Summary
GLI-similar (Glis)1–3 proteins constitute a subfamily of the Krüppel-like zinc finger transcription factors that are closely related to the Gli family. Glis1–3 play critical roles in the regulation of a number of physiological processes and have been implicated in several pathologies. Mutations in GLIS2 have been linked to nephronophthisis, an autosomal recessive cystic kidney disease. Loss of Glis2 function leads to renal atrophy and fibrosis that involves epithelial-mesenchymal transition (EMT) of renal tubule epithelial cells. Mutations in human GLIS3 have been implicated in a syndrome characterized by neonatal diabetes and congenital hypothyroidism (NDH) and in some patients accompanied by polycystic kidney disease, glaucoma, and liver fibrosis. In addition, the GLIS3 gene has been identified as a susceptibility locus for the risk of type 1 and 2 diabetes. Glis3 plays a key role in pancreatic development, particularly in the generation of β-cells and in the regulation of insulin gene expression. Glis2 and Glis3 proteins have been demonstrated to localize to the primary cilium, a signaling organelle that has been implicated in several pathologies, including cystic renal diseases. This association suggests that Glis2/3 are part of primary cilium-associated signaling pathways that control the activity of Glis proteins. Upon activation in the primary cilium, Glis proteins may translocate to the nucleus where they subsequently regulate gene transcription by interacting with Glis-binding sites in the promoter regulatory region of target genes. In this review, we discuss the current knowledge of the Glis signaling pathways, their physiological functions, and their involvement in several human pathologies.
Keywords: Diabetes, Polycystic kidney disease, Primary cilium, Pancreatic β-cells, Glis Krüppel-like zinc finger protein
Introduction
Krüppel-like zinc finger proteins constitute one of the largest families of transcription factors. Typically, these proteins contain two or more Cys2-His2-type zinc fingers that are separated by the conserved consensus sequence, T/SGEKPY/F. Krüppel-like zinc finger proteins are divided into several subfamilies based on the number of zinc finger motifs, sequence homology between the zinc-finger domains, and the presence of specific repressor and activation domains (Williams et al., 1995; Poncelet et al., 1998; Agata et al., 1999; Dang et al., 2000; Peng et al., 2000).
Gli-similar 1–3 (HUGO nomenclature Glis1–3) form a subfamily of Krüppel-like zinc finger proteins that are most closely related to members of the Gli and Zic subfamilies (Kinzler et al., 1988; Ruppert et al., 1988; Aruga et al., 1994; Lamar et al., 2001; Zhang and Jetten, 2001; Kim et al., 2002; Nakashima et al., 2002; Zhang et al., 2002; Kim et al., 2003; Aruga, 2004; Kim et al., 2005; Kasper et al., 2006; Merzdorf, 2007; Riobo and Manning, 2007). While the Gli signaling pathways have been well studied (Matise and Joyner, 1999; Ruiz i Altaba et al., 2002; Gill and Rosenblum, 2006; Kasper et al., 2006; Rubin et al., 2006; Oro et al., 2007; Riobo et al., 2007; Rohatgi et al., 2007; Ruiz i Altaba et al., 2007), the physiological functions of Glis proteins are only beginning to emerge. Human genome linkage studies and analyses of Glis-null mice have implicated Glis2 and Glis3 in several diseases, including cystic kidney disease, fibrosis, diabetes, and hypothyroidism, and have revealed key roles for Glis proteins in several biological processes, including pancreatic development and the maintenance of normal renal functions (Senee et al., 2006; Attanasio et al., 2007; Kim et al., 2008; Kang et al., 2009a; Kang et al., 2009b; Watanabe et al., 2009). This review provides a summary of our current knowledge of the mechanism of action and biological functions of this protein subfamily and their roles in disease.
Structural relationship between Gli, Zic, and Glis family members
Glis1 was identified in a yeast two-hybrid screen using the ligand-binding domain of the retinoid-related orphan receptor RORγ, a member of the nuclear receptor superfamily, as bait (Kim et al., 2002; Jetten, 2009). Subsequently, two additional proteins, closely related to Glis1, were identified and referred to as Glis2 and Glis3 (Zhang and Jetten, 2001; Zhang et al., 2002; Kim et al., 2003; Kim et al., 2005). Glis1 and Glis2 were independently identified by two other laboratories and referred to as Gli homolog 1 (GliH1) and neuronal Krüppel-like (NKL), respectively (Lamar et al., 2001, Nakashima et al., 2002). The human GLIS1–3 genes map to chromosome 1p32.3, 16p13.3 and 9p24.2, span about 248, 44.8, and 495 kb, and encode for proteins 65.9, 55.7, and 90 kD in size, respectively. The GLIS3 gene has been reported to generate several alternative transcripts (Senee et al., 2006), but it has yet to be established whether the proteins generated by these transcripts are associated with any distinct physiological function.
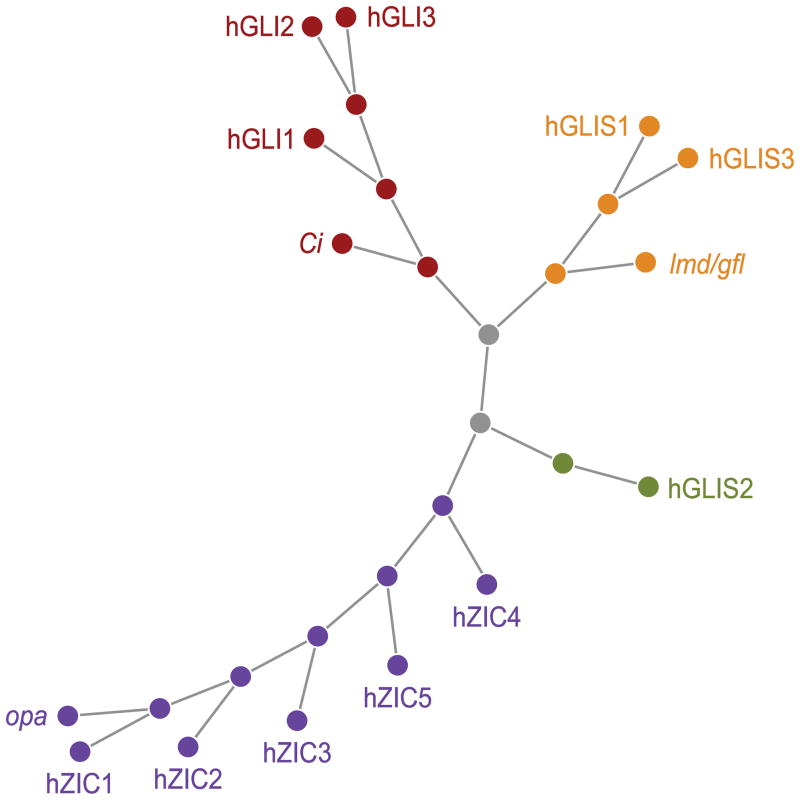
Glis1–3 exhibit a high degree of homology across species (e.g., about 99% between human and mouse) and share a highly homologous zinc finger domain (ZFD) containing a tandem repeat of five Cys2-His2-type zinc finger (ZF) motifs with members of the Gli and Zic family (Kinzler et al., 1988; Ruppert et al., 1988; Zhang and Jetten, 2001; Kim et al., 2002; Zhang et al., 2002; Kim et al., 2003; Aruga, 2004, Kasper et al., 2006, Merzdorf, 2007). Cubitus interruptus (Ci), odd-paired (opa), and lame duck (lmd), also named gleeful (gfl), are the Drosophila homologues of Gli, Zic, and Glis, respectively (Benedyk et al., 1994; Aruga et al., 1996; Kuo et al., 1998; Lessing and Nusse, 1998; Aza-Blanc and Kornberg, 1999; Duan et al., 2001, Furlong et al., 2001). A Glis3 homolog was recently identified in Oryzias latipes (Japanese ricefish or medaka) (Hashimoto et al., 2009). Fig. 1 shows the phylogenic relationship between the Glis, Gli, and Zic family members. Glis and Gli family members show little sequence homology outside their ZFD, while Zic1–5 proteins exhibit a moderate degree of conservation outside their ZFD. The ZFDs of Glis1 and Glis3 are 93% identical and exhibit a 68–71%, 59%, and 52% identity with ZFDs of Gli, Glis2, and Zic proteins, respectively (Kim et al., 2002, Kim et al., 2003). The locations of the exon-intron junctions in the region encoding the zinc finger motifs are conserved between Glis1 and Glis3, but are different from those in Glis2 suggesting that Glis1 and Glis3 are evolutionarily derived from the same parental gene, while Glis2 is more distantly related.
Figure 1.
Phylogenetic relationship between members of the GLIS, GLI, and ZIC family. Ci, opa, and lmd (or gfl), are the Drosophila homologues of GLI, ZIC, and GLIS, respectively.
Transcriptional regulation
Although the mechanisms by which Glis proteins regulate gene transcription are not fully understood, recent studies have provided considerable insights into several critical elements of Glis-mediated transcriptional regulation. These include identification and characterization of specific domains required for their nuclear translocation and transcriptional activity, the recognition of specific Glis-binding sites (Glis-BS), the interaction with co-activators and co-repressors, and the discovery of several putative target genes.
