Abstract
The nature of the different kinesin family members that function in a single, specific neuron type has not been systematically investigated. Here, we used quantitative real-time PCR to analyze the developmental expression patterns of kinesin family genes in cultured mouse hippocampal neurons, a highly homogeneous population of nerve cells. For purposes of comparison, we also determined the set of kinesins expressed in embryonic and adult hippocampal tissue. Twenty kinesins are expressed at moderate-to-high levels in mature hippocampal cultures. These include 9 plus-end directed kinesins from the Kinesin-1, -2, and -3 families that are known to mediate organelle transport and 6 other members of the Kinesin-3 and -4 families that are candidate organelle motors. Hippocampal cultures express high levels of a Kinesin-13, which regulates microtubule depolymerization, and moderate-to-high levels of Kinesin-9 and -14 family members, whose functions are not understood. Twelve additional kinesins, including 10 known mitotic kinesins, are expressed at moderate levels in embryonic hippocampus but at very low levels in mature cultures and the adult hippocampus. Collectively, our findings suggest that kinesins subserve diverse functions within a single type of neuron.
Keywords: Kinesin, motor protein, gene expression, neuronal culture, hippocampus
Introduction
Kinesins are mechanoenzymes that use ATP hydrolysis to power their translocation along microtubule tracks. The human and mouse genomes contain more than 40 kinesin genes, which are categorized into 14 subfamilies based on phylogenetically conserved similarities in their motor domains and in other parts of the proteins (Lawrence et al. 2004; Miki et al. 2005). Most kinesins translocate toward the plus ends of microtubules, with the notable exception of members of the minus-end directed Kinesin-14 family, the only family where the motor domain resides at the C-terminal end of the proteins. In terms of their cellular functions, kinesins perform two major tasks. Some kinesins function principally during mitosis and meiosis, where they mediate the movements of microtubules and chromosomes that occur during metaphase and anaphase (Sharp et al. 2000; Zhu et al. 2005). Other kinesins function principally in the microtubule-mediated long-range transport of membranous organelles, RNA-protein complexes, and signaling complexes and their scaffolding proteins (Hirokawa and Takemura 2004a; Hirokawa and Takemura 2004b; Hirokawa and Takemura 2005; Hirokawa 2006). A few kinesins serve other functions. For example, some members of the Kinesin-13 family regulate microtubule depolymerization (Wordeman 2005; Moores and Milligan 2006), while some Kinesin-2 and -3 family members regulate intraciliary transport (Silverman and Leroux 2009).
The role of kinesins in fast organelle transport is particularly crucial in nerve cells, because of the long distances that separate the cell body from the distal regions of the dendrites and axon. In axons, where nearly all microtubules are oriented plus-end out (Baas et al. 1988), kinesins are responsible for all microtubule-based anterograde organelle transport. In dendrites, which contain microtubules of mixed orientation (Baas et al. 1988), plus-end directed kinesins could move cargoes in either direction. In accord with the particular importance of microtubule-based transport in neurons, the nervous system displays a great diversity of kinesin gene expression (Goldstein and Yang 2000; Miki et al. 2001; Miki et al. 2003; Hirokawa and Takemura 2005). Nineteen kinesins have been detected in adult brain by Northern blotting (Miki et al. 2001); of these, 8 are expressed predominantly or exclusively in the nervous system.
Does the diversity of kinesin expression reflect the remarkable neuronal heterogeneity within the nervous system or the diversity of roles played by different kinesins within a single population of neurons? To address this question, we investigated kinesin expression using RT-PCR in cultures derived from the embryonic hippocampus, which are highly enriched for a single type of neuron, hippocampal pyramidal cells. As cultured hippocampal neurons have become the accepted model for studying neuronal polarity, we were particularly interested in identifying the full complement of candidate organelle motors that might play a role in protein trafficking to axons and dendrites. Thus we focused on the following questions: Do hippocampal pyramidal neurons express most of the kinesins found in brain or only a small subset of these? Which kinesins show a pattern of expression similar to the well-characterized organelle motors in the Kinesin-1 -2, and -3 families? Is there a subset of kinesins whose expression increases in parallel with dendritic development, suggesting that they might encode motors that mediate dendritic transport?
