Abstract
THI6 is a bifunctional enzyme found in the thiamin biosynthetic pathway in eukaryotes. The N-terminal domain of THI6 catalyzes the ligation of the thiamin thiazole and pyrimidine moieties to form thiamin phosphate and the C-terminal domain catalyzes the phosphorylation of 4-methyl-5-hydroxyethylthiazole in a salvage pathway. In prokaryotes, thiamin phosphate synthase and 4-methyl-5-hydroxyethylthiazole kinase are separate gene products. Here we report the first crystal structure of a eukaryotic THI6 along with several complexes that characterize the active sites responsible for the two chemical reactions. THI6 from Candida glabrata is a homohexamer in which the six protomers form a cage-like structure. Each protomer is composed of two domains, which are structurally homologous to their monofunctional bacterial counterparts. Two loop regions not found in the bacterial enzymes provide interactions between the two domains. The structures of different protein-ligand complexes define the thiazole and ATP binding sites of the 4-methyl-5-hydroxyethylthiazole kinase domain, and the thiazole phosphate and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate binding sites of the thiamin phosphate synthase domain. Our structural studies reveal that the active sites of the two domains are 40 Å apart and are not connected by an obvious channel. Biochemical studies show 4-methyl-5-hydroxyethylthiazole phosphate is a substrate for THI6; however, adenosine diphospho-5-β-ethyl-4-methylthiazole-2-carboxylic acid, the product of THI4, is not a substrate for THI6. This suggests that unidentified enzyme is necessary to produce the substrate for THI6 from the THI4 product.
Thiamin (vitamin B1) is an essential component of all living systems. The active form of thiamin, thiamin pyrophosphate (ThDP), acts as a cofactor for several important enzymes in carbohydrate and amino acid metabolism. The mechanistic role of the cofactor is stabilization of an acyl carbanion intermediate (1). Thiamin biosynthetic pathways are found in prokaryotes and some eukaryotes (yeast, fungi and plants); however, vertebrates cannot synthesize thiamin. Therefore, thiamin is an essential component of the human diet with a daily requirement of 1.0–1.2 mg (2). Deficiency of thiamin leads to diseases such as beriberi and Wernicke-Korsakoff syndrome (3, 4). The absence of thiamin biosynthetic enzymes in mammals makes them potential targets for new antimicrobial and antifungal drug design.
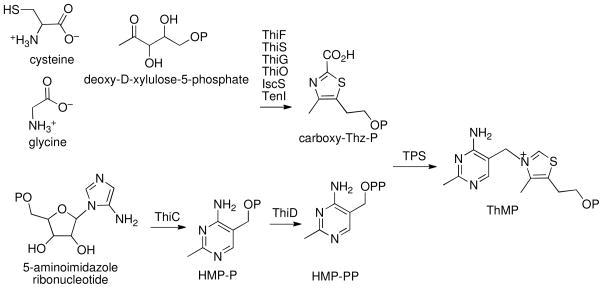
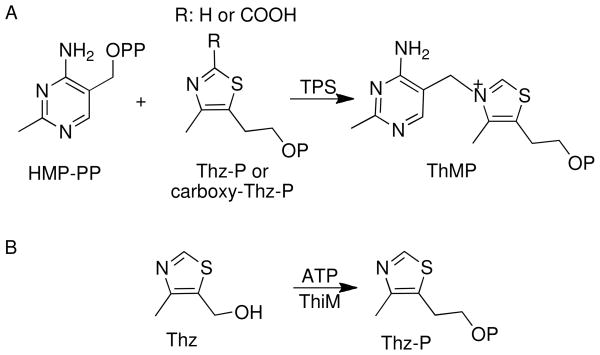
Thiamin is composed of a thiazole and a pyrimidine moiety, which are synthesized separately and joined to form thiamin phosphate (ThMP). Prokaryotes and eukaryotes use different strategies for the production of thiamin. The details of thiamin biosynthesis are well characterized both structurally and mechanistically in prokaryotes (Scheme 1) (5–7). Thiazole formation in bacteria requires six gene products and involves the oxidative condensation of 1-deoxy-D-xylulose-5-phosphate, glycine (in Bacillus subtilis) or tyrosine (in Escherichia coli) and cysteine (8, 9). Formation of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) occurs by a complex rearrangement of 5-aminoimidazole ribonucleotide catalyzed by ThiC, a member of the radical SAM superfamily of enzymes (10, 11). HMP-P is further phosphorylated by ThiD to form 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate (HMP-PP) (12). The thiazole and pyrimidine modules are then coupled by the enzyme thiamine phosphate synthase (TPS) to form ThMP (Scheme 2A) (13–15), which is finally phosphorylated to ThDP by the enzyme ThiL (16, 17).
Scheme 1.
Scheme 2.
In addition to the biosynthetic pathway, many organisms contain salvage pathways. The enzyme 4-methyl-5-hydroxyethylthiazole kinase (ThiM) catalyzes the adenosine 5′-triphosphate ATP-dependent phosphorylation of 4-methyl-5-hydroxyethylthiazole (Thz) to form 4-methyl-5-hydroxyethylthiazole phosphate (Thz-P) (Scheme 2B) (18). ThiD is responsible for the phosphorylation of 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) (12).
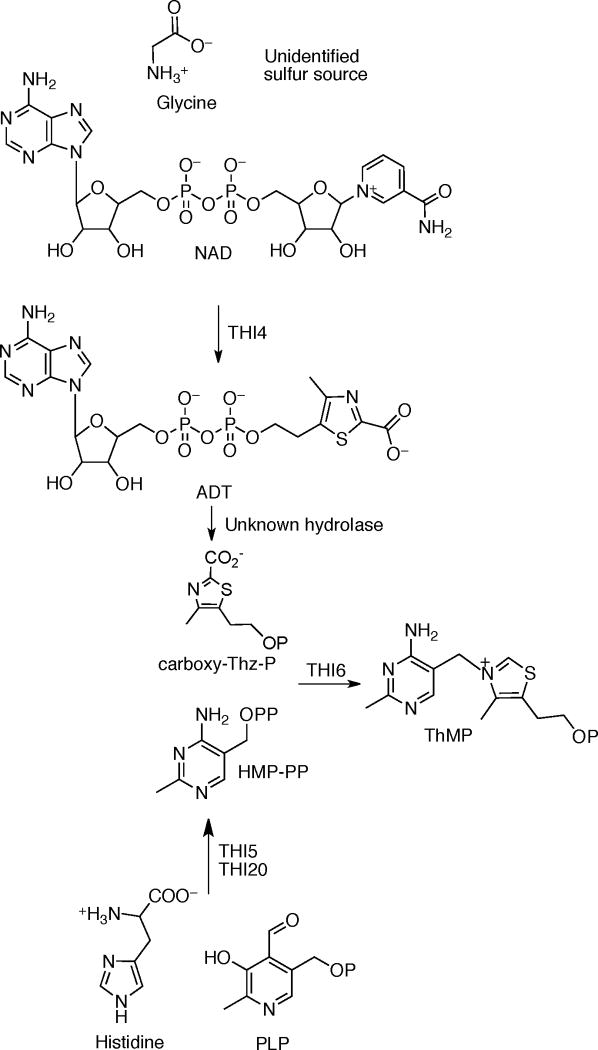
In contrast to the prokaryotic system, the mechanistic and structural understanding of thiamin biosynthesis is still at an early stage in the eukaryotes (Scheme 3). Currently, only four enzymes are known to catalyze the biosynthesis of ThMP. THI4 converts nicotinamide adenine dinucleotide, glycine and a yet unknown sulfur source to form the adenosine diphospho-5-β-ethyl-4-methyl-thiazole-2-carboxylic acid (ADT) (19, 20). THI5 is responsible for HMP-P formation; however, the mechanistic and structural details of this process remain unknown (21). THI20 catalyzes the phosphorylation of HMP-P (22). The coupling of Thz-P and HMP-PP in eukaryotes is catalyzed by THI6 (23). This bifunctional enzyme contains a TPS domain and a ThiM domain. Earlier studies on THI6 from Saccharomyces cerevisiae showed that the N-terminal domain is responsible for the TPS activity and the C-terminal domain is responsible for ThiM activity (24). The recent identification of the product of the S. cerevisiae thiazole synthase as ADT (19, 20) suggests that Thz-P utilization is a salvage reaction and that carboxy-Thz-P is the biosynthetic substrate.
Scheme 3.