Subcellular localization
Confocal microscopy utilizing EGFP-tagged Glis proteins revealed that the proteins were predominantly localized to the nucleus of exponentially growing cells. Although Glis proteins contain putative nuclear localization signals (NLS), deletion and mutation analysis showed that nuclear localization did not require these motifs, but instead depended on the zinc finger domain (Kim et al., 2002; Zhang et al., 2002; Kim et al., 2003; Beak et al., 2008). Detailed study of Glis3 demonstrated that a putative bipartite NLS overlapping with ZF5 was not required for its nuclear localization. When the tetrahedral configuration of each of the Glis3 zinc fingers was disrupted, only loss of the tetrahedral configuration of ZF4 or deletion of ZF4 was found to profoundly affect the accumulation of Glis3 in the nucleus (Beak et al., 2008). These data suggest that ZF4 is critical for the nuclear localization of Glis3. Further studies are required to determine the mechanism by which this region directs the protein to the nucleus.
Recent studies have provided evidence showing that both Glis2 and Glis3 localize to the primary cilium suggesting that they are part of a primary cilium-associated signaling pathway (Attanasio et al., 2007; Hashimoto et al., 2009; Kang et al., 2009a; Kang et al., 2009b). This will be discussed below in more detail.
Interaction with Glis binding sites
The ZFD, the most distinct domain within Glis proteins, plays a role in DNA sequence recognition, protein-protein interaction, and nuclear localization. Crystal structure analysis of the Gli ZFD has indicated that ZF2 through ZF5 bind in the major groove and wrap around the DNA (Pavletich and Pabo, 1993; Klug and Schwabe, 1995). While ZF1 does not appear to contact the DNA, ZF4 and ZF5 were shown to make extensive base contacts within the conserved nine base-pair Gli binding site (GBS). Circular dichroism (CD) spectroscopy has indicated that the ZFDs of Glis, Zic, and Gli proteins form an α-helical conformation in solution and that binding to the GBS increases the α-helix content and stabilizes the ZFD (Sakai-Kato et al., 2009). Due to the high homology between the ZFDs of Glis and Gli proteins, it is not surprising that Glis proteins also recognize the consensus GBS, 5′-TGGGTGGTC (Ruppert et al., 1988; Lamar et al., 2001; Kim et al., 2002; Nakashima et al., 2002; Zhang et al., 2002; Kim et al., 2003). An in vitro approach was utilized to determine the optimal Glis-BS and (G/C)TGGGGGG(A/C) was identified as the consensus binding sequence for Glis3 (Beak et al., 2008). The tetrahedral configuration in each of the zinc finger motifs was required for optimal binding of Glis3 to the Glis-BS.
Glis3 binds the consensus Glis-BS with a higher affinity than the consensus GBS (Beak et al., 2008). Although Glis, Gli and Zic proteins bind similar DNA response elements, they exhibit different affinities for distinct sequences (Kinzler and Vogelstein, 1990; Vortkamp et al., 1995; Mizugishi et al., 2001; Beak et al., 2008). These different affinities are likely due, in part, to differences in the amino acid sequences within their ZFDs. Moreover, in vivo, the promoter context of the Glis-BS and the recruitment of different cofactors by Glis likely play a critical role in determining the binding affinity of Glis proteins to Glis-BS sequences in target genes. The overlap in binding specificity between Glis, Gli, and Zic proteins suggests that these proteins might compete for the same DNA binding sites in cells in which they are co-expressed and thus provide a mechanism for crosstalk between Glis, Gli, and possibly Zic, signaling pathways. This hypothesis is supported by data showing that Glis1 and Glis2 can indeed inhibit GBS-dependent transcriptional activation of a reporter by Gli1 (Kim et al., 2002; Nakashima et al., 2002; Kim et al., 2003). Future identification and characterization of Glis target genes and functions will be required to determine whether such cross-talk between Glis and Gli proteins has any physiological significance.
Glis proteins as transcriptional activators and repressors
Analysis of the transcriptional activity of Glis proteins in several cell lines showed that, in contrast to full-length Glis3, full-length Glis1 and Glis2 did not enhance (GBS)- or (Glis-BS)-dependent transcriptional activation of a reporter gene (Kim et al., 2002; Nakashima et al., 2002; Zhang et al., 2002; Kim et al., 2003; Beak et al., 2007; Kim et al., 2007; Beak et al., 2008; Kang et al., 2009a). Regulation of gene expression by Glis1–3 likely involves the recruitment of transcriptional mediators that interact with specific repressor and/or activation domains within the respective Glis proteins. Sequence analysis of Glis proteins, however, did not identify any known evolutionary-conserved activation or repressor domains present in many other Krüppel-like zinc finger proteins, such as the SCAN box or Krüppel-associated box (KRAB) (Williams et al., 1995; Peng et al., 2000).
Although full-length Glis1 exhibits little transcriptional activity, study of several Glis1 deletion mutants identified a strong activation domain at its C-terminus (Kim et al., 2002; Nakashima et al., 2002). The lack of transcriptional activity by full-length Glis1 indicates the presence of a repressor domain in its N-terminus and suggests that Glis1 might function as a repressor as well as an activator of transcription. It was further shown that Ca2+-dependent calmodulin kinase IV was able to enhance the transcriptional activity of an N-terminal truncated Glis1 (Kim et al., 2002). Although potential repressor and activation domains have been identified in the N-terminal half of Glis2, exogenous expression of full-length Glis2 in several cell lines did not result in increased transcriptional activation of a reporter gene (Zhang et al., 2002; Kim et al., 2005; Kim et al., 2007). Analysis of various Glis3 deletion mutants identified a strong transactivation domain at its C-terminus (Kim et al., 2003; Beak et al., 2008; Kang et al., 2009a). Although full-length Glis3 is able to induce transcriptional activation, deletion of a 300 amino acid N-terminal region greatly increased its transcriptional activity suggesting the presence of a N-terminal repressor domain (Kim et al., 2003; Kang et al., 2009b).
Protein-protein interactions, post-translational modifications, and proteolytic processing play a critical role in regulating the activity of many transcription factors and, as has been demonstrated for Gli proteins (Kasper et al., 2006, Rubin and de Sauvage, 2006), may also be critical in modulating the transcriptional activity of Glis1–3. Future studies are required to identify the signaling pathways that control the transcriptional activity of Glis proteins.
Interaction with transcriptional mediators
Interactions with other proteins are not only important in regulating the subcellular localization and activation of Glis proteins, but also in mediating Glis-dependent transcriptional activation. The latter involves recruitment of co-activator complexes that through their histone acetylase activity induce relaxation of chromatin and increased transcription of target genes (Glass and Rosenfeld, 2000; Hager et al., 2009). Conversely, transcriptional repression involves interaction with co-repressor complexes containing histone deacetylase activity that promotes the compaction of chromatin. Although little is known about the transcriptional modulators involved in Glis-mediated regulation of target genes, several Glis-interacting proteins have been identified that modulate Glis activity.
Yeast two-hybrid analysis using Glis2 as bait identified several putative Glis2 interacting proteins, including C-terminal binding protein 1 (CtBP1), β-catenin, G-protein modulator 2 (GPSM2), and WNK lysine deficient protein kinase 1 (WNK1) (Kim et al., 2005; Kim et al., 2007). CtBP1 predominantly functions as a co-repressor by interacting with a large number of transcriptional repressors through PXDLS-like motifs (Chinnadurai, 2003). CtBP1 does not bind Glis1 or Glis3, but is able to interact with Glis2 by binding several PXDLS-like motifs (Kim et al., 2005). In addition to CtBP1, Glis2 can recruit histone deacetylase 3 (HDAC3). It has been suggested that both CtBP1 and HDAC3 are part of a Glis2 silencing complex that mediates transcriptional repression by Glis2 (Kim et al., 2005).
Interaction of Glis2 with β-catenin has been shown to involve ZF1 of Glis2 and the armadillo repeats of β-catenin (Kim et al., 2007). The 12 armadillo repeats, each of which consists of three α-helices, form a superhelical structure with a long positively charged groove (Huber et al., 1997). Interestingly, the second loop in ZF1 of Glis2 consists of a negatively charged α-helix containing 3 Glu/Asp residues that could facilitate the interaction of Glis2 with the armadillo repeats. β-catenin is a multifunctional protein that, when bound to cadherin, regulates cell adhesion and migration (Perez-Moreno and Fuchs, 2006). Cytoplasmic β-catenin functions as a transducer in the Wnt signaling pathway where it interacts with and regulates the transcriptional activity of Tcf/Lef (Akiyama, 2000). Glis2 was shown to inhibit β-catenin-Tcf/Lef mediated transcriptional activation as well as the induction of the β-catenin-Tcf target gene, cyclin D1. These observations suggest crosstalk between the Glis2 and Wnt/β-catenin/TCF signaling pathways and indicate that Glis2 might function as a repressor of Wnt/β-catenin/TCF induced gene expression. Given the important role of β-catenin in cancer (Gavert et al., 2007), reduced Glis2 expression might enhance β-catenin-dependent transcription and promote tumorigenesis. Future studies will determine whether Glis2 has any role in tumor development.
