S100B and the Mob proteins, thus provide novel candidate genes for hereditary retinal disorders of humans and other species.
Supplementary Material
The canine STK38L gene: sequence of the mRNA and its predicted protein for both the normal and erd-mutant allele.
A1. Canine STK38L mRNA sequence, normal allele. Upper case indicates coding sequence; the first methionine and the stop codon are boxed; exon 4 (which is missing from the erd-mutant allele) is in bold; the polyadenylation signals are in underlined italic; # indicates the alternative poly A tail location.
A2. Canine STK38L predicted protein, normal allele. In bold are the 41 amino acids missing from the predicted protein in the erd-mutant allele.
B1. Sequence of the coding region of STK38L mRNA in erd-affected dogs. Boxed are the first methionine and the stop codon. ◊ indicates the location of exon 4 missing in the affecteds.
B2. Canine STK38L, predicted erd-mutant protein. ◊ indicates the location of the 41 amino acids missing in affected dogs.
Canine STK38L putative protein sequence conservation analysis. The putative STK38L canine protein sequence was aligned with human, mouse, zebrafish and chicken proteins. The S100B domain (KKL…GLD) is highlighted in bold letters. Three autophosphorylation residues are each labeled with an asterisk. Boxed are the 41 amino acids missing from the putative protein in erd-affected dogs. Amino acids that differ from dog protein are boxed. The protein kinase domain starts at position 90 and ends at position 383.. Alignment was undertaken using MegAlign 6.1 software (Lasergene, DNAstar, Inc.).
Primer pair sequences used in this study.
A. Gene associated markers used for Linkage and RH mapping of the erd interval on canine chromosome 27.
B. Primer sequences used to screen the coding regions of candidate genes within the final LD region.
C. STK38L screening primers: primers sequences used to screen the STK38L coding region, and to further characterize the gene and mutation.
Population screening for the 4 bases deletion in intron 3, and the SINE insertion in exon 4 of canine STK38L, that both cosegregate with disease in erd-informative pedigrees. Identification of dogs and breeds evaluated.
Acknowledgments
Technical assistance from Jennifer Johnson, Julie Jordan, Amanda Nickle, and the animal care staff at the RDSF is gratefully acknowledged. Ewen Kirkness (The Institute for Genome Research) kindly provided prepublication information for several canine genomic sequences. Supported by NIH Grants EY006855, EY13132, and EY17549; The Foundation Fighting Blindness; the Morris Animal Foundation; and the Van Sloun Fund for Canine Genetic Research.
Footnotes
Disclosure: Orly Goldstein, None; A.V. Kukekova, None; G. D. Aguirre, None; G. M. Acland, None.
References
- 1.Acland GM, Aguirre GD. Retinal degenerations in the dog: IV. Early retinal degeneration (erd) in Norwegian elkhounds. Exp. Eye Res. 1987;44:491–521. doi: 10.1016/s0014-4835(87)80160-4. [DOI] [PubMed] [Google Scholar]
- 2.Acland GM, Ray K, Mellersh CS, Langston AA, Rine J, Ostrander EA, Aguirre GD. A novel retinal degeneration locus identified by linkage and comparative mapping of canine early retinal degeneration. Genomics. 1999;59:134–142. doi: 10.1006/geno.1999.5842. [DOI] [PubMed] [Google Scholar]
- 3.Tamaskovic R, Bichsel SJ, Rogniaux H, Stegert MR, Hemmings BA. Mechanism of Ca2+mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J Biol Chem. 2003;278:6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- 4.Stork O, Zhdanov A, Kudersky A, Yoshikawa T, Obata K, Pape HC. Neuronal functions of the novel serine/threonine kinase Ndr2. J Biol Chem. 2004;279:45773–45781. doi: 10.1074/jbc.M403552200. [DOI] [PubMed] [Google Scholar]
- 5.Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J Biol Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein O, Zangerl B, Pearce-Kelling S, Sidjanin DJ, Kijas JW, Felix J, Acland GM, Aguirre GD. Linkage disequilibrium mapping in the domestic dog breeds narrows the progressive rod-cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics. 2006;88:541–550. doi: 10.1016/j.ygeno.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, Parker HG, et al. A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci U S A. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukekova AV, Aguirre GD, Acland GM. Cloning and characterization of canine SHARP1 and its evaluation as a positional candidate for canine early degeneration (erd) Gene. 2003;312:335–343. doi: 10.1016/s0378-1119(03)00630-9. [DOI] [PubMed] [Google Scholar]