We detected moderate-to-high levels of expression of mRNAs encoding 20 different kinesins in cultured hippocampal neurons. Of particular note are 15 members of the Kinesin-1, -2, and -3 -4 families that likely mediate organelle transport in axons or dendrites and one member of the Kinesin-13 family that induces microtubule depolymerization. One Kinesin-9 and three members of the Kinesin-14 family, which translocate to microtubule minus-ends but whose functions are unknown, are also expressed at moderate-to-high levels. Only two kinesins, KIF17 and KIFC2, displayed a significant increase in expression that paralleled dendritic development.
Experimental Methods
Hippocampal cell cultures and RNA extraction
Neuronal cultures were prepared from hippocampi dissected from the brains of embryonic day 16 (E16) C57/BL6 mice, as described by Kaech and Banker (Kaech and Banker 2006). Following trypsin treatment, the dissociated cells were plated into 60 mm Petri dishes containing five 18 mm coverslips that had been treated with poly-L-lysine; 300,000–500,000 cells were plated per dish. After the neurons had attached, the coverslips were transferred to dishes containing a monolayer of rat astroglia and maintained in Neurobasal Medium with B27 supplement. After 3 days in culture, cytosine arabinoside was added to reduce the proliferation of non-neuronal cells.
RNA was obtained from cultured hippocampal neurons at three stages of development—4, 7, and 14 days in vitro (DIV). To harvest RNA from neuronal cultures, the culture medium was removed and coverslips were extracted with lysis buffer (Qiagen, Valencia, CA). One aliquot of lysis buffer was used to extract RNA from multiple coverslips. For each data point, RNA was extracted from 25 to 35 coverslips, in a total volume of 600 μl. The RNA was then purified from the lysate using RNAeasy columns (Qiagen), according to the manufacturer’s directions. This method yielded 4–6 μg of total RNA per time point. Two or three independently prepared cultures were analyzed at each developmental stage. Kinesin expression was also measured in RNA prepared from hippocampi dissected from the brains of E16 mouse embryos (the starting material used for preparing neuronal cultures) and adult mice.
Total RNA was reverse transcribed using 2 μg oligo-dT, 200 units Superscript II Reverse Transcriptase, 1x First-Strand Buffer, 10 mM dithiothreitol and 0.5 mM dNTPs (all from Invitrogen, Carlsbad, CA) in a total volume 20 μl and incubated at 37°C for 60 minutes, then at 45°C for 30 minutes.
Immunostaining and Microscopy
We also assessed the cellular composition of the cultures used for RNA isolation by performing double-label, indirect immunofluorescence staining using appropriate cell-type specific markers (for details, see Kaech and Banker, 2006). A monoclonal antibody against MAP2 (clone AP-20, Sigma, St. Louis, MO, # M4403) was used as a neuronal marker and polyclonal anti-GFAP (DAKO, Carpinteria, CA, # Z0334) antibodies served as a marker for astrocytes. Following immunostaining, coverslips were mounted in medium containing 4′,6-diamidino-2-phenylindole (DAPI) at 50 μg/ml to stain nuclei. Low-magnification images of randomly selected fields (chosen using predetermined stage coordinates) were acquired using a Princeton Instruments Micromax cooled CCD camera controlled by Metamorph Software (Molecular Devices, Downingtown, PA). Every cell present within the field (identified by DAPI staining) was scored as MAP2-positive, GFAP-positive, or negative for both markers.
Primer and Probe Design
PCR primers and probes were designed using Primer Express V.1.0 software (Perkin Elmer, Waltham, MA) based on expressed sequence tags (EST) for mouse kinesin superfamily transcripts identified by Miki et al. (2003) or deposited on the Kinesin Home Page (http://www.cellbio.duke.edu/kinesin/). The ESTs were assigned to specific kinesin genes in the Entrez Gene database using a BLAST homology search. Sequences for primers and probes can be found in the supplemental tables S1 and S2. For the KIF1B gene, we probed for both of the well-characterized splice variants (KIF1Bα and 1Bβ), which encode functionally distinct proteins (Nangaku et al. 1994; Conforti et al. 1999; Gong et al. 1999). All kinesin fluorogenic probes contained a reporter dye (FAM) covalently attached at the 5′ end and a quencher dye (TAMRA) covalently attached at the 3′ end. 18S ribosomal RNA (Accession #K01364), the internal standard, was labeled by the reporter dye VIC.