Another study provided evidence for an interaction between Glis2 and p120 catenin (p120ctn), which functions as a substrate for Src kinase and has been shown to regulate RhoGTPase activity (Hosking et al., 2007). p120ctn is associated with cadherins and microtubules and plays an important role in regulating cell adhesion, motility, morphology, and proliferation (Perez-Moreno et al., 2006). Complete loss, down-regulation or mislocalization of p120ctn correlates with tumor progression in several human cancers (van Hengel and van Roy, 2007). The region of Glis2 between amino acids 35–174 and the C-terminus of p120ctn, including the armadillo repeats 6–10, were required for this interaction. Glis2 was found to promote the nuclear translocation of p120ctn, while p120ctn induced cleavage of Glis2 by a Src kinase-dependent mechanism. The dissociation of p120ctn from the E-cadherin complex or from microtubules is required for the cleavage of Glis2 as indicated by data showing that increased expression of E-cadherin reduced cleavage, whereas nocodazole treatment, which depolymerizes microtubules, promoted cleavage (Hosking et al., 2007). The physiological significance of the Glis2-p120ctn interaction has yet to be determined.
A recent study showed that Glis3, but not Glis1 or Glis2, is able to interact with the transcriptional mediator, WW domain-containing protein Wwtr1, also referred to as TAZ (Kang et al., 2009a). Wwtr1 interacts with proteins through its WW domain that recognizes P/LPXY motifs (Hong et al., 2005; Hong and Yaffe, 2006). Wwtr1 has been reported to interact with several transcription factors and to function either as a co-activator or co-repressor. Although Glis3 contains 4 P/LPXY motifs, only the most C-terminal motif is required for its interaction with Wwtr1. Wwtr1 was able to significantly increase the transcriptional activity of Glis3 in reporter gene assays suggesting that it can function as a co-activator of Glis3-mediated transcription. Mutation within the C-terminal Glis3 P/LPXY motif greatly inhibited Glis3 transcriptional activity supporting the importance of this site in the transactivation function of Glis3. Interestingly, loss in either Glis3 or Wwtr1 function has been reported to lead to the development of renal cysts (Hossain et al., 2007; Makita et al., 2008; Kang et al., 2009a). Thus, one could speculate that the development of polycystic kidney disease in Glis3 or Wwtr1 null mice might be due, in part, to the disruption of Glis3-Wwtr1 interactions.
Patterns of Glis expression
Glis1–3 are expressed in a tissue- and cell type-specific manner and in a variety of mammalian cell lines. In adult mice, all three Glis family members are most abundantly expressed in the kidney. Glis1 is further expressed at low levels in testis, thymus, colon, brain, and adipose tissue, and at high levels in the placenta (Kim et al., 2002). Glis2 mRNA was present at low levels in a number of tissues, including heart, brain, lung, intestine, and prostate (Zhang and Jetten, 2001; Zhang et al., 2002). Glis3 mRNA is widely expressed, abundantly in pancreas, thyroid gland, thymus, and uterus and at low levels in ovary, brain, lung, and liver (Kim et al., 2003; Senee et al., 2006).
GLIS3 has been reported to be highly expressed in a number of different cancer cell types. Overexpression of GLIS3 in human ependymomas was found to correlate with a poor survival (Lukashova-v Zangen et al., 2007). High levels of GLIS3 expression have also been observed in human chromophobe renal carcinoma (Yusenko and Kovacs, 2009). These observations suggest a link between elevated GLIS3 expression and cancer progression.
Expression of Glis1–3 during development
During mouse embryonic development, Glis1–3 are expressed in a temporal and spatial pattern. Glis1 mRNA expression was first detected in extra-embryonic tissues and lateral mesoderm in E8 embryos (Kim et al., 2002, Nakashima et al., 2002). At E10, Glis1 is primarily expressed in the ventral regions of the mesenchyme of the limb buds and tail. Additional expression was observed at E10.5 in the developing maxillary process, mandibular and hyoid arches. Glis1 was detected during limb development as early as E9.5 in the anterior-proximal mesenchyme. At E14.5, Glis1 expression is restricted to ventral limb surface and the joint interzone of the digits (Kim et al., 2002). At E15.5 of eye development, Glis1 was expressed throughout the basal layer of the anterior lens epithelium and strongly expressed in the presumptive iris and ciliary body (Nakashima et al., 2002). In skin, Glis1 was highly expressed in the dermal cells surrounding the vibrissae. At 16.5, Glis1 mRNA was most abundantly expressed in the kidney. During tooth development, Glis1 was highly expressed in a region of the dental papilla. Further studies are needed to understand the function of Glis1 in these tissues.
At E9.5 of mouse neural development, Glis2 (NKL) is expressed in the cranial and dorsal root ganglia and neural tube (Lamar et al., 2001) and at E10.5 is broadly expressed in the intermediate zone of the hindbrain and spinal cord. At the neural plate stage in Xenopus development, Glis2 expression corresponds to two midline regions that correspond to precursors of primary sensory and motoneurons. These patterns of expression are similar to that of NeuroD, but temporally later than that of neurogenin 1 (Xngn1). Furthermore, exogenous expression of Xngn1 in two-cell Xenopus embryos induced Glis2 expression suggesting that it may act downstream of Xngn1. These patterns of expression suggest that Glis2 might play a role in neurogenesis. This is supported by observations that exogenous expression of Glis2 in chick neuronal precursors induced cell cycle arrest and expression of neuronal differentiation markers (Lamar et al., 2001).
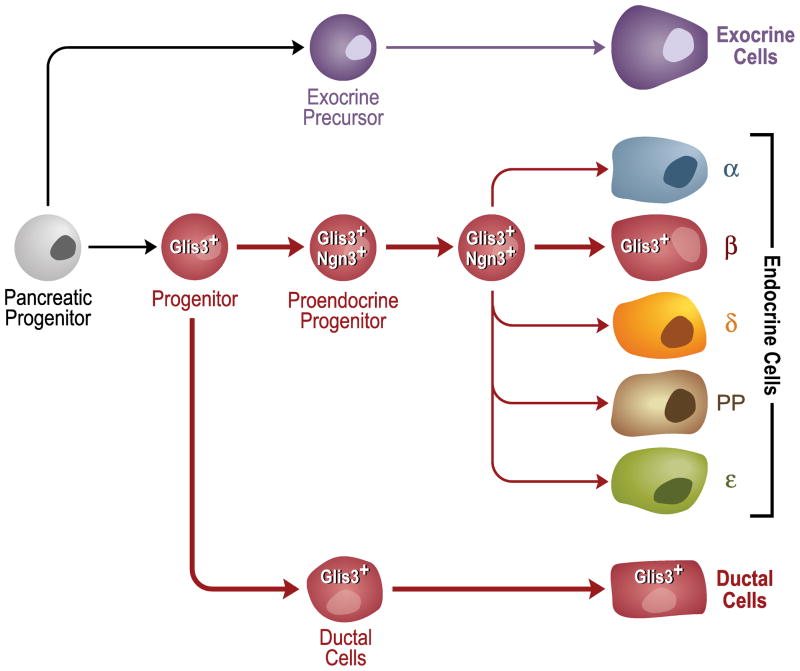
Analysis of embryos from stage E6.5–E14.5 revealed that Glis3 mRNA is expressed in specific regions of the developing kidney, testes, and lung, and in a highly dynamic pattern during neurulation and eye development (Kim et al., 2003). Glis3 transcripts were first detected at E8.0 in the node which gives rise to the embryonic notochord. During limb development, Glis3 expression was restricted to specific mesenchymal regions. At E11.5–12.5, Glis3 is expressed in the inter-digital regions that are fated to undergo apoptosis. Analysis of Glis3 mRNA expression during mouse pancreas development showed low levels of Glis3 at E11.5 followed by a significant increase at E12.5–13.5 (Kang et al., 2009b). By E16.5–E18.5 and in the postnatal pancreas, Glis3 expression is mostly restricted to the ductal epithelial cells and islets where its expression is largely confined to β-cells (Senee et al., 2006; Kang et al., 2009b). During early branching (E11.5–13.5), Sox9+HNF1β+ cells of the trunk compartment are precursors of exocrine, endocrine, and ductal lineages, while between E13.5–E16.5 the Sox9+HNF1β+ precursors generate both endocrine and ductal lineages, but not acinar cells (Solar et al., 2009). Taken together with observations showing that Glis3 is present in both ductal and β-cells and is increased at E12.5, this suggests that Glis3 is not expressed in early multipotent pancreatic progenitors, but in the Sox9+HNF1β+ precursors that have a more limited differentiation potential (Ackermann and Gannon, 2007; Jorgensen et al., 2007; Murtaugh, 2007; Zhou et al., 2007; Solar et al., 2009)(Fig. 2). Further studies on the precise timing of Glis3 induction are required to corroborate this hypothesis.
Figure 2.