Here we report the crystal structures of unliganded Candida glabrata THI6 (CgTHI6) and of several complexes that map out the two active sites. These include ThMP; ThMP and pyrophosphate (PP); Thz and β,γ-methylene adenosine 5′-diphosphate (AMP-PCP); ThMP and AMP-PCP and 4-amino-5-hydroxymethyl-2-trifluoromethylpyrimidine pyrophosphate (CF3HMP-PP) and carboxy-Thz-P. The THI6 structure reveals a cage-like homohexamer in which the two active sites are separated by about 40 Å. The two active sites are not connected by an obvious channel. We also report biochemical studies that show that the TPS domain accepts Thz-P as a substrate, but not ADT, the product of THI4.
MATERIALS AND METHODS
Chemical Reagents
Kanamycin, ThMP, sodium pyrophosphate, Thz, AMP-PCP, ATP, magnesium chloride and the buffer components were all purchased from Sigma-Aldrich. Ni-NTA resin was obtained from Qiagen (Valencia, CA). HPLC grade solvents were obtained from Fisher Scientific. Thz-P and HMP-PP for the activity assays were synthesized as previously described (25). TEV protease was provided by Cornell Protein Production Facility. ADT was prepared as previously described (20). Carboxy-Thz-P was prepared as previously described (26).
Expression and Purification of CgTHI6
The CgTHI6 gene was cloned into a modified pET28 overexpression vector encoding an N-terminal hexahistidine tag with a TEV protease cleavage site. The plasmid CgTHI6.XF1 was transformed into Escherichia coli cell line BL21(DE3). A 10 mL overnight culture, grown in Luria-Bertani media at 37 °C supplemented with 100 μg/mL ampicillin, was used to start a 1 L culture. Growth at 37 °C was allowed until the OD600 reached 0.5 at which point the temperature was reduced to 15 °C. After 45 min of cooling, the culture was induced with 0.1 mM isopropyl-1-β-D thiogalactopyranoside and allowed to grow overnight at 15 °C. Cells were harvested by centrifugation at 9000 g for 15 min at 4 °C using an Avanti J-251 centrifuge (Beckman) and frozen at −20 °C for later use.
The purification was carried out at 4 °C. The frozen cell pellet was thawed and resuspended in 50 mL of lysis buffer [50 mM Tris (pH 8.0), 300 mM NaCl, 10 mM imidazole and 3 mM β-mercaptoethanol (BME)] and lysed by sonication. Cellular debris was removed by centrifugation at 35000 g for 50 min at 4 °C. The clarified lysate was loaded onto a Ni-NTA column (Qiagen) pre-equilibriated with 50 mL of the lysis buffer. The column was then washed with 100 mL of wash buffer [50 mM Tris (pH 8.0), 300 mM NaCl, 60 mM imidazole and 3 mM BME] to remove the nonspecific binding proteins. CgTHI6 was eluted with elution buffer [50 mM Tris (pH 8.0), 300 mM NaCl, 250 mM imidazole and 3 mM BME]. The elution was monitored by measuring protein concentration via Coomassie Plus Protein Assay Reagent (Pierce).
To remove the N-terminal hexahistidine tag, the eluted protein was incubated with TEV protease, in dialysis tubing (Pierce) with a 10 kDa molecular weight cutoff (MWCO) (Millipore) for 12 h at 4 °C in a buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol (DTT). The proteolysis was monitored by SDS-PAGE analysis. After complete proteolysis, the reaction mixture was reloaded onto a Ni-NTA column and the flow through was collected. The non-binding material was buffer exchanged into 10 mM Tris (pH 7.5), 50 mM NaCl. The protein was then concentrated to 8 mg/mL using a 30 kDa MCWO Amicon Ultracentrifugal filter (Millipore) and stored at −80 °C for future use. Protein concentration was determined by the Bradford method (27) with bovine serum albumin as standard. The purity of the protein was determined by Coomassie-stained SDS-PAGE analysis and was found to be greater that 95% (data not shown).
Crystallization of CgTHI6 in Unliganded and Complex Forms
The crystallization experiments were conducted at 22 °C using the hanging-drop vapor diffusion method by combining 1 μL of protein solution and 1 μL of reservoir solution. Initial crystallization conditions of unliganded CgTHI6 were determined using sparse matrix screens, Crystal Screen 1 and 2 (Hampton Research). The final optimized condition contained 0.1 M HEPES (pH 7.5), 0.2–0.25 M MgCl2 and 25%–32% polyethylene glycol 400 (PEG400). Rod shaped crystals, 200–300 μm long, appeared in 2–3 days.
Crystals of the THI6/ThMP complex were obtained by cocrystallization under similar conditions. The enzyme was incubated with 2.5 mM of ThMP at 4 °C for 1 h prior to crystallization. The crystals grew after 2–3 days to the maximum size of 400 μm.
Additional complexes were prepared by soaking CgTHI6 crystals with the appropriate ligands. The THI6/ThMP/PP complex was prepared by transferring CgTHI6 crystals into a solution containing the mother liquor with 5 mM ThMP and 5 mM sodium pyrophosphate and soaking for 1 h at 22 °C. The THI6/Thz/AMP-PCP was obtained by soaking CgTHI6 crystals in a solution containing the mother liquor with 20 mM Thz and 20 mM AMP-PCP for 12 h at 22 °C. The THI6/ThMP/AMP-PCP complex was obtained by soaking the CgTHI6 crystals in mother liquor solution containing 20 mM each of ThMP, Thz and AMP-PCP for 2 h at 22 °C. However Thz was not bound in the active site. The THI6/CF3HMP-PP/carboxy-Thz-P complex was obtained by soaking CgTHI6 crystals in a solution containing 10 mM each of the ligands for 1 h at 22 °C.
The high concentration of PEG400 in the mother liquor acted as a cryo-protectant. Crystals were flash frozen directly from the drop by plunging them into liquid nitrogen.
Data Collection and Processing
All data were collected at the NE-CAT beamlines 24-ID-E or 24 -ID-C at the Advanced Photon Source at Argonne National Laboratory using a Quantum 315 X-ray detector (Area Detector Systems Corporation). Crystals of CgTHI6 initially diffracted to ~ 3 Å but were very sensitive to radiation damage. As a result, a highly attenuated beam with 10 sec exposure time, 1° oscillation and 400 mm of crystal to detector distance were used to record the diffraction images. To obtain a complete dataset, multiple segments of data were collected by exposing the same rod-shaped crystal at different spots and merged together. The data were indexed, integrated and scaled using the HKL2000 program suite (28). The data collection and processing statistics are shown in Table 1.
Table 1.
Data collection statistics. Values for the highest resolution shell are given in parentheses.
| THI6 | THI6/ThMP | THI6/ThMP–PP | THI6/Thz/AMP-PCP | THI6/ThMP/AMP-PCP | THI6/CF3HMP-PP/carboxy-Thz-P | |
|---|---|---|---|---|---|---|
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 |
| Resolution (Å) | 3.2 | 3.0 | 3.1 | 3.3 | 2.6 | 3.2 |
| Space group | P21 | P21 | P21 | C2 | C2 | P21 |
| Unit Cell | ||||||
| a (Å) | 102.4 | 104.5 | 105.2 | 161.3 | 163.2 | 104.9 |
| b (Å) | 153.0 | 154.3 | 154.6 | 153.9 | 153.5 | 154.2 |
| c (Å) | 146.7 | 148.6 | 148.6 | 108.6 | 109.6 | 148.7 |
| β (°) | 102.3 | 102.5 | 102.1 | 117.6 | 117.9 | 102.1 |
| Matthews coefficient | 3.1 | 3.3 | 3.4 | 3.4 | 3.5 | 3.2 |
| % solvent | 60.1 | 63.3 | 63.7 | 64.1 | 64.7 | 62.3 |
| mol/asym | 6 | 6 | 6 | 3 | 3 | 6 |
| Reflections | 75076 | 273270 | 188896 | 163837 | 143976 | 159587 |
| Unique reflections | 38571 | 84611 | 72303 | 32447 | 69157 | 74009 |
| Completeness (%) | 93.8 (68.9) | 93.2 (61.0) | 86.6 (71.3) | 91.5 (58.7) | 96.5 (82.3) | 98.5 (95.9) |
| Rsym *(%) | 10.6 (32.8) | 10.3 (26.9) | 11.0 (34.8) | 14.8 (39.7) | 6.3 (35.5) | 8.1 (42.6) |
| I/σ | 13.7 (2.3) | 9.5 (1.8) | 8.5 (1.4) | 7.8 (1.6) | 11.6 (1.8) | 12.8 (1.9) |
| Redundancy | 2.1 (2.1) | 3.2 (2.1) | 2.6 (1.9) | 5.0 (2.8) | 2.1 (1.9) | 2.2 (2.0) |
Rsym = ΣΣi| Ii − <I> | /Σ<I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.