Quantitative Real-Time PCR
Real-time PCR reactions were performed using the ABI-PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA). Each reaction was performed in triplicate in a total volume of 25 μl. 5 μl of a 1:25 dilution of cDNA from the reverse transcription reaction was combined with 300 nM forward and reverse primers and 100 nM probe specific for a given KIF, along with 100 nM 18S forward and reverse primers and 100 nM 18S probe and 1x Taqman universal PCR Master Mix (Applied Biosystems). All reactions were performed under the following thermal cycling conditions: cycle 1 – 2 min at 50 °C and 10 min at 95 °C; cycles 2 through 40 – 95 °C for 15 sec, followed by 60 °C for 1 min. Because of the large number of probe sets required to assay such a large number of different transcripts, we did not attempt to optimize individual PCR reactions for each.
Data Analysis and Statistics
ABI-PRISM software was used to calculate the cycle threshold (CT) for the target sequence and for 18S rRNA; the relative cycle threshold (ΔCT) is the difference between these values. The threshold for 18S rRNA varied between 15 and 17 cycles. Supplemental Table S3 shows relative cycle thresholds (Mean ± standard error) for 38 kinesin messages based on samples (assayed in triplicate) obtained from 2–3 independent experiments. Two probes (for KIF 6 and KIF20B) failed to give reliable data (not reported in Table S3). Transcripts for 3 kinesin genes, KIF4, KIF16B, and KIF26A, were detected at very low levels in all samples-– at least 1000-fold lower than transcripts of KIF5B. We do not illustrate data for these kinesins in the figures since we cannot rule out the possibility that they were not amplified efficiently using the standard primer selection and reaction conditions we employed.
Cycle threshold is inversely proportional to the copy number of the target template; the higher the template concentration, the lower the cycle threshold measured. Theoretically, amplification follows a log2 logarithmic scale, with each unit difference in CT corresponding to a 2-fold difference in message abundance (Johnson et al. 2000; Bustin et al. 2005). To simplify interpretation, we have converted relative cycle threshold values to relative abundance (2−ΔCT) and normalized these values to the abundance of the Kinesin-1 family member KIF5B in 14 DIV cultures, which was arbitrarily assigned a value of 10,000. The figures illustrate values of normalized relative abundance.
To determine those genes whose expression changed significantly during development in culture, we compared expression in E16 hippocampus (the starting material used to prepare cultures) and in hippocampal cultures at 4, 7, and 14 days in culture, using a mixed-effects analysis of variance. We also determined those kinesins whose level of expression differed significantly in E16 versus adult hippocampus. Mixed effect models were fit using R (Ihaka and Gentleman 1999) and the associated nlme package (Pinheiro et al. 2002). Resulting p-values for changes in expression level were adjusted using the MULTI program (Brown and Russel 1996), which applied the method of Schweder and Spjøtvoll (Schweder and Spjøtvoll 1982) to identify the number of genuinely significant effects.
Immunoblotting of KIFs
E18 hippocampi, hippocampal cultures at 4 DIV, 7 DIV, 14 DIV, and adult rat brain homogenate were lysed in RIPA buffer, which consists of 50 mM Tris HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, Complete Protease Inhibitor Cocktail EDTA-free (Roche, Mississauga, ON, Canada), and Halt™ Phosphatase Inhibitor Cocktail (Thermo Scientific, Rockford, IL). Protein concentrations were assessed using a BCA protein assay kit (Thermo Scientific). Rabbit polyclonal antibodies against KIF17 (Abcam), KIFC2 (Sigma), KIF5C, a kind gift from Larry Goldstein (UC-San Diego), and a mouse monoclonal antibody against tubulin (DM1a; Sigma) were used for immunoblot analysis. Secondary antibodies against mouse and rabbit IgGs coupled to horseradish peroxidase were from Bio-Rad (Hercules, CA).