Schematic presentation of the putative role of Glis3 in pancreas development. Between E11.5–13.5, as the pancreas undergoes extensive branching morphogenesis, multipotent progenitors cells are precursors of exocrine, endocrine, and ductal lineages (Zhou et al., 2007; Solar et al., 2009). Between E13.5-E16.5, during the so-called secondary transition, Sox9+HNF1β+ progenitors with a more limited differentiation potential in the trunk compartment are able to generate the endocrine and ductal lineages, but not acinar cells. The timing of Glis3 induction during pancreatic development and its expression in ductal and β-cells appears consistent with a role for Glis3 in the lineage determination of those Sox9+HNF1β+ precursors generate the ductal and endocrine, but not the exocrine lineage (Solar et al., 2009). In addition to its role in β-cell development, Glis3 acts as a regulator of insulin expression and ductal epithelial cell function.
Expression of Glis1–3 in kidney
Although Glis1–3 genes are most highly expressed in the kidney, similarities and differences in their pattern of expression have been observed during renal development (Kim et al., 2002; Nakashima et al., 2002; Zhang et al., 2002; Kim et al., 2003). During kidney development, the ureteric bud emerges from the Wolffian duct and invades into the metanephric mesenchyme (Perantoni, 2003; Yu et al., 2004). The ureteric bud undergoes a stereotypical pattern of branching morphogenesis and subsequently gives rise to the collecting ducts. The branching is controlled by a complex series of cell-cell interactions involving metanephric mesenchymal, ureteric bud epithelial, and stromal or interstitial mesenchymal cells. Signals from the ureteric bud promote the aggregation of metanephric mesenchymal cells around the ureteric bud tips. These aggregates become polarized through a process referred to as mesenchymal-epithelial transition (MET) thereby giving rise to tubular structures, called comma- and S-shaped bodies, and subsequently the epithelia that form the nephron (Davies, 1996; Perantoni, 2003; Yu et al., 2004).
In the mouse kidney, Glis1 expression is first observed at E13.5 in the mesenchymal condensations and comma-shaped nodes, but is absent from the ureteric bud epithelium (Nakashima et al., 2002). At E14 and E15.5, Glis1 is present in the S-shaped bodies and at postnatal day (PND) 15 continues to be expressed in renal tubules, but not in glomeruli and collecting ducts (Nakashima et al., 2002). In contrast to Glis1, Glis2 is highly expressed in the ureteric bud at E16 of murine metanephric development, whereas mesenchymal tissue, comma- and S-shaped bodies were devoid or expressed low levels of Glis2 (Zhang et al., 2002; Attanasio et al., 2007). In adult kidneys Glis2 was detected in epithelial cells of all segments of the renal tubule and in epithelial cells of Bowman’s capsule, but not in glomeruli, endothelial, or mesenchymal cells (Attanasio et al., 2007). This pattern of expression suggests that Glis2 is induced during epithelialization of the metanephric mesenchyme and continues to be expressed in epithelial cells of the adult kidney. Like Glis2, Glis3 is highly expressed in the branches of the ureteric bud of E14.5 metanephros (Kim et al., 2003). In the adult mouse kidney and in medaka, Glis3 mRNA is expressed in the epithelia of the collecting ducts, renal tubules, and Bowman’s capsule (Hashimoto et al., 2009; Kang et al., 2009a). Because Glis1–3 have overlapping patterns of expression in the kidney and bind similar DNA sequences, it is quite possible that they share some regulatory functions.
Physiological functions of Glis proteins and their roles in disease
Recent studies have linked the loss of GLIS2 and GLIS3 function in humans to several pathologies (Senee et al., 2006; Attanasio et al., 2007). Furthermore, the study of Glis2- and Glis3-null mice has revealed important clues about the biological and physiological functions of these proteins (Attanasio et al., 2007; Kim et al., 2008; Kang et al., 2009a; Kang et al., 2009b; Watanabe et al., 2009). In contrast, analysis of Glis1-null mice has thus far not revealed any particular phenotype nor have mutations in GLIS1 been linked to a specific disease (Nakashima et al., 2002). However, expression of GLIS1 was found to be elevated in psoriatic epidermis suggesting a possible role for GLIS1 in this pathology (Nakanishi et al., 2006). Table 1 shows a summary of the physiological functions of Glis proteins and their roles in disease.
Table 1.
Tissue-specific Physiological Roles of GLIS1–3 in Disease.
| Protein | Tissue | Pathology | Biological or Physiological Role | Ref. |
|---|---|---|---|---|
| Glis1 | Skin | Psoriasis | • Highly expressed in psoriatic epidermis | Nakanishi et at. 2006 |
| • Promotes epidermal differentiation | Nakanishi et at. 2006 | |||
| Glis2 | Kidney | NPHP | • Mutation associated with NPHP in humans | Attanasio et al. 2007 |
| • KO mice exhibit impaired renal function and renal atrophy, fibrosis, and cysts | Attanasio et al. 2007, Kim et al. 2008 | |||
| • Modulates expression of several EMT-related genes | Attanasio et al. 2007, Kim et al. 2008 | |||
| Neurons | Neuropathies | • Expressed in primary neurons and promotes neuronal differentiation in vertebrates | Lamar et al. 2001 | |
| Glis3 | Pancreas | Diabetes | • Identified as a risk locus for both type-1 and type-2 diabetes in humans | Barrett et al. 2009, Dupuis et al 2010 |
| • Mutation associated with NDH, characterized by neonatal diabetes, in humans | Senee et al 2006 | |||
| • KO mice exhibit hyperglycemia, hypoinsulinemia, and lack fully differentiated β-cells | Kang et al 2009b, Watanabe et al. 2009 | |||
| • KO mice exhibit dilated and cystic pancreatic ducts | Kang et al. 2009b | |||
| • Positively regulates the expression of insulin in mature β-cells | Kang et al. 2009b, Yang et al. 2009 | |||
| Thyroid | Hypothyroidism | • Mutation associated with NDH, characterized by hypothyroidism, in humans | Senee et al. 2006 | |
| Kidney | PKD | • Mutation associated with polycystic kidneys in humans | Senee et al. 2006 | |
| • KO mice exhibit renal cysts and dilation of tubules and collecting ducts | Kang et al. 2009a | |||
| • Mutation in the medaka ortholog linked to the development of PKD | Hashimoto et al. 2009 | |||
| Bone | Osteoporosis | • Promotes osteoblast and inhibits adipocyte differentiation in MSCs | Beak et al. 2007 | |
| • Enhances the expression of FGF18, a regulator of osteogenesis and chondrogenesis | Beak et al. 2007 | |||
| Tumors | Cancer | • Expressed in malignant ependymomas and renal carcinomas | Lukashova-v. Zangen et al. 2007, Yusenko et al. 2009 |
Glis1
Potential role of Glis1 in skin disease
In E15.5 mouse skin, Glis1 mRNA was expressed at high levels in the dermal cells surrounding the vibrissae and the first wave of pelage hairs (Kim et al., 2002; Nakashima et al., 2002). At PND1, Glis1 was weakly expressed in the ectodermal cells at the base of the hair follicle and in the dermal papillae. Although GLIS1 is not expressed in normal human adult interfollicular epidermis, its expression is significantly induced in psoriatic epidermis and in mouse skin upon treatment with the tumor promoter phorbol-12-myristate-13-acetate (PMA) (Nakanishi et al., 2006). The expression of GLIS1 mRNA is restricted to the differentiated, suprabasal layers of psoriatic skin (Nakanishi et al., 2006). Similarly, GLIS1 expression is induced in cultured normal human epidermal keratinocytes (NHEK) upon PMA- or interferon γ-induced differentiation, but not in squamous carcinoma cells that are unable to undergo differentiation. Overexpression of GLIS1 in NHEK cells enhanced the expression of several markers of epidermal differentiation, including S100A9, KLK7, small proline-rich protein (SPRR or cornifin), involucrin, and transglutaminase 1, many of which have been reported to be increased in psoriatic skin (Iizuka et al., 2004). These observations suggest that increased expression of GLIS1 promotes certain aspects of epidermal differentiation and may have a role in psoriasis.
Glis2
Glis2 is critical in maintaining normal renal functions
Nephronophthisis (NPHP), an autosomal recessive disease, is the most common genetic cause of end-stage renal disease (ESRD) in the first 30 years of life (Attanasio et al., 2007; Hildebrandt et al., 2009). The disorder is relatively rare with about 1 patient per million affected in the USA. NPHP is characterized by tubular atrophy, fibrosis, and in some instances renal cyst formation (Fig. 3). Mutations in the GLIS2 gene have been recently linked to NPHP (Attanasio et al., 2007). In three affected individuals examined, a homozygous transversion within the GLIS2 gene was identified leading to the loss of the 5′-splice donor site of exon 5 and as a result the synthesis of a nonfunctional protein. To date, mutations in nine different genes (NPHP1–9) have been implicated in NPHP accounting for about 35% of the NPHP cases (Hildebrandt et al., 2009). In this context, GLIS2 has also been referred to as NPHP7.