Structure Determination and Refinement
BLAST searches (29) showed that the N-terminal domain of CgTHI6 is about 30% identical to Bacillus subtilis TPS (BsTPS) and the C-terminal domain is about 30% identical to Bacillus subtilis ThiM (BsThiM). As a result, molecular replacement was used for the structure determination of unliganded CgTHI6. The Matthews number (30) suggested that the asymmetric unit contains six monomers with solvent content of 60.1%. A homology model containing three C-terminal domains of THI6 was generated using trimeric BsThiM (PDB entry 1C3Q) as a template. Only conserved side chains, based on sequence alignments of CgTHI6 with multiple bacterial thiazole kinases using ClustalW (31), were retained in the homology model. Molecular replacement using CNS (32) revealed the orientations and positions of two trimeric assemblies. After fixing the assemblies from the C-terminal domains, the six N-terminal domains were found by molecular replacement using MOLREP (33) with a monomer of BsTPS (PDB entry 2TPS) as the search model.
All model building was performed using the computer program COOT (34). The initial refinement was done using CNS and involved successive rounds of rigid body refinement, simulated annealing, temperature factor refinement, energy minimization and model rebuilding. Tight non-crystallographic symmetry restraints (NCS) were used throughout the refinement. The sixfold NCS averaged maps were generated using the RAVE suit of programs (35). NCS averaged electron density maps and averaged composite omit maps were used for all stages of model building. Final rounds of refinements were performed using the PHENIX suit of programs (36) with tight NCS restraints.
Structure determinations of the CgTHI6-ligand complexes were carried out using the unliganded structure as a starting point. The THI6/Thz/AMP-PCP complex crystallized in space group C2 with three CgTHI6 chains per asymmetric unit. The structure was readily determined by molecular replacement using half of a hexamer as the search model. Successive rounds of rigid body refinement, simulated annealing, temperature factor refinement and energy minimization were carried out, followed by model rebuilding. After few rounds of refinement the NCS averaged electron density map was used to position the ligands in the active site. PRODRG was used to generate the topology and parameter files of ligands, required for refinement (37). The final refinement statistics are summarized in Table 2.
Table 2.
Refinement statistics.
| THI6 | THI6/ThMP | THI6/ThMP/PP | THI6/Thz/AMP-PCP | THI6/ThMP/AMP-PCP | THI6/CF3HMP-PP/carboxy-Thz-P | |
|---|---|---|---|---|---|---|
| Resolution (Å) | 3.2 | 3.0 | 3.1 | 3.3 | 2.6 | 3.2 |
| No of protein atoms | 22510 | 22678 | 22967 | 11429 | 11967 | 22584 |
| No of ligand atoms | - | 144 | 192 | 128 | 162 | 222 |
| RMSD | ||||||
| Bonds (Å) | 0.009 | 0.008 | 0.007 | 0.006 | 0.008 | 0.010 |
| Angles (deg) | 1.165 | 1.012 | 1.115 | 1.084 | 1.120 | 1.227 |
| R factor a (%) | 21.38 | 21.25 | 20.50 | 20.44 | 21.23 | 20.1 |
| Rfree b (%) | 24.48 | 24.18 | 25.10 | 24.29 | 24.40 | 23.2 |
| Ramachandran plot | ||||||
| Most favored (%) | 86.1 | 88.4 | 85.2 | 81.6 | 88.0 | 85.7 |
| Add allowed (%) | 12.3 | 10.0 | 12.8 | 15.8 | 11.2 | 12,2 |
| Gen allowed (%) | 1.6 | 1.4 | 1.7 | 2.3 | 0.8 | 1.8 |
| Disallowed (%) | 0.0 | 0.2 | 0.3 | 0.3 | 0.0 | 0.2 |
R factor = Σhkl||Fobs|−k|Fcal|/Σhkl|Fobs|, where Fobs and Fcal are observed and calculated structure factors, respectively.
In bRfree the sum is extended over a subset of reflections that were excluded from all stages of refinement.
Biochemical Assay of CgTHI6 Activity
Reaction mixtures of the following composition were incubated at room temperature for 2 h. ThiM activity assay: 1.2 mM Thz, 2 mM ATP, 2 mM MgSO4 and 33 μM purified enzyme in 50 mM Tris-HCl containing 150 mM NaCl and 2 mM DTT. TPS activity assay: 1.2 mM Thz, 2 mM ATP, 2 mM MgSO4, 1 mM HMP-PP and 33 μM purified enzyme in 50 mM Tris-HCl containing 150 mM NaCl and 2 mM DTT. ADT cleavage activity assay: 100 μM ADT, 2 mM MgSO4 and 33 μM purified enzyme in 50 mM Tris-HCl containing 150 mM NaCl and 2 mM DTT. A reaction in which the enzymes were replaced by the reaction buffer served as a control in each case.
HPLC Analysis of the CgTHI6 Catalyzed Reactions
Analytical HPLC was carried out using a Supelco LC-18-T (150×4.6 mm, 3 μm ID) reverse phase column. HPLC analyses were performed in an Agilent 1100 instrument equipped with an inline diode-array detector. The enzyme was denatured by heat (100 °C, 40 s) and the reaction mixtures were rapidly cooled on ice. The precipitated protein was removed by centrifugation and the supernatant was filtered through a 10 kDa MWCO filter (Microcon YM-10, Millipore). 100 μL of the filtrate was analyzed by reversed phase HPLC. The following linear gradient was used at 1 mL/min flow rate (solvent A is water, solvent B is 100 mM KPi, pH 6.6, solvent C is methanol): 0 min 100% B, 5 min 10% A 90% B, 8 min 25% A 60% B 15% C, 14 min 25% A 60% B 15% C, 19 min 30% A 40% B 30% C, 21 min 100% B, 30 min 100% B. The absorbance at 261 nm was monitored. Different compounds were identified in the chromatogram by comigration with authentic standards.
Figure Preparation
All figures were prepared using ClustalW (31), ESPript (38), PyMOL (39) or ChemDraw (CambridgeSoft).
RESULTS
Overall Structure of CgTHI6
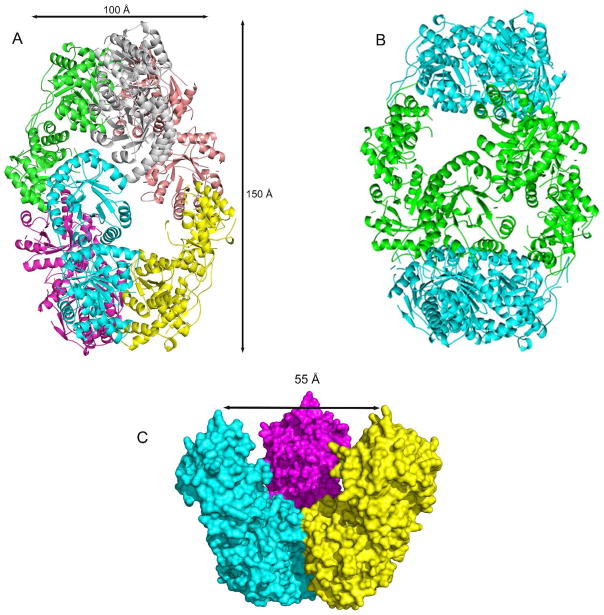
The crystal structure shows that the enzyme is a hexamer composed of six identical protomers with 32 point symmetry (Figure 1A, B). The cage-like hexamer has the shape of a prolate spheroid with a diameter along the twofold axis of about 100 Å and a length along the threefold axis of about 150 Å. The inner cavity of the cage has a maximum diameter of about 55 Å (Figure 1C). The presence of the cavity contributes to the high solvent content (~60%) of the crystals, and explains previous results from analytical size exclusion chromatography, which suggested THI6 was an octamer (23). The caps of the prolate spheroid are formed by trimeric assemblies of the C-terminal (ThiM) domains and the equator is formed by dimeric assemblies of the N-terminal (TPS) domains. The trimeric caps contact each other only through the TPS dimers; TPS dimers contact the trimers, but not each other.
Figure 1.
(A) Cartoon representation of the quaternary structure of CgTHI6 hexamer color coded by subunits. (B) Cartoon representation of the quaternary structure of CgTHI6 hexamer color coded by domains; N-terminal domains are colored in cyan and C-terminal domains are green. (C) Surface diagram showing the interior cavity of the prolate-spheroid shaped hexamer. One half of the hexamer is shown with the central cavity.