- 10.Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 11.Kukekova AV, Goldstein O, Johnson JL, Richardson MA, Pearce-Kelling SE, Swaroop A, Friedman JS, Aguirre GD, Acland GM. Canine RD3 mutation establishes rod cone dysplasia type 2 (rcd2) as ortholog of human and murine rd3. Mammalian Genome. 2009;20:109–123. doi: 10.1007/s00335-008-9163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider I, Houston DW, Rebagliati MR, Slusarski DC. Calcium fluxes in dorsal forerunner cells antagonize beta-catenin and alter left-right patterning. Development. 2008;135:75–84. doi: 10.1242/dev.004713. [DOI] [PubMed] [Google Scholar]
- 13.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Kirkness EF. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minnick MF, Stillwell LC, Heineman JM, Stiegler GL. A highly repetitive DNA sequence possibly unique to canids. Gene. 1992;110:235–238. doi: 10.1016/0378-1119(92)90654-8. [DOI] [PubMed] [Google Scholar]
- 16.Bentolila S, Bach JM, Kessler JL, Bordelais I, Cruaud C, Weissenbach J, Panthier JJ. Analysis of major repetitive DNA sequences in the dog (Canis familiaris) genome. Mammalian Genome. 1999;10:699–705. doi: 10.1007/s003359901074. [DOI] [PubMed] [Google Scholar]
- 17.Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM, Venter JC. The dog genome: Survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- 19.Pele M, Tiret L, Kessler J-L, Blot S, Panthier J-J. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14:1417–1427. doi: 10.1093/hmg/ddi151. [DOI] [PubMed] [Google Scholar]
- 20.Clark LA, Wahl JM, Rees CA, Murphy KE. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA. 2005;103:1376–1381. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J Biol Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- 22.Aguirre GD, Rubin LF, Bistner SI. Development of the Canine Eye. Amer. J. Vet. Res. 1972;33:2399. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The canine STK38L gene: sequence of the mRNA and its predicted protein for both the normal and erd-mutant allele.
A1. Canine STK38L mRNA sequence, normal allele. Upper case indicates coding sequence; the first methionine and the stop codon are boxed; exon 4 (which is missing from the erd-mutant allele) is in bold; the polyadenylation signals are in underlined italic; # indicates the alternative poly A tail location.
A2. Canine STK38L predicted protein, normal allele. In bold are the 41 amino acids missing from the predicted protein in the erd-mutant allele.
B1. Sequence of the coding region of STK38L mRNA in erd-affected dogs. Boxed are the first methionine and the stop codon. ◊ indicates the location of exon 4 missing in the affecteds.
B2. Canine STK38L, predicted erd-mutant protein. ◊ indicates the location of the 41 amino acids missing in affected dogs.
Canine STK38L putative protein sequence conservation analysis. The putative STK38L canine protein sequence was aligned with human, mouse, zebrafish and chicken proteins. The S100B domain (KKL…GLD) is highlighted in bold letters. Three autophosphorylation residues are each labeled with an asterisk. Boxed are the 41 amino acids missing from the putative protein in erd-affected dogs. Amino acids that differ from dog protein are boxed. The protein kinase domain starts at position 90 and ends at position 383.. Alignment was undertaken using MegAlign 6.1 software (Lasergene, DNAstar, Inc.).
Primer pair sequences used in this study.
A. Gene associated markers used for Linkage and RH mapping of the erd interval on canine chromosome 27.
B. Primer sequences used to screen the coding regions of candidate genes within the final LD region.
C. STK38L screening primers: primers sequences used to screen the STK38L coding region, and to further characterize the gene and mutation.
Population screening for the 4 bases deletion in intron 3, and the SINE insertion in exon 4 of canine STK38L, that both cosegregate with disease in erd-informative pedigrees. Identification of dogs and breeds evaluated.