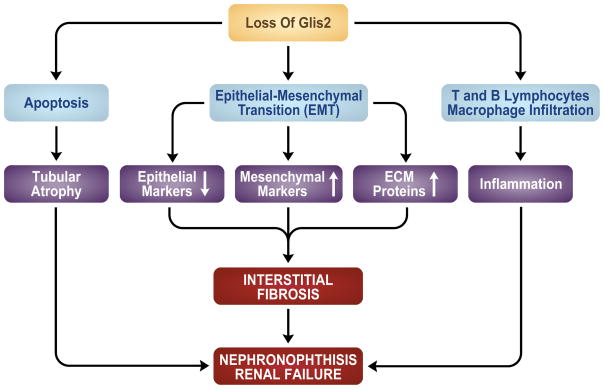
Figure 3.
Loss of Glis2 function leads to nephronophthisis. Loss of Glis2 function in humans and mice leads to renal atrophy and interstitial fibrosis, characteristics of nephronophthisis. This involves increased apoptosis of tubular epithelial cells, induction of epithelial-mesenchymal transition, and infiltration of macrophages and lymphocytes that ultimately results in end-stage renal disease.
Glis2-null mice have a normal appearance and exhibit normal behavior and motor coordination (Attanasio et al., 2007; Kim et al., 2008). However, Glis2-null mice progressively developed severe renal atrophy characterized by tubulo-interstitial fibrosis and reduced kidney size, the hallmarks of NPHP (Fig. 3). Moreover, kidneys of Glis2-null mice showed increased apoptosis of tubular epithelial cells and infiltration of inflammatory cells (Attanasio et al., 2007; Kim et al., 2008). At 6–12 months, severe glomerular abnormalities, including thickening of the basement membrane of Bowman’s capsule, and glomerular cysts, are observed. Loss of Glis2 leads to impaired kidney functions and end-stage renal disease as indicated by elevated levels of serum creatinine and blood urea nitrogen (BUN), and the development of proteinuria.
EMT connections with Glis2
Fibrosis is an important characteristic of NPHP (Hildebrandt et al., 2009). Interstitial fibrosis is caused by increased deposition of the extracellular matrix (ECM) by myofibroblasts that, in turn, can be the result of transformation of activated fibroblasts into myofibroblast, epithelial mesenchymal transition (EMT), and/or infiltration of fibrocytes (Zeisberg et al., 2004; Wada et al., 2007; Wynn, 2007). EMT, a process by which epithelial cells undergo transition into matrix-producing fibroblasts and myofibroblasts, is increasingly recognized as playing a critical role in the development of renal fibrosis (Liu, 2009). Gene expression profile analysis in Glis2-null mouse kidneys revealed that the expression of many EMT signature genes was enhanced, including transforming growth factor β (Tgfb), vimentin, Snail, Slug, connective tissue growth factor (Ctgf), and matrix metalloproteinase 14 (Mmp14), suggesting that loss of Glis2 promotes EMT (Attanasio et al., 2007; Kim et al., 2008)(Fig. 3). Conversely, Glis2 appears to act as a repressor of EMT and the expression of EMT-related genes in tubule epithelial cells. This concept is consistent with the expression pattern of Glis2; expression of Glis2 was observed in renal tubule and Bowman’s capsule epithelial cells, but not in endothelial and mesenchymal cells. Whether Glis2 regulates the EMT signature genes directly or whether the induction of EMT is indirectly related to the loss of Glis2, requires further study.
In addition to EMT-related genes, the expression of a number of inflammatory genes was significantly induced in Glis2-null kidneys and accompanied by increased infiltration of macrophages and lymphocytes (Attanasio et al., 2007; Kim et al., 2008)(Fig. 3). These results suggest that inflammation may be part of the progression of the NPHP phenotype in Glis2-null mice.
Glis3
Loss of GLIS3 function leads to development of NDH
Genetic aberrations in the GLIS3 gene have been recently associated with a rare syndrome characterized by neonatal diabetes and congenital hypothyroidism (NDH) (Senee et al., 2006). Additional evidence for a role of GLIS3 in diabetes comes from genome-wide association studies identifying the GLIS3 gene as a locus affecting the risk of type 1 and type 2 diabetes (Barrett et al., 2009; Boesgaard et al., 2010; Dupuis et al., 2010). NDH patients develop hyperglycemia and hypoinsulinemia, elevated blood levels of thyroid stimulating hormone (TSH), and reduced levels of thyroid hormones T3 and T4. Furthermore, depending on the nature of the mutation, NDH may be accompanied by facial dysmorphology, polycystic kidney disease, hepatic fibrosis, glaucoma, and mild mental retardation (Taha et al., 2003; Senee et al., 2006). One family of NDH patients (NDH1) harbored a homozygous insertion (2067insC) that resulted in a frame shift and the expression of a truncated, inactive GLIS3 protein. These patients had the most severe abnormalities and a greatly reduced lifespan (from 10 days to 1.5 years). However, in patients with an incomplete syndrome (NDH2 and NDH3), the deletion does not affect the coding region of GLIS3, but causes the apparent loss of a critical regulatory region and consequently in a greatly diminished GLIS3 expression (Senee et al., 2006). These studies, together with evidence that GLIS3 is expressed in the pancreatic β-cells and ducts and in the renal tubules and collecting ducts in the kidney, suggest that GLIS3 is part of a transcription network controlling pancreatic and renal functions.
As observed in NDH patients, mice with disrupted Glis3 function developed neonatal diabetes, hypothyroidism, and polycystic kidney disease (Kang et al., 2009a; Kang et al., 2009b; Watanabe et al., 2009). Glis3-null pups were hyperglycemic and died prematurely within 10 days after birth. Although Glis3 pups are smaller, they do not exhibit any gross anatomical abnormalities.
Role of Glis3 in pancreatic β-cell development
Histological analysis revealed that there was no significant difference in the size of the pancreas between Glis3-null and wild type (WT) mice; however, the islets were greatly reduced in size and morphologically distinct in the null mice (Kang et al., 2009b; Watanabe et al., 2009). The reduced islet size is likely largely due to the decline in the number of fully differentiated β-cells as indicated by the low levels of insulin, Pdx1, MafA, and Glut2 positive cells. This loss in β-cells is likely responsible for the development of neonatal diabetes in the Glis3-null mice.
The absence of pancreatic β-cells observed in Glis3-null mice was not due to increased apoptosis, but related to a defect in β-cell generation (Kang et al., 2009b; Watanabe et al., 2009). These observations suggest that Glis3 plays a critical role in cell lineage specification particularly in the development of β-cells. Lineage determination in the pancreas is regulated by several transcriptional networks and signaling pathways (Gradwohl et al., 2000; Gu et al., 2002; Murtaugh and Melton, 2003; Jensen, 2004; Jorgensen et al., 2007; Oliver-Krasinski and Stoffers, 2008; Gittes, 2009). Early multipotent pancreatic progenitors, marked by the expression of Pdx1, Ptf1a, Nkx2.2, Sox9, Hnf1β, and Cpa1, are the source of all differentiated cells of the exocrine, ductal, and endocrine lineages (Zhou et al., 2007; Burlison et al., 2008)(Fig. 2). Between E11.5–13.5, as the pancreas undergoes extensive branching morphogenesis, Sox9+HNF1β+ precursors in the trunk compartment give rise to exocrine, endocrine, and ductal lineages, while between E13.5-E16.5, during the so-called secondary transition, the Sox9+HNF1β+ precursors are only able to generate endocrine and ductal lineages, but not acinar cells (Solar et al., 2009). The differentiation of these precursors into pro-endocrine-progenitors is marked by the induction of neurogenin 3 (Ngn3). Loss of Glis3 function does not appear to affect the early pancreatic progenitors; however, between E13.5 and E16.5, it greatly diminishes the number of Ngn3-positive cells (Kang et al., 2009b). Given the important role of Ngn3 in the generation of pro-endocrine progenitors, these results suggest that Glis3 may regulate the transition of embryonic Sox9+Hnf1β+ duct precursors into endocrine progenitors and/or the maintenance of endocrine progenitors (Fig. 2). Future studies are required to determine the role of Glis3 in the transcriptional network that regulates β-cell development.
Similar to β-cells, the number of somatostatin-positive γ-cells was also diminished in Glis3-null pancreas, whereas that of glucagon-positive α-cells was only slightly affected. The development of pancreatic acini and the expression of acini markers were normal in Glis3-null mice (Kang et al., 2009b; Watanabe et al., 2009). However, as observed in renal tubules, pancreatic ducts were dilated and formed cysts, suggesting that Glis3 also plays a role in regulating the function and maintenance of ductal epithelial cells (Kang et al., 2009b). The latter is consistent with the expression of Glis3 in pancreatic ducts.