CgTHI6 Protomeric Structure
The protomer consists of two domains (Figure 1B). The N-terminal domain is composed of residues 1–236 and the C-terminal domain is composed of residues 248–540. The domains are connected by a linker consisting of 11 amino acid residues 237–247. The N-terminal domain has a (βα)8 barrel fold, also known as a TIM barrel fold. The structural fold is characterized by eight parallel β strands tilted at an angle of 45° from the barrel axis, surrounded by eight α helices. In addition to those eight α helices, flanking the barrel, there are two 310 helices. The first one, α0, forms a cap near the N-terminal end (residues 7–10) and the second one, α80 formed by residues 210–213, connects β8 and α8. This helix is tilted at an angle of approximately 20° to the axis of the TIM barrel helices. The β strands are 4–6 residues long, whereas the α helices consist of 4–14 residues, α8 being the longest one. There are eight loops connecting the alternative helical and strand segments. The active site loop formed by residues 143–153 is disordered in the unliganded structure.
The C-terminal domain has an α/β fold. The centrally located, nine stranded mixed β sheet is flanked by 11 α helices. The β sheet has a topology: β′2↑ β′1↑ β′3↑ β′4↑ β′5↑ β′6 ↑ β′7↓ β′8↓ β′9↓. Helices α′3, α′4, α′5, α′6, and α′7 are packed on one side of the sheet and are antiparallel to the strands of the sheets. The remaining six α helices form a bundle on the opposite side of the sheet. The strands contain 4–6 residues and the helices are made up of 6–14 residues. The C-terminal domains assemble to form a trimer in the top and bottom of the egg shaped structure.
Domain Interactions: Interface between N-terminal and C-terminal Domains
Within the protomer, 2630 Å2 is buried at the interface of the TPS and ThiM domains (40). The interface is formed by several interactions (Figure 2, Table 3). The first interaction is between residues 193–203, a loop joining α7 and β8 of the TPS domain and a portion of the loop containing residues 432–438 of the ThiM domain. The second interaction occurs between residues 230–236 of the TPS domain and the loop, consisting of residues 428–432 from the ThiM domain. The third interaction is between part of α7 (residues 188–190) of the TPS domain and a portion of α′5 (residues 359–361) from the ThiM domain. Finally, the loop connecting α′10 and α′11 (residues 479–486) of the ThiM domain interacts with a portion of the linker (residues 238–241) between the TPS and ThiM domains.
Figure 2.
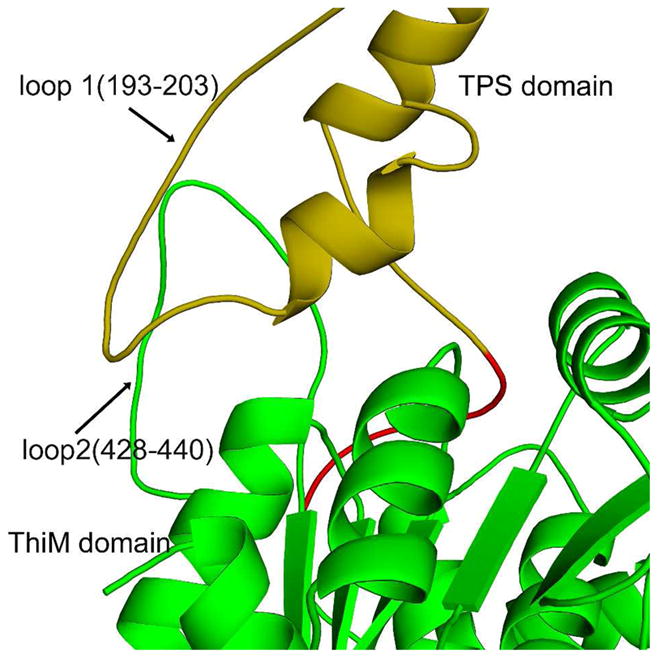
Ribbon diagram showing interactions between the two domains within CgTHI6 protomer. The N-terminal domain (residues 1–236) is colored in golden yellow, C-terminal (residues 248–540) in green and the linker region (residues 237–247) in red.
Table 3.
Hydrogen bonds between the N-terminal and C-terminal domains.
| N-terminal domain | C-terminal domain | H-bonding atoms |
|---|---|---|
| Ser202 | Ser432 | Oγ—N |
| Leu203 | Ser434 | O—N |
| Lys200 | Ala435 | O—N |
| Tyr234 | Gly429 | N—O |
| Arg189 | Thr359 | NH1—O |
Domain Interactions: Interface between N-terminal Domains
The six TPS domains of CgTHI6 have (βα)8 barrel folds and pair up to form three dimers. The individual domains are cis to each other with respect to the barrel axis. The total surface area buried at each dimeric interface is approximately 1500 Å2 (40). The major interactions at dimer interfaces include helices α3 (residue 78–84) and α4 (residue 98–105). Helix α3 of one monomer is oriented almost perpendicular to the barrel axis of the other monomer, interacting with α3 and α4. A second interaction occurs between 49–52 from one domain and 94–97 from the other. The interface is mostly hydrophobic residues with only one intermolecular hydrogen (Asp76 of one monomer and Arg77 of another).
Domain Interactions: Interface between C-terminal Domains
The ThiM domains group together to form two trimers, which are twisted by about 20° with respect to each other. The trimers are about 78 Å along the edge and about 45 Å thick. The total surface area buried between two ThiM domains is approximately 1600 Å2 (40) and the interface is formed by thee main interactions. The first interaction occurs between part of α′2 (residues 276–279) from one domain and a portion of α′2 residues (273–275) from an adjacent domain. The second interaction occurs between a loop region from one domain (residues 297–303) and part of α′5 (347–350) from an adjacent domain. The side chain of Glu298 forms hydrogen bonds with the side chains of Thr348 and Arg350. The third interaction occurs between part of helix α′10 (residues 515–523) and the loop between α′9 and α′10 (residues 507–513).
Active Site of the TPS Domain
The active site of the TPS (N-terminal) domain is located in a cavity surrounded by loops 2 (residues 45–51), 4 (residues 92–97) and 6 (residues 138–154) of the (βα)8 barrel. Loop 6 is the longest one and is partly disordered in the unliganded TgTHI6 structure.
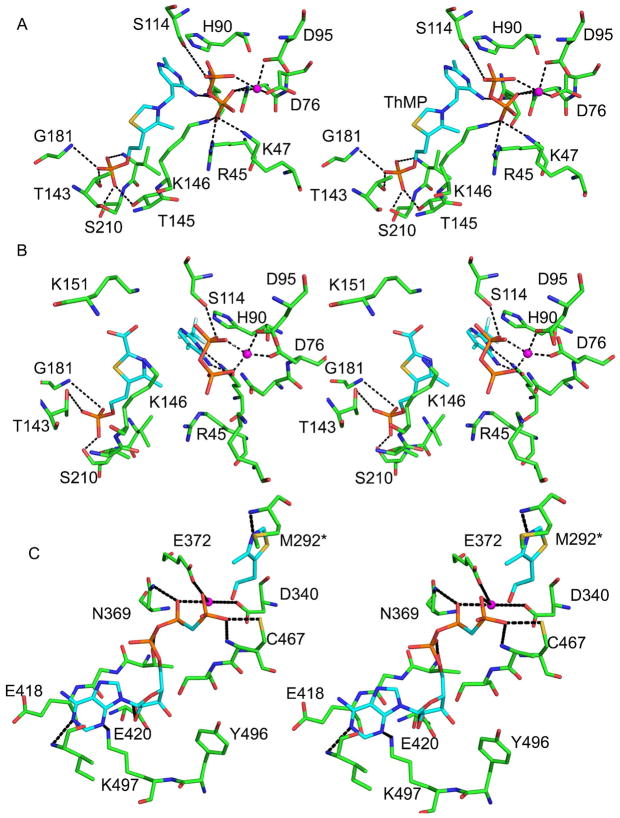
The binding geometry of ThMP in the CgTHI6/ThMP/PP complex is similar to that of the BsTPS structure (13). The main interactions between the protein and the ligands are shown in Figure 3A. ThMP displays a V-shaped conformation with the pyrimidine ring more exposed and the thiazole ring more buried. The thiazolium moiety binds in an extended conformation lying under loop 6. The side chain of Lys146, running parallel to the hydroxyethyl group of the thiazolium ring provides van der Waals interactions. The phosphate group of ThMP forms an extensive network of hydrogen bonds with Thr143, Thr145, Gly181, Val 209 and Ser210; these residues come mostly from loop 6 and α8. Ile79 interacts with both the pyrimidine and thiazole rings. The mostly hydrophobic pyrimidine binding is located near the entrance of the β-barrel and is formed by Tyr13, Val15, His90, Tyr134, Ile179 and Cys207 (not shown). The only hydrogen bond occurs between N3 of the pyrimidine and the side chain of Gln43 (not shown).
Figure 3.
The active sites of CgTHI6. (A) Stereo-diagram showing active site of the N-terminal domain of the enzyme. The product ThMP, pyrophosphate and the active site residues are shown in ball-and-stick representation. (B) Stereo-diagram showing the carboxy-thiazole binding in the active site of the N-terminal domain of the enzyme. The substrates, carboxy-Thz-P, CF3HMP-PP, pyrophosphate and the active site residues are shown in ball-and-stick representation. (C) Stereo-diagram showing active site of the C-terminal domain of the enzyme. The substrate Thz, AMP-PCP and the active site residues are shown in ball-and-stick representation. In all panels, carbon atoms are shown in cyan for ligand and green for protein, nitrogen in blue, oxygen in red, sulfur in yellow, phosphorus in orange, and Mg2+ is marked by a magenta sphere.
The pyrophosphate binding site in the CgTHI6/ThMP/PP complex is located near the opening of the active site and is more solvent exposed than the ThMP. The pyrophosphate and the thiazole moiety are present on the opposite faces of the pyrimidine ring of ThMP. Interactions with the pyrophosphate include both hydrogen bond and electrostatic interactions with a Mg2+ ion. Hydrogen bonds occur between the pyrophosphate oxygen atoms and side-chains of Asn75, Asp76, Asp95 and Ser114. Salt bridges are formed with Arg45 and Lys47 from loop 1, and Lys146 from loop 6. Loop 6 is partly disordered in the THI6/ThMP structure (residues 146–149 are missing in the electron density) and becomes ordered after pyrophosphate binding.
In the CgTHI6/ThMP/PP complex structure, the Mg2+ binding site is located on the outside edge of the active site cavity. The Mg2+ is octahedrally coordinated by the side-chains of Asp76 and Asp95 and the oxygen atoms of pyrophosphate. However in absence of pyrophosphate, the Mg2+ binding site is slightly shifted and the cation is tetrahedrally coordinated by side-chains of Asp76 and Asp95, the main-chain of Ala49 and the side-chain of Thr51 from the neighboring monomer.
In the CgTHI6/CF3HMP-PP/carboxy-Thz-P complex (Figure 3B), density for carboxy-Thz-P was found only in the TPS active site. Similar interactions are observed between the thiazole moiety and the protein as for the product analogue complexes, CgTHI6/ThMP and CgTHI6/ThMP/PP. Two lysine residues are positioned near the 2-carboxy group of the thiazole ring; Lys146 and Lys151, both of them coming from loop 6. Separate, well-defined electron densities were observed for the CF3HMP and PP moieties suggesting that bond cleavage had occurred. A similar bond cleavage was observed for the B. subtilis TPS/CF3HMP-PP/Thz-P complex (see discussion below). The bond cleavage requires only small movements of the ligands and the interactions between the protein and the pyrimidine moiety and pyrophosphate remain essentially the same as for the CgTHI6/ThMP/PP complex.
Active Site of the ThiM Domain
The overall active site of the ThiM domain in the CgTHI6/Thz/AMP-PCP complex is similar to that of BsThiM (18). The ATP binding site is inferred from the complex containing the non-hydrolizable ATP analogue, AMP-PCP (Figure 3C). The site is located near the C-terminal end of the central β-sheet and includes β′5, β′6, β′7 and β′8. The adenine moiety of the nucleotide is positioned under two loops, one containing a β-turn connecting β ′6 and β ′7 (residues 417–419), and the other containing a loop preceding α′8 (residues 451–457). The region containing residues 456–465 becomes ordered upon AMP-PCP binding. The N1 and N3 atoms of the adenine form hydrogen bonds with the Ile455 amide nitrogen and the Lys497 side chain, respectively. Both hydroxyl groups of the ribose moiety are hydrogen bonded to the side chain of Glu420. One oxygen atom of the α-phosphate interacts with the hydroxyl group of Thr416. The side-chain of Lys367 (not shown) forms a hydrogen bond with the β-phosphate, which is connected to Mg2+. The γ-phosphoryl of the nucleotide analogue is oriented towards the hydroxyl group of thiazole and is hydrogen bonded with the side chain of Cys467 and is coordinated with Mg2+. A second Mg2+ interacts with the β-phosphate and γ-phosphoryl of the nucleotide analog and the side-chains of the strictly conserved residues Asp340 and Glu372.
The Thz binding site in the CgTHI6/Thz/AMP-PCP structure is located at the interface of two domains and is rich in hydrophobic residues. The main interactions with the thiazole occur between Asn272, Val274 and Thr462 from one protomer and Ala280, Leu284, Pro290 and Met292 from an adjacent protomer (not shown). The only hydrogen bond is formed between the nitrogen atom of thiazole ring and the amide nitrogen of Met292. The side chain of Cys467 is positioned near the hydroxyl group of thiazole.
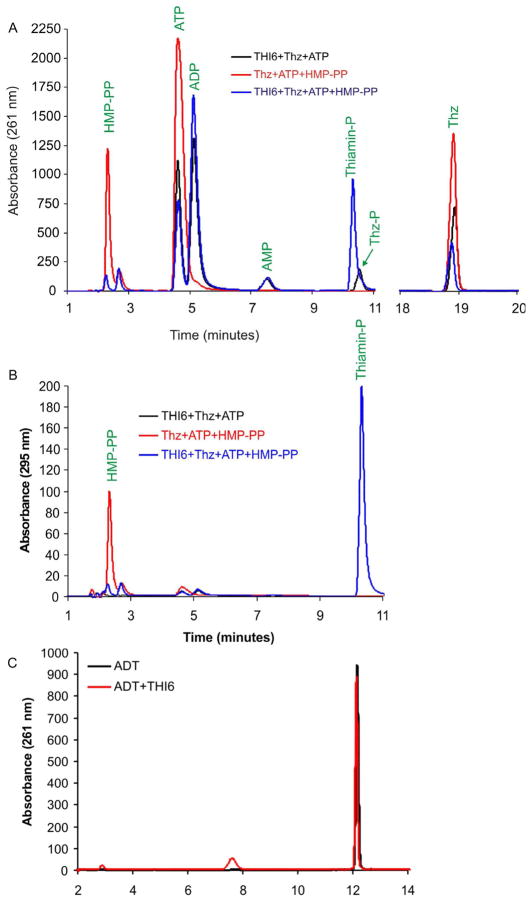
Biochemical Activities of CgTHI6
Recombinant CgTHI6 exhibited both ThiM and TPS activities (Figure 4A, B). When the purified protein was incubated with Thz and ATP, formation of Thz-P was noted. Incorporation of HMP-PP in the same reaction resulted in the production of ThMP. Consumption of Thz in the latter reaction was higher, presumably due to the removal of the Thz-P, product of the ThiM reaction. However, incubation of the enzyme with the THI4 product ADT did not show any reaction suggesting that ADT is not a substrate for THI6 (Figure 4C).
Figure 4.
Activity assays for CgTHI6. (A) HPLC analyses of the TPS and ThiM activities, absorbance measured at 261 nm. The red trace shows the control where the enzyme is not added. The black trace shows the Thz-P formation in presence of the enzyme showing the ThiM activity. The blue trace shows the ThMP formation in presence of the enzyme showing the TPS activity. (B) HPLC analyses of the TPS and ThiM activities, absorbance measured at 261 nm. The red trace shows the control where the enzyme is not added. The black trace shows the Thz-P formation in presence of the enzyme showing the ThiM activity. The blue trace shows the ThMP formation in presence of the enzyme showing the TPS activity. (C) HPLC analysis of ADT cleavage activity, absorbance measured at 261 nm. The red trace shows the control where the enzyme is not added. The black trace shows that there is no reaction in presence of the enzyme.
DISCUSSION
Biochemical Assays on THI6
We have confirmed that THI6 catalyzes the conversion of Thz to Thz-P and the coupling of Thz-P and HMP-PP to give TMP. In addition, we have demonstrated that ADT, the product of the S. cerevisiae thiazole synthase, is not a substrate for THI6. This observation strongly suggests that an as yet unidentified nudix hydrolase is required for the conversion of ADT to carboxy-Thz-P.
Reliability of the Medium Resolution Structure
The resolutions of the reported unliganded and complex structures range from 2.6 Å to 3.3 Å, a range bordering on medium resolution in protein crystallography. However, the current structures represent a favorable scenario. The determination of these structures involved molecular replacement using high-resolution BsThiM (1.5 Å) and BsTPS (1.25 Å) structures as starting models making the current structures very reliable. Furthermore, the presence of six monomers in the asymmetric unit allowed the use of a six-fold NCS averaged map for model building and tight NCS restraints for refinement, which further strengthened the reliability of the structural information and resulted in a small difference between R and R-free (3–4 %).