Glis3-mediated insulin regulation
In addition to its role in the generation of β-cells, Glis3 has been shown to positively regulate the expression of the insulin gene in mature β-cells (Senee et al., 2006; Kang et al., 2009b; Yang et al., 2009). Thus, Glis3 has dual role in regulating both the development and maintenance of mature β-cells, as been reported for several other transcription factors, such as Pdx1 (Ahlgren et al., 1998; Holland et al., 2005). Knockdown of Glis3 in rat insulinoma INS1 (832/13) cells by corresponding siRNAs reduced the expression of endogenous insulin 2 (Ins2), while over-expression of murine Glis3 increased Ins2 (Kang et al., 2009b; Yang et al., 2009). The transcriptional regulation and β-cell specific expression of the human insulin (INS) and murine Ins2 genes have been reported to involve a 600 bp regulatory region upstream of the transcriptional start site (Ohneda et al., 2000; Melloul et al., 2002). Pdx1, β2/NeuroD1, and MafA through their binding to A, E, and C-boxes within this region, are key activators of insulin gene expression. These transcriptional activator complexes interact with each other to synergistically activate insulin gene expression (Peers et al., 1994; Peshavaria et al., 1997; Ohneda et al., 2000). The proximal regulatory regions of the mouse Ins2 and human INS genes contain two Glis-BS sequences that can bind Glis3 efficiently. Optimal activation of the mouse Ins2 and human INS promoters by Glis3 was found to require both Glis-BS sequences (Kang et al., 2009b; Yang et al., 2009). Furthermore, a Glis3 mutant containing the NDH1 mutation that results in the loss of the Glis3 transactivation domain at the C-terminus, was unable to activate the Ins2 promoter. A Glis3 mutant containing a N-terminal truncation of 300 amino acids was a significantly more potent activator of the Ins2 promoter than full-length Glis3 suggesting the presence of a N-terminal regulatory domain. Glis3 was also found to enhance the expression of MafA and to interact with Pdx1, MafA, and NeuroD1 complexes (Yang et al., 2009). These interactions may help to stabilize the Glis3-DNA binding complex and enhance transcriptional up-regulation of the Ins gene. Given the critical regulatory role of Glis3 in β-cell generation and Ins expression, the Glis3 pathway might offer a potential target and opportunities for the development of new therapeutic strategies in the treatment of diabetes.
Roles of Glis3 in Kidney
As observed in NDH1 patients, Glis3-null mice develop autosomal recessive polycystic kidney disease (Taha et al., 2003; Senee et al., 2006). At PND3, Glis3-null mice show major cysts arising from glomeruli and extensive dilation of renal tubules and collecting ducts. Dilation of Bowman’s spaces in Glis3-null kidneys can be first observed around E14.5 to E15.5 of kidney development (Hashimoto et al., 2009; Kang et al., 2009a). This phenotype becomes more prominent at later stages of development.
Recently, a mutation in the medaka pc gene, an ortholog of mammalian Glis3, has also been linked to the development of polycystic kidney disease in the medaka pc mutant (Hashimoto et al., 2009). This mutant contains an insertion of a large transposon in the 4th intron causing alternative splicing and expression of a truncated pc/Glis3 protein lacking the normal C-terminus, including the last four ZFs. This suggests that the critical role of Glis3 in regulating normal kidney functions is well conserved across species.
Glis3 in mesenchymal stem cell differentiation
Glis3 is highly expressed in human osteoblasts and certain osteosarcoma cell lines, and is induced during osteoblast differentiation (Beak et al., 2007). Glis3 has been reported to promote osteoblast differentiation in multipotent mesenchymal C3H10T1/2 cells in synergy with bone morphogenic protein 2 (BMP2) and Shh as measured by the induction of alkaline phosphatase activity and osteocalcin (OCN) and osteopontin (OPN) mRNA expression. In contrast, Glis3 expression inhibits adipocyte differentiation in multipotent C3H10T1/2 cells. Gene profiling analysis revealed that Glis3 enhanced the expression of several additional genes, including fibroblast growth factor 18 (Fgf18), a regulator of osteogenesis and chondrogenesis (Liu et al., 2002; Beak et al., 2007). EMSA and reporter gene analysis further revealed that Glis3 regulates the Fgf18 gene directly (Beak et al., 2007). Two putative Glis-BS were identified in the upstream promoter region of Fgf18, one of which, located at −1406, was shown to mediate the transactivation by Glis3. These observations are consistent with the positive regulatory role of Glis3 in osteoblast differentiation. The latter suggests that Glis3 might provide a new therapeutic target for the treatment of osteoporosis.
Role of the primary cilium in the Glis signaling pathway
The mechanisms by which Glis2- and Glis3-deficiencies lead to NPHP and polycystic kidney disease are not yet fully understood. However, there are several indications of a relationship between Glis function, the primary cilium, and renal cystic disease. First, both Glis2 and Glis3 have been reported to localize to the primary cilium (Attanasio et al., 2007; Hashimoto et al., 2009; Kang et al., 2009a). Interestingly, the closely related Gli proteins also localize to the primary cilium which has been proven indispensable for at least part of Shh-Gli signaling (Corbit et al., 2005; Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005; Rohatgi et al., 2007; Kiprilov et al., 2008). Second, deficiency in Glis2 and Glis3 function lead to NPHP and polycystic kidney disease (PKD), which belong to a group of disorders with abnormalities in the structure and/or function of the primary cilium, referred to as ciliopathies (Saunier et al., 2005; Bisgrove and Yost, 2006; Torres and Harris, 2006; Fliegauf et al., 2007; Hildebrandt et al., 2009). Many of the proteins implicated in NPHP and PKD are linked to ciliary function and localize to the primary cilia, basal body and/or centrosome. These observations strongly suggested that the primary cilium plays a critical role in regulating the activity and function of Glis proteins.
Primary cilia are immotile hair-like structures that extend from the cell surface into the extracellular space (Bisgrove and Yost, 2006; Fliegauf et al., 2007; Berbari et al., 2009; Gerdes et al., 2009; Veland et al., 2009). The primary cilium consists of an axoneme formed by 9 doublet microtubules surrounded by a specialized membrane that is continuous with the plasma membrane but is enriched for certain specific receptors and channels. Ciliary proteins are assembled into transport rafts at the base of the cilium and are transported in and out the primary cilium along the length of the axoneme by intraflagellar transport (IFT) (Rosenbaum and Witman, 2002; Bisgrove and Yost, 2006; Fliegauf et al., 2007; Berbari et al., 2009; Gerdes et al., 2009; Veland et al., 2009). The primary cilium functions as a sensory organelle that processes a variety of mechano-, chemo-, photo-, and osmosensors, as well as Shh-, Wnt-, and PDGFα-dependent signaling pathways (Oro, 2007; Rohatgi et al., 2007; Corbit et al., 2008; Veland et al., 2009). Both the formation of the primary cilium and the control of cilium-associated signaling pathways are dependent on IFT. Although it has not yet been demonstrated, the localization of Glis2 and Glis3 to the primary cilium is likely dependent on a functional IFT. Although the percentage of renal tubule epithelial cells with a primary cilium was decreased in mice with disrupted Glis2 or Glis3 function, many cells were still able to form a normal primary cilium. These observations suggest that Glis2 or Glis3 are not essential for the formation of the primary cilium structure (Kim et al., 2008; Hashimoto et al., 2009; Kang et al., 2009a).
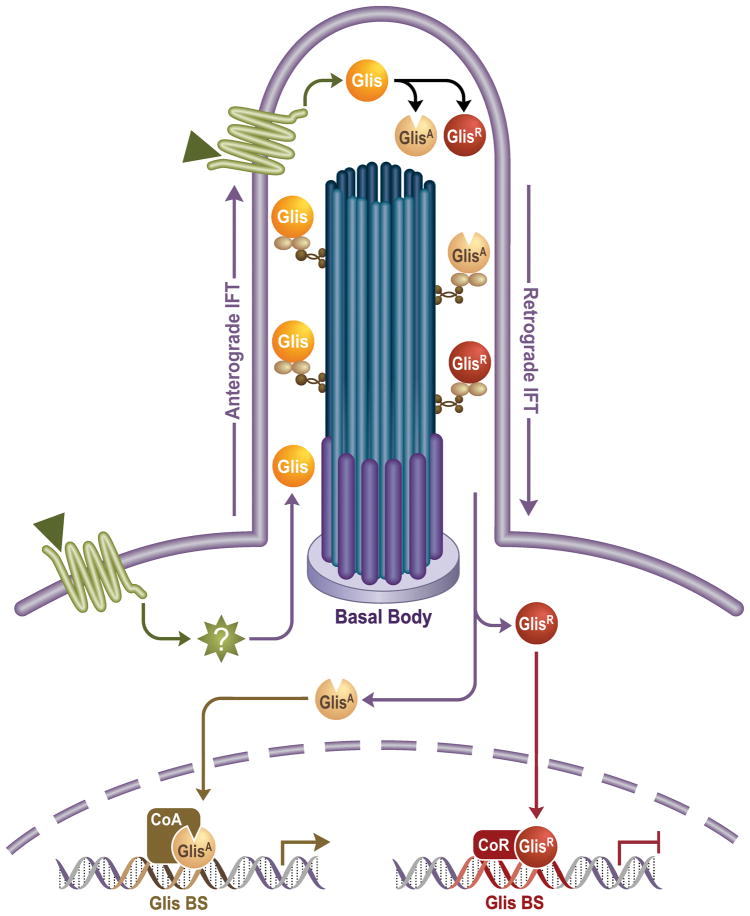
Based on their localization to the primary cilium, it is likely that Glis proteins are part of a primary cilium-associated signaling pathway as has been demonstrated for Shh-Gli (Rohatgi et al., 2007; Hashimoto et al., 2009; Attanasio et al., 2007; Kang et al., 2009a; Kang et al., 2009b). Comparable to models of the primary cilium-associated Gli pathways (Haycraft et al., 2005; Oro, 2007; Rohatgi et al., 2007; Kim et al., 2009), activation of a membrane-bound protein (e.g. G-protein-coupled receptors) by an external signal (e.g., protein or chemo-, osmo-, or mechanosensors) may control Glis accumulation in the primary cilium and the activation of Glis proteins through phosphorylation and/or proteolytic processing (Fig. 4). Upon activation and their subsequent translocation into the nucleus, Glis proteins repress or activate the transcription of target genes (Kang et al., 2009a). The identification of repressor and activation domains in Glis proteins, the greatly increased transactivation activity of N-terminal truncated Glis3, and the reported proteolytic processing of Glis2 are consistent with this model (Kim et al., 2002; Zhang et al., 2002; Kim et al., 2003; Hosking et al., 2007; Kang et al., 2009b). Future studies are required to identify the various molecular components that are part of these exciting new signaling pathways.