Structural Comparison with Bacterial TPS
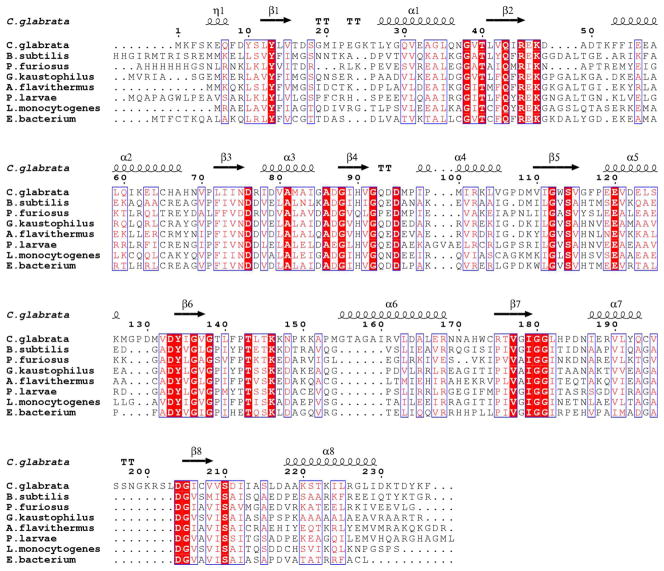
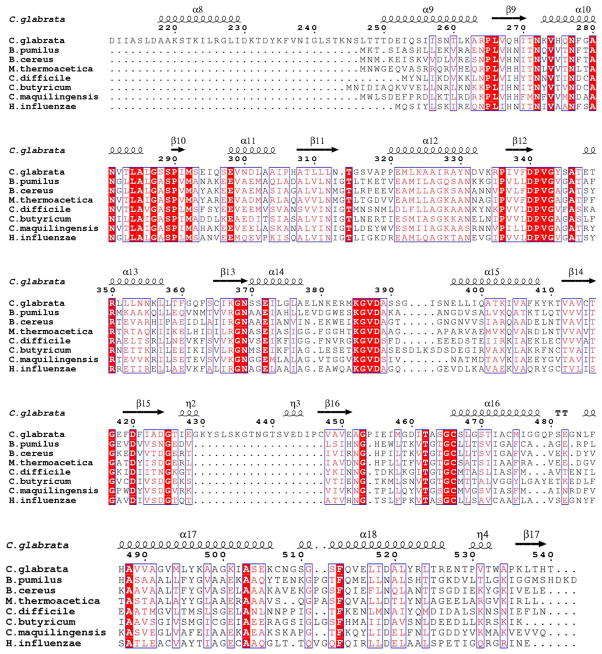
TPS catalyzes the formation of ThMP from HMP-PP and Thz-P or carboxy-Thz-P. High resolution structures of this enzyme have been reported in unliganded and complex forms from B. subtilis (41) and P. furiosus (PDB 1XI3). An initial structural homology search using the DALI server (42) indicated that BsTPS is structurally homologous to CgTHI6 having a Z score of 26.5 and RMSD 1.9 Å, based on alignment of 236 residues of the N-terminal domain of CgTHI6. A sequence alignment for the N-terminal domain of CgTHI6 and bacterial TPS homologues is shown in (Figure 5).
Figure 5.
Sequence alignment for the N-terminal (TPS) domain of CgTHI6 and representative monofunctional bacterial homologues. Numbering and secondary structural elements refer to CgTHI6. Absolutely conserved residues are highlighted with red blocks and similar residues are shown in pink font.
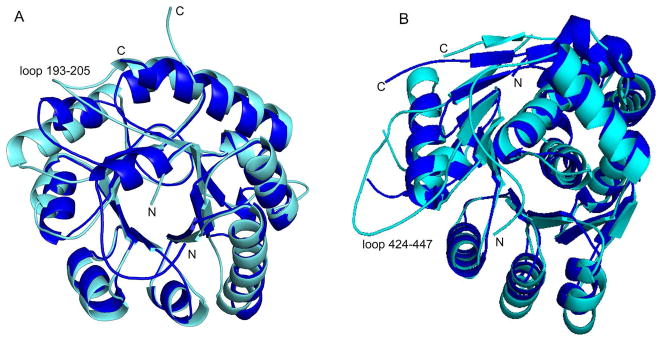
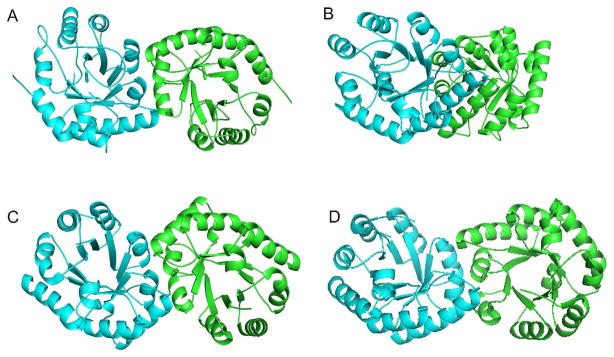
Although the N-terminal domain of CgTHI6 utilizes the same (βα)8 barrel fold as BsTPS (Figure 6A), a detailed structural comparison suggests that there are changes between the eukaryotic enzyme and its bacterial homologue. The significant differences occur mainly in the loop regions. The most interesting one is the presence of a long loop (193–203) in the bifunctional enzyme holding the two domains together.
Figure 6.
Comparison between the bifunctional CgTHI6 and its monofunctional bacterial homologues. (A) Ribbon diagram of the N-terminal domain of CgTHI6 superimposed on BsTPS monomer. CgTHI6 is colored in cyan and BsTPS in blue. (B) Ribbon diagram of the C-terminal domain of CgTHI6 superimposed on BsThiM monomer. CgTHI6 is colored in cyan and BsThiM in blue.
The active site residues of the two enzymes are mostly conserved with the same binding conformation of ThMP, PP and Mg2+. A comparison of the active site residues has been listed in Table 4.
Table 4.
Comparison of the active site residues of BsTPS and CgTHI6.
| BsTPS | CgTHI6 | |
| Gln 57 | Gln 43 | |
| Thr 156 | Thr 143 | |
| ThMP binding residues | Thr 158 | Thr 145 |
| Gly 168 | Gly 181 | |
| Ser 209 | Ser 210 | |
| Ile 208 | Val 209 | |
| Pyrophosphate binding residues | Ser 130 | Ser 114 |
| Arg 59 | Arg 45 | |
| Lys 159 | Lys 146 | |
| Asn 92 | Asn 75 | |
| Lys 61 | Lys 47 | |
| Mg2+ binding residues | Asp 93 | Asp 76 |
| Asp 112 | Asp 95 |
Comparison of the Dimer Interfaces of CgTHI6 with Bacterial TPS and TenI
The N-terminal domain of CgTHI6 and BsTPS both exist as dimers of (βα)8 barrels. However in CgTHI6 the barrel axes are cis to each other unlike in BsTPS, where they are in trans orientation (Figure 7). In the BsTPS dimer, the major interactions occur between two symmetry related α3 helices running parallel to each other. Interestingly the dimeric interface of the bifunctional enzyme is similar to PfTPS, the available structure of archaebacterial TPS. The CgTHI6 dimer is also similar to BsTenI (43), which is an enzyme in bacterial thiamin biosynthetic pathway, that shows a significant structural homology to TPS. Previous gene identification studies have suggested that tenI genes are paralogues of thiE, the gene encoding TPS (44). All three dimers have the barrel axes cis to each other (Figure 7). The formation of a different dimeric subunit in BsTPS is not yet well understood. One possible reason can be considered as an artifact of the crystal packing. The occurrence of the same dimeric subunit in PfTPS (archebacterium), BsTenI (eubacterium) and CgTHI6 (eukaryote) also suggests the possibility of a divergent evolution, all three enzymes originating from a common ancestor.
Figure 7.
Comparison of the dimeric subunit of CgTHI6 with other enzymes. (A) Ribbon diagram showing the dimer of the N-terminal domain of CgTHI6. (B) Ribbon diagram showing the dimer of BsTPS. (C) Ribbon diagram showing the dimer of BsTenI. (D) Ribbon diagram showing the dimer of PfTPS. In all figures the monomers are labeled by different colors.