Figure 4.
Schematic representation of the role of the primary cilium in Glis signaling. The localization of Glis proteins to the primary cilium suggests that these proteins are part of a primary cilium-associated signaling pathway. A hypothetical upstream signal (e.g., peptide, mechano-, osmo, or chemosensor)(triangle) through plasma membrane-associated receptors (e.g., G-protein coupled receptors) may regulate the primary cilium localization and activity of Glis. Intraflagellar transport (IFT) proteins may direct anterograde and retrograde movement of Glis proteins within the primary cilium. Upon activation, Glis proteins may undergo post-translational modifications or proteolytic processing resulting in modified Glis proteins that function as transcriptional activators (GlisA) or repressors (GlisR). After their translocation to the nucleus, Glis proteins regulate the transcription of target genes through interactions with co-activators (CoA) or co-repressors (CoR). Glis-BS, Glis-binding site.
The molecular mechanisms underlying cyst formation are not yet completely understood. Several studies have indicated a connection between cyst formation, planar cell polarity (PCP), and the primary cilium (Torres and Harris, 2006; Bacallao and McNeill, 2009; Fischer and Pontoglio, 2009; Hildebrandt et al., 2009). PCP is defined as the polarization of cells perpendicular to their apical-basal axis and has been implicated in the regulation of the cell division axis. During elongation, renal tubules cells undergo oriented cell division (OCD) in which most cells divide along the axis of the tubule. In cystic disease, cells undergo random cell division thereby leading to dilation of the tubules and subsequently cysts (Kolb and Nauli, 2008; Fischer and Pontoglio, 2009; McNeill, 2009). Loss of primary cilia or ciliary function lead to defective PCP and misoriented cell division suggesting that primary cilia play a critical role in regulating PCP and OCD. However, the precise molecular mechanisms that connect the primary cilium with OCD, are not yet clear and await further study. The formation of cysts in Glis2-null and Glis3-null mice might also be due to defective PCP and misoriented cell division. Thus, Glis2 and Glis3 could be part of a link between primary cilium function and OCD.
Conclusions
Glis transcription factors play a key role in the regulation of a variety of physiological processes and have been implicated in several pathologies. Glis3 is essential for the development of pancreatic β-cells and is critical in the regulation of insulin expression, while both Glis2 and Glis3 are important for the maintenance of normal renal functions. Mutations in GLIS2 have been implicated in nephronophthisis, while mutations in GLIS3 have been linked to the development of neonatal diabetes, hypothyroidism, and polycystic kidney disease. In addition, elevated overexpression of GLIS3 in various cancer cells suggests a role for GLIS3 in tumorigenesis.
Further analysis is required to determine the molecular mechanisms by which Glis transcription factors regulate gene expression. The identification of additional Glis target genes will not only aid in the understanding of the molecular mechanisms of these proteins, but will also provide insights into their physiological functions in the pancreas and kidney, and their roles in disease. The association of Glis proteins with the primary cilium suggests that these proteins are part of a primary cilium-associated signaling cascade. The latter is consistent with studies showing a connection between primary cilium and cystic kidney disease. Elucidation of the Glis signaling pathways may provide new opportunities for the development of therapeutic strategies in the treatment of diabetes, cancer, and cystic renal disease.
Acknowledgments
The authors would like to thank Drs. Christina Teng and Thomas Eling for their comments on the manuscript and Sue Edelstein for the artwork. This research was supported by the Intramural Research Program of the NIEHS, NIH (Z01-ES-100485).
References
- Ackermann AM, Gannon M. Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. J Mol Endocrinol. 2007;38:193–206. doi: 10.1677/JME-06-0053. [DOI] [PubMed] [Google Scholar]
- Agata Y, Matsuda E, Shimizu A. Two novel Kruppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1beta/KRIP-1) J Biol Chem. 1999;274:16412–16422. doi: 10.1074/jbc.274.23.16412. [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman VM, Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J Biol Chem. 1996;271:1043–1047. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- Aruga J, Yokota N, Hashimoto M, Furuichi T, Fukuda M, Mikoshiba K. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem. 1994;63:1880–1890. doi: 10.1046/j.1471-4159.1994.63051880.x. [DOI] [PubMed] [Google Scholar]
- Attanasio M, Uhlenhaut NH, Sousa VH, O’Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, Seelow D, Nürnberg G, Becker C, Chudley AE, Nürnberg P, Hildebrandt F, Treier M. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Kornberg TB. Ci: a complex transducer of the hedgehog signal. Trends Genet. 1999;15:458–462. doi: 10.1016/s0168-9525(99)01869-7. [DOI] [PubMed] [Google Scholar]
- Bacallao RL, McNeill H. Cystic kidney diseases and planar cell polarity signaling. Clin Genet. 2009;75:107–117. doi: 10.1111/j.1399-0004.2008.01148.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS The Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009 [Google Scholar]
- Beak JY, Kang HS, Kim YS, Jetten AM. Kruppel-like zinc finger protein Glis3 promotes osteoblast differentiation by regulating FGF18 expression. J Bone Miner Res. 2007;22:1234–1244. doi: 10.1359/jbmr.070503. [DOI] [PubMed] [Google Scholar]
- Beak JY, Kang HS, Kim YS, Jetten AM. Functional analysis of the zinc finger and activation domains of Glis3 and mutant Glis3(NDH1) Nucleic Acids Res. 2008;36:1690–1702. doi: 10.1093/nar/gkn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Boesgaard TW, Grarup N, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O. Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia. doi: 10.1007/s00125-010-1753-5. In Press. [DOI] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP family proteins: more than transcriptional corepressors. Bioessays. 2003;25:9–12. doi: 10.1002/bies.10212. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA. Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat (Basel) 1996;156:187–201. doi: 10.1159/000147846. [DOI] [PubMed] [Google Scholar]
- Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128:4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I DIAGRAM Consortium GIANT Consortium, Global BPgen Consortium, Wilson J.F. Anders Hamsten on behalf of Procardis Consortium MAGIC investigators. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010 doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E, Pontoglio M. Planar cell polarity and cilia. Semin Cell Dev Biol. 2009;20:998–1005. doi: 10.1016/j.semcdb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- Gavert N, Ben-Ze’ev A. beta-Catenin signaling in biological control and cancer. J Cell Biochem. 2007;102:820–828. doi: 10.1002/jcb.21505. [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill PS, Rosenblum ND. Control of murine kidney development by sonic hedgehog and its GLI effectors. Cell Cycle. 2006;5:1426–1430. doi: 10.4161/cc.5.13.2928. [DOI] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Miyamoto R, Watanabe N, Shiba D, Ozato K, Inoue C, Kubo Y, Koga A, Jindo T, Narita T, Naruse K, Ohishi K, Nogata K, Shin-I T, Asakawa S, Shimizu N, Miyamoto T, Mochizuki T, Yokoyama T, Hori H, Takeda H, Kohara Y, Wakamatsu Y. Polycystic kidney disease in the medaka (Oryzias latipes) pc mutant caused by a mutation in the Gli-Similar3 (Glis3) gene. PLoS One. 2009;4:e6299. doi: 10.1371/journal.pone.0006299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto N, Kido Y, Uchida T, Asahara S, Shigeyama Y, Matsuda T, Takeda A, Tsuchihashi D, Nishizawa A, Ogawa W, Fujimoto Y, Okamura H, Arden KC, Herrera PL, Noda T, Kasuga M. Ablation of PDK1 in pancreatic beta cells induces diabetes as a result of loss of beta cell mass. Nat Genet. 2006;38:589–593. doi: 10.1038/ng1774. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54:2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Hong JH, Yaffe MB. TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle. 2006;5:176–179. doi: 10.4161/cc.5.2.2362. [DOI] [PubMed] [Google Scholar]