Conformational Change upon Ligand Binding in the TPS Domain
A comparison between the structures of the unliganded enzyme and the THI6/ThMP and THI6/ThMP/PP complexes suggests that there are some conformational changes upon ligand binding. The active site C loop composed of residues 143–153 becomes ordered upon product binding. Residues Thr143 and Thr145 from the C loop take part in the phosphate binding of the product ThMP. The side-chain of Lys146 is involved in pyrophosphate binding. This longest loop acts as a gate near the active site, which closes upon the product binding and opens when the reaction is complete and the product is dissociated from the enzyme surface. The other active site loops A and B also show some minor changes upon product binding.
Structural Comparison with Bacterial Thz Kinase
ThiM catalyzes the conversion of Thz to Thz-P, in the presence of ATP and Mg2+. This is a salvage enzyme that provides an alternative to the regular thiazole biosynthetic pathway and enables the cell to use the recycled Thz. The crystal structures of the unliganded ThiM, ThiM/Thz and ThiM/ThiZP/ATP complexes have been reported from B. subtilis, in monoclinic and rhombohedral forms (18). Structural similarity between these two enzymes was found using a DALI search, which showed a Z score of 31.6 and RMSD 1.9 Å, for 270 superimposed Cα atoms. A sequence alignment for the C-terminal domain of CgTHI6 and bacterial ThiMs is shown in (Figure 8). The trimeric assembly remains conserved in the eukaryotic homologue.
Figure 8.
Sequence alignment for the C-terminal (ThiM) domain of CgTHI6 and representative monofunctional bacterial homologues. Numbering and secondary structural elements refer to CgTHI6. Absolutely conserved residues are highlighted with red blocks and similar residues are shown in pink font.
The overall fold of the C-terminal domain of CgTHI6 is similar to BsThiM, having a centrally located β sheet flanked by several α helices on both sides. The secondary structural elements are mostly conserved between the two enzymes (Figure 6B). The major structural differences are observed in the loop regions. The presence of the insertion (residues 426–446) in the bifunctional enzyme is in the form of a single turn helix, α′80 (426–429) followed by an extended loop, interacting with the N-terminal domain. The loop connecting α′9 and α′10 is longer in the bifunctional enzyme (residues 479–486) and interacts with a portion of the linker between the two domains (residues 238–241). The C-terminal helix α′12, composed of residues 256–264 in BsThiM, is substituted by a single turn helix containing residues 530–533 in CgTHI6. A very interesting observation is that the loop containing residues 379–393 is disordered in our current structures. The corresponding loop containing residues 135–150 is present in the ThiM/Thz (rhombohedral crystal form) structure but disordered in the ThiM/Thz-P/ATP complex structure (monoclinic crystal form) (18). However the structural significance of this loop is still not known.
A comparison of the active site residues between the two enzymes is shown in Table 5. The active sites are mostly conserved with similar binding orientations of Thz, AMP-PCP and Mg2+. However, there are some minor modifications observed in the eukaryotic enzyme. The thiazole-binding loop containing residues 314–316 is slightly shifted away from the ligand in CgTHI6 compared to the loop containing residues 66–69 in BsThiM. The ATP binding site also shows some differences in the eukaryotic enzyme. A very interesting shift of ~8 Å is observed in the ATP binding loop containing residues 451–455, compared to the loop containing residues 184–188 in BsThiM. The adenine base remains more solvent exposed in the bacterial enzyme with no hydrogen bond between the adenine base and the protein. In the bifunctional enzyme the adenine ring is shielded by the loop 451–455. There are two hydrogen bonds between the ring and the protein. The N1 of the adenine base makes one hydrogen bond with the main chain nitrogen of Ile455 and the other one is between N3 of the adenine base and the side-chain of Lys497, which is replaced by Gly226 in BsThiM.
Table 5.
Comparison of the active site residues of BsThiM and CgTHI6.
| BsThiM | CgTHI6 | |
| Mg2+ Binding residues | Asp 94 | Asp 340 |
| Glu 126 | Glu 372 | |
| Arg 121 | Lys 497 | |
| Asn 123 | Asn 369 | |
| AMP-PCP binding residues | Thr 168 | Thr 416 |
| Gly 169 | Gly 417 | |
| Glu 170 | Glu 418 | |
| Thr 191 | Gly 459 | |
| Gly 195 | Ala 463 | |
| Ala 196 | Ser 464 | |
| Tyr 225 | Tyr 496 | |
| Asp 172 | Asp 420 | |
| Cys 198 | Cys 466 | |
| Thiazole binding residues | * Met 45 | * Met 292 |
Residues from the other chain
Conformational Changes upon Thz and AMP-PCP Binding
Thz binding in the C-terminal domain induces an interesting change of space group from primitive monoclinic P21 to C-centered monoclinic C2, having three monomers in the asymmetric unit, composing half of the hexamer. However the whole hexamer is generated by the crystallographic 2-fold axis. The active site loop, preceded by the single turn helix containing residues 456–464, also becomes ordered upon ligand binding. This observation is consistent with the fact that several residues in this region make contact with the ligand.
Relationship of the Two CgTHI6 Active Sites
Substrate channeling is a common phenomenon in most multifunctional enzymes (45). The product of the C-terminal domain, carboxy-ThZ-P, is the substrate of the N-terminal domain in bifunctional CgTHI6. Our biochemical studies reveal that the consumption of ThZ was higher in the presence of HMP-PP, possibly due to removal of carboxy-ThZ-P, suggesting cooperative interaction between two domains and raising the possibility of a channel between the two domains. However, in the crystal structure of CgTHI6, the two active sites are ~ 40 Å apart with no apparent channel. The possibility of interchain channeling was also considered; however, in this case the two active sites are even farther apart. Therefore it appears that the chemically unstable product, carboxy-ThZ-P, is transferred from one domain to the other by diffusion before condensation with HMP-PP to form product.
Trapping of an Intermediate in the THI6/CF3HMP-PP Complex
The active site of this complex surprisingly showed three fragments of density for carboxy-ThZ-P, the pyrimidine moiety and PP, providing a snapshot of the enzyme-intermediate complex. The distance between the C7′ of pyrimidine and the closest oxygen of PP was 2.7 Å, much longer than an average C-O single bond length, which is 1.43 Å. The distance between the C7′ and the nitrogen of the thiazole ring was 3 Å, also longer than a standard C-O single bond, which is 1.47 Å. This suggests clearly that C7′ of the pyrimidine is not bonded to either nucleophile. The pyrimidine moiety was modeled in the active site as iminemethide resulting from the deprotonation of the carbenium ion. The strongly electron withdrawing trifluoromethyl substituent in the pyrimidine ring would destabilize the carbenium ion favoring the formation of iminemethide by deprotonation. The active site contents support a dissociative mechanism where the ionization of HMP-PP is followed by the nucleophilic attack by the thiazole ring. A similar trapping of intermediate was also observed in case of bacterial TPS (41).
Catalytic Mechanism
The study of the THI6/ThMP and THI6/ThMP/PP complexes shows that all the active site residues are conserved between CgTHI6 and BsTPS. The binding conformations of the products, ThMP, PP and Mg2+ also remain the same in the two active sites, indicating the involvement of a similar dissociative mechanism with the formation of pyrimidine carbocation (14, 15, 41). The binding of HMP-PP and carboxy-Thz-P to the active site is followed by the closing of the C-loop, forming a stable enzyme-substrate ternary complex. The ionization of HMP-PP is facilitated by the stabilization of pyrophosphate as a leaving group and the delocalization of the positive charge in the pyrimidine ring. This intermediate is trapped by the nucleophilic thiazole moiety to form thiamin phosphate, followed by the decarboxylation. The decarboxylation is facilitated by the positive charge on the nitrogen atom of the thiazole ring.
The structural comparison of the THI6/Thz-P/AMP-PCP complex and the ThiM/Thz-P/AMP-PCP complex shows that the active site residues are mostly conserved between the two enzymes with a similar kind of ligand binding. This provides an insight to the similar mechanism of action between the two enzymes. The positioning of the γ-phosphoryl of AMP-PCP and the Thz indicates an inline phosphate transfer in the presence of ATP. Cys467 is the probable candidate for the base to deprotonate the thiazole alcohol and facilitate phosphorylation. Interestingly this cysteine residue is replaced by aspartate in the other closely related small molecule kinases, ribokinase and adenosine kinase (46, 47).
The substrates for the bacterial and S. cerevisiae TPS are carboxy-Thz-P or Thz-P and HMP-PP. The mechanism of the decarboxylation is not yet established. The active site of the THI6/CF3HMP-PP/carboxy-Thz-P complex indicates that there are two lysine residues (Lys151 and Lys146) positioned near the thiazole carboxylate group. Lys146 is strictly conserved in THI6 orthologues as well as in the bacterial TPS homologues. On the other hand, Lys151 does not remain conserved in the eukaryotes and is replaced by a histidine residue (His132) in BsTPS. This suggests that Lys146 is likely to be involved in binding the thiazole carboxylate.