- Hosking CR, Ulloa F, Hogan C, Ferber E, Figueroa A, Gevaert K, Birchmeier W, Briscoe J, Fujita Y. The Transcriptional Repressor Glis2 Is a Novel Binding Partner for p120 Catenin. Mol Biol Cell. 2007 doi: 10.1091/mbc.E06-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Iizuka H, Takahashi H, Honma M, Ishida-Yamamoto A. Unique keratinization process in psoriasis: late differentiation markers are abolished because of the premature cell death. J Dermatol. 2004;31:271–276. doi: 10.1111/j.1346-8138.2004.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Kang HS, Beak JY, Kim YS, Herbert R, Jetten AM. Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol Cell Biol. 2009a;29:2556–2569. doi: 10.1128/MCB.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, Sosa-Pineda B, Jensen J, Foley J, Jetten AM. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009b;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Kim YS, Jetten AM. Kruppel-like zinc finger protein Gli-similar 2 (Glis2) represses transcription through interaction with C-terminal binding protein 1 (CtBP1) Nucleic Acids Res. 2005;33:6805–6815. doi: 10.1093/nar/gki985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kang HS, Herbert R, Beak JY, Collins JB, Grissom SF, Jetten AM. Kruppel-like zinc finger protein Glis2 is essential for the maintenance of normal renal functions. Mol Cell Biol. 2008;28:2358–2367. doi: 10.1128/MCB.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kang HS, Jetten AM. The Kruppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/beta-catenin signaling pathway. FEBS Lett. 2007;581:858–864. doi: 10.1016/j.febslet.2007.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J Biol Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- Kim YS, Nakanishi G, Lewandoski M, Jetten AM. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–5525. doi: 10.1093/nar/gkg776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprilov EN, Awan A, Desprat R, Velho M, Clement CA, Byskov AG, Andersen CY, Satir P, Bouhassira EE, Christensen ST, Hirsch RE. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- Kolb RJ, Nauli SM. Ciliary dysfunction in polycystic kidney disease: an emerging model with polarizing potential. Front Biosci. 2008;13:4451–4466. doi: 10.2741/3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JS, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- Lessing D, Nusse R. Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched- mediated repression. Development. 1998;125:1469–1476. doi: 10.1242/dev.125.8.1469. [DOI] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Liu Y. New Insights into Epithelial-Mesenchymal Transition in Kidney Fibrosis. J Am Soc Nephrol. 2009;21:212–222. doi: 10.1681/ASN.2008121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashova-v Zangen I, Kneitz S, Monoranu CM, Rutkowski S, Hinkes B, Vince GH, Huang B, Roggendorf W. Ependymoma gene expression profiles associated with histological subtype, proliferation, and patient survival. Acta Neuropathol (Berl) 2007;113:325–337. doi: 10.1007/s00401-006-0190-5. [DOI] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, Nakagawa O, Small EV, Cobo-Stark P, Igarashi P, Murakami M, Tominaga J, Sato T, Asano T, Kurihara Y, Kurihara H. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol. 2008;294:F542–553. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Gli genes in development and cancer. Oncogene. 1999;18:7852–7859. doi: 10.1038/sj.onc.1203243. [DOI] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- McNeill H. Planar cell polarity and the kidney. J Am Soc Nephrol. 2009;20:2104–2111. doi: 10.1681/ASN.2008111173. [DOI] [PubMed] [Google Scholar]
- Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276:2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC. Pancreas and beta-cell development: from the actual to the possible. Development. 2007;134:427–438. doi: 10.1242/dev.02770. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Melton DA. Genes, signals, and lineages in pancreas development. Annu Rev Cell Dev Biol. 2003;19:71–89. doi: 10.1146/annurev.cellbio.19.111301.144752. [DOI] [PubMed] [Google Scholar]
- Nakanishi G, Kim YS, Nakajima T, Jetten AM. Regulatory role for Kruppel-like zinc-finger protein Gli-similar 1 (Glis1) in PMA-treated and psoriatic epidermis. J Invest Dermatol. 2006;126:49–60. doi: 10.1038/sj.jid.5700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Tanese N, Ito M, Auerbach W, Bai C, Furukawa T, Toyono T, Akamine A, Joyner AL. A novel gene, GliH1, with homology to the Gli zinc finger domain not required for mouse development. Mech Dev. 2002;119:21. doi: 10.1016/s0925-4773(02)00291-5. [DOI] [PubMed] [Google Scholar]
- Ohneda K, Ee H, German M. Regulation of insulin gene transcription. Semin Cell Dev Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro AE. The primary cilia, a ‘Rab-id’ transit system for hedgehog signaling. Curr Opin Cell Biol. 2007;19:691–696. doi: 10.1016/j.ceb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Peers B, Leonard J, Sharma S, Teitelman G, Montminy MR. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJI. Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000;275:18000–18010. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- Perantoni AO. Renal development: perspectives on a Wnt-dependent process. Semin Cell Dev Biol. 2003;14:201–208. doi: 10.1016/s1084-9521(03)00022-3. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Fuchs E. Catenins: keeping cells from getting their signals crossed. Dev Cell. 2006;11:601–612. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshavaria M, Henderson E, Sharma A, Wright CV, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncelet DA, Bellefroid EJ, Bastiaens PV, Demoitie MA, Marine JC, Pendeville H, Alami Y, Devos N, Lecocq P, Ogawa T, Muller M, Martial JA. Functional analysis of ZNF85 KRAB zinc finger protein, a member of the highly homologous ZNF91 family. DNA Cell Biol. 1998;17:931–943. doi: 10.1089/dna.1998.17.931. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Manning DR. Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem J. 2007;403:369–379. doi: 10.1042/BJ20061723. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sanchez P, Dahmane N. GLI and hedgehog in cancer: tumours, embryos and stem cells. Nature Rev. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, Seuanez HN, O’Brien SJ, Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai-Kato K, Umezawa Y, Mikoshiba K, Aruga J, Utsunomiya-Tate N. Stability of folding structure of Zic zinc finger proteins. Biochem Biophys Res Commun. 2009;384:362–365. doi: 10.1016/j.bbrc.2009.04.151. [DOI] [PubMed] [Google Scholar]
- Saunier S, Salomon R, Antignac C. Nephronophthisis. Curr Opin Genet Dev. 2005;15:324–331. doi: 10.1016/j.gde.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Senee V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, Bougnères P, Taha D, Julier C. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- Solar M, Cardalda C, Houbracken I, Martin M, Maestro MA, De Medts N, Xu X, Grau V, Heimberg H, Bouwens L, Ferrer J. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Taha D, Barbar M, Kanaan H, Williamson Balfe J. Neonatal diabetes mellitus, congenital hypothyroidism, hepatic fibrosis, polycystic kidneys, and congenital glaucoma: a new autosomal recessive syndrome? Am J Med Genet A. 2003;122A:269–273. doi: 10.1002/ajmg.a.20267. [DOI] [PubMed] [Google Scholar]
- Torres VE, Harris PC. Mechanisms of Disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol. 2006;2:40–55. doi: 10.1038/ncpneph0070. quiz 55. [DOI] [PubMed] [Google Scholar]
- van Hengel J, van Roy F. Diverse functions of p120ctn in tumors. Biochim Biophys Acta. 2007;1773:78–88. doi: 10.1016/j.bbamcr.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 2009;111:39–53. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A, Gessler M, Grzeschik KH. Identification of optimized target sequences for the GLI3 zinc finger protein. DNA Cell Biol. 1995;14:629–634. doi: 10.1089/dna.1995.14.629. [DOI] [PubMed] [Google Scholar]
- Wada T, Sakai N, Matsushima K, Kaneko S. Fibrocytes: a new insight into kidney fibrosis. Kidney Int. 2007;72:269–273. doi: 10.1038/sj.ki.5002325. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Hiramatsu K, Miyamoto R, Yasuda K, Suzuki N, Oshima N, Kiyonari H, Shiba D, Nishio S, Mochizuki T, Yokoyama T, Maruyama S, Matsuo S, Wakamatsu Y, Hashimoto H. A murine model of neonatal diabetes mellitus in Glis3-deficient mice. FEBS Lett. 2009;583:2108–2113. doi: 10.1016/j.febslet.2009.05.039. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Khachigian LM, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J Biol Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117:524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chang BH, Samson SL, Li MV, Chan L. The Kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–2538. doi: 10.1093/nar/gkp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, McMahon AP, Valerius MT. Recent genetic studies of mouse kidney development. Curr Opin Genet Dev. 2004;14:550–557. doi: 10.1016/j.gde.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Yusenko MV, Kovacs G. Identifying CD82 (KAI1) as a marker for human chromophobe renal cell carcinoma. Histopathology. 2009;55:687–695. doi: 10.1111/j.1365-2559.2009.03449.x. [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Kalluri R. The role of epithelial-to-mesenchymal transition in renal fibrosis. J Mol Med. 2004;82:175–181. doi: 10.1007/s00109-003-0517-9. [DOI] [PubMed] [Google Scholar]
- Zhang F, Jetten AM. Genomic structure of the gene encoding the human GLI-related, Kruppel-like zinc finger protein GLIS2. Gene. 2001;280:49–57. doi: 10.1016/s0378-1119(01)00764-8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Nakanishi G, Kurebayashi S, Yoshino K, Perantoni A, Kim YS, Jetten AM. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcription factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J Biol Chem. 2002;277:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]