Acknowledgments
We thank NE-CAT beamlines 24-ID-C and 24-ID-E for the use of beamtime. We thank Cynthia Kinsland for the preparation of the overexpression plasmids and Leslie Kinsland for assistance in the preparation of this manuscript. S.E.E. is indebted to the W. M. Keck Foundation and the Lucille P. Markey Charitable Trust.
ABBREVIATIONS
- CgTHI6
Candida glabrata THI6
- TEV
tobacco etch virus
- BME
β-mercaptoethanol
- NCS
noncrystallographic symmetry
- DTT
dithiothreitol
- TPS
thiamin phosphate synthase
- BsTPS
Bacillus subtilis thiamin phosphate synthase
- PfTPS
Pyrococcus furiosus thiamin phosphate synthase
- ThiM
4-methyl-5-hydroxyethylthiazole kinase
- BsThiM
Bacillus subtilis thiazole kinase
- ThMP
thiamin monophosphate
- ThDP
thiamin pyrophosphate
- PP
pyrophosphate
- HMP-P
4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate
- HMP-PP
4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate
- CF3HMP-PP
4-amino-5-hydroxymethyl-2-trifluoromethylpyrimidine pyrophosphate
- Thz
4-methyl-5-hydroxyethylthiazole
- Thz-P
4-methyl-5-hydroxyethylthiazole phosphate
- carboxy-Thz-P
2-carboxy-4-methyl-5-hydroxyethylthiazole phosphate
- ATP
adenosine 5′-triphosphate
- AMP-PCP
β,γ-methylene-adenosine 5′-diphosphate
- ADT
adenosine diphospho-5-β-ethyl-4-methylthiazole-2-carboxylic acid
- PEG400
polyethylene glycol 400
Footnotes
This work was supported by NIH grant DK44083 to T.P.B. and DK67081 to S.E.E.
This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The coordinates of the THI6 structures have been deposited in the Protein Data Bank under accession number 3NL2, 3NL3, 3NM3, 3NL5, 3NL6 and 3NM1 for THI6, THI6/ThMP, THI6/ThMP/PP, THI6/Thz/AMP-PCP, THI6/ThMP/AMP-PCP and THI6/CF3HMP-PP/carboxy-Thz-P, respectively.
References
- 1.Jordan F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat Prod Rep. 2003;20:184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- 2.Monsen ER. The 10th edition of the Recommended Dietary Allowances: what’s new in the 1989 RDAs? J Am Diet Assoc. 1989;89:1748–1752. [PubMed] [Google Scholar]
- 3.Butterworth RF. Thiamin deficiency and brain disorders. Nutr Res Rev. 2003;16:277–284. doi: 10.1079/NRR200367. [DOI] [PubMed] [Google Scholar]
- 4.Martin PR, Singleton CK, Hiller-Sturmhofel S. The role of thiamine deficiency in alcoholic brain disease. Alcohol Res Health. 2003;27:134–142. [PMC free article] [PubMed] [Google Scholar]
- 5.Begley TP, Ealick SE. Thiamin Biosynthesis. In: Lew M, Hung-Wen L, editors. Comprehensive Natural Products II. Elsevier; Oxford: 2010. pp. 547–559. [Google Scholar]
- 6.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurgenson CT, Ealick SE, Begley TP. Chapter 3.6.3.7 Biosynthesis of Thiamine Pyrophosphate. In: Böck A, Curtiss R III, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, Squires CL, editors. EcolSal – Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C: 2009. [Google Scholar]
- 8.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chem Biol. 2004;11:1373–1381. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Kriek M, Martins F, Leonardi R, Fairhurst SA, Lowe DJ, Roach PL. Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J Biol Chem. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol. 2008;4:758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Gomez NC, Downs DM. ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry. 2008;47:9054–9056. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G, Bennett EM, Begley TP, Ealick SE. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure. 2002;10:225–235. doi: 10.1016/s0969-2126(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 13.Chiu HJ, Reddick JJ, Begley TP, Ealick SE. Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 Å resolution. Biochemistry. 1999;38:6460–6470. doi: 10.1021/bi982903z. [DOI] [PubMed] [Google Scholar]
- 14.Hanes JW, Ealick SE, Begley TP. Thiamin phosphate synthase: the rate of pyrimidine carbocation formation. J Am Chem Soc. 2007;129:4860–4861. doi: 10.1021/ja0679634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddick JJ, Nicewonger R, Begley TP. Mechanistic studies on thiamin phosphate synthase: evidence for a dissociative mechanism. Biochemistry. 2001;40:10095–10102. doi: 10.1021/bi010267q. [DOI] [PubMed] [Google Scholar]
- 16.McCulloch KM, Kinsland C, Begley TP, Ealick SE. Structural studies of thiamin monophosphate kinase in complex with substrates and products. Biochemistry. 2008;47:3810–3821. doi: 10.1021/bi800041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb E, Downs D. Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J Biol Chem. 1997;272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 18.Campobasso N, Mathews II, Begley TP, Ealick SE. Crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry. 2000;39:7868–7877. doi: 10.1021/bi0000061. [DOI] [PubMed] [Google Scholar]
- 19.Jurgenson CT, Chatterjee A, Begley TP, Ealick SE. Structural insights into the function of the thiamin biosynthetic enzyme Thi4 from Saccharomyces cerevisiae. Biochemistry. 2006;45:11061–11070. doi: 10.1021/bi061025z. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. Thiamin biosynthesis in eukaryotes: characterization of the enzyme-bound product of thiazole synthase from Saccharomyces cerevisiae and its implications in thiazole biosynthesis. J Am Chem Soc. 2006;128:7158–7159. doi: 10.1021/ja061413o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeidler J, Sayer BG, Spenser ID. Biosynthesis of vitamin B1 in yeast. Derivation of the pyrimidine unit from pyridoxine and histidine Intermediacy of urocanic acid. J Am Chem Soc. 2003;125:13094–13105. doi: 10.1021/ja030261j. [DOI] [PubMed] [Google Scholar]
- 22.Haas AL, Laun NP, Begley TP. Thi20, a remarkable enzyme from Saccharomyces cerevisiae with dual thiamin biosynthetic and degradation activities. Bioorg Chem. 2005;33:338–344. doi: 10.1016/j.bioorg.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki Y. Copurification of hydroxyethylthiazole kinase and thiamine-phosphate pyrophosphorylase of Saccharomyces cerevisiae: characterization of hydroxyethylthiazole kinase as a bifunctional enzyme in the thiamine biosynthetic pathway. J Bacteriol. 1993;175:5153–5158. doi: 10.1128/jb.175.16.5153-5158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosaka K, Nishimura H, Kawasaki Y, Tsujihara T, Iwashima A. Isolation and characterization of the THI6 gene encoding a bifunctional thiamin-phosphate pyrophosphorylase/hydroxyethylthiazole kinase from Saccharomyces cerevisiae. J Biol Chem. 1994;269:30510–30516. [PubMed] [Google Scholar]
- 25.Lawhorn BG, Mehl RA, Begley TP. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org Biomol Chem. 2004;2:2538–2546. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- 26.Hazra A, Chatterjee A, Begley TP. Biosynthesis of the thiamin thiazole in Bacillus subtilis: identification of the product of the thiazole synthase-catalyzed reaction. J Am Chem Soc. 2009;131:3225–3229. doi: 10.1021/ja806752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput Appl Biosci. 1994;10:19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project-Number 4. The CCP-4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN-programs for reformatting, analysis, and manipulation of biomacromolecular electron-density maps and reflection datasets. Acta Crystallogr D. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 38.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Peapus DH, Chiu HJ, Campobasso N, Reddick JJ, Begley TP, Ealick SE. Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase. Biochemistry. 2001;40:10103–10114. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]
- 42.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 43.Toms AV, Haas AL, Park JH, Begley TP, Ealick SE. Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II. Biochemistry. 2005;44:2319–2329. doi: 10.1021/bi0478648. [DOI] [PubMed] [Google Scholar]
- 44.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 46.Mathews II, Erion MD, Ealick SE. Structure of human adenosine kinase at 1.5 Å resolution. Biochemistry. 1998;37:15607–15620. doi: 10.1021/bi9815445. [DOI] [PubMed] [Google Scholar]
- 47.Sigrell JA, Cameron AD, Jones TA, Mowbray SL. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 Å resolution: insights into a new family of kinase structures. Structure. 1998;6:183–193. doi: 10.1016/s0969-2126(98)00020-3. [DOI] [PubMed] [Google Scholar]