Lysates were mixed with sample buffer and boiled for 5 min. Equal proteins amounts were separated by SDS-PAGE (8% gel) under reducing conditions, and transferred to a PVDF membrane (Bio-Rad). Membranes were blocked with 5% non-fat dry milk powder in Tris-buffered saline, pH 8.4 (50 mM Tris base and 150 mM NaCl) with 0.05% Tween 20 and then incubated with primary antibodies overnight at 4°C. Primary antibodies were detected with corresponding HRP-conjugated secondary antibodies and visualized using chemiluminescence (Thermo Scientific). Quantitative analysis of protein band intensities was performed using ImageJ software (National Institutes of Health, Bethesda, MD). Statistical significance of changes in protein expression was determined by use of a one-way ANOVA with a post-hoc Tukey-Kramer test in Prism 5.0 (GraphPad).
Results
Characterization of Mouse Hippocampal Cultures
Hippocampal cultures prepared according to the method used here consist principally of a single type of neuron, the hippocampal pyramidal cell (Benson et al. 1994). A variety of different interneurons are also present, but collectively they account for only about 6% of the neuronal population (Benson et al. 1994). To confirm the purity of the cultures used for our experiments, we immunostained coverslips taken from the same culture preparations used for RNA analysis using cell type-specific antibodies. Figure 1 shows an example of a 14 day-old culture stained with a nuclear dye (DAPI) to identify every cell present, and with markers for neurons (MAP2) and astrocytes (GFAP). The vast majority of cells in this field are neurons; one astrocyte and one other, unidentified non-neuronal cell (DAPI-positive, but unstained with either antibody) are also present. The table in Figure 1 quantifies the percentage of neurons and nonneuronal cells present in cultures at 4, 7, and 14 days in vitro (DIV). The cellular composition of the cultures changed little during development; at all stages, neurons accounted for 92–96% of all cells present. Astrocytes accounted for 2–3% and other types of non-neuronal cells (likely including fibroblasts and oligodendroglial progenitors) accounted for another 3%. Thus the cultures used for RNA analysis were highly homogeneous.
Figure 1.
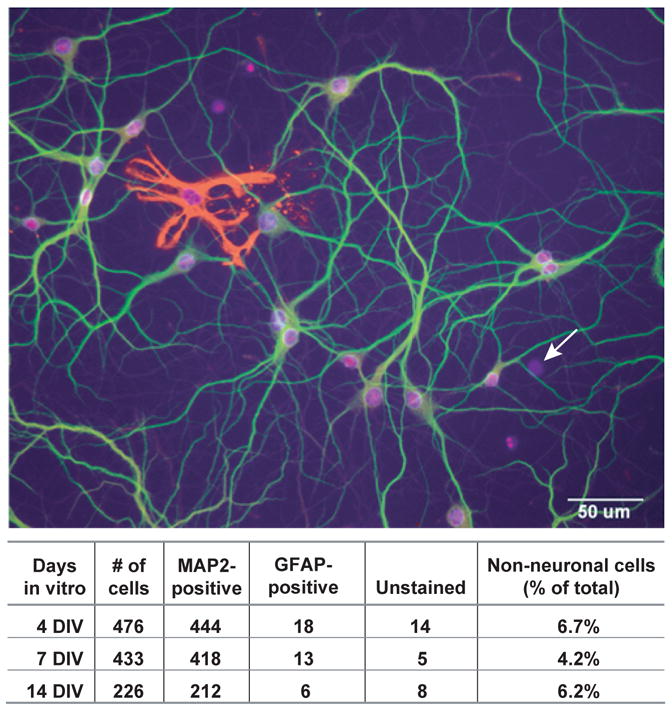
Hippocampal Cultures Consist Almost Exclusively of Neurons. Three-color image of a 14DIV hippocampal culture illustrating the method used to quantify cell types. The number of all cells in the field was determined by staining with DAPI, a DNA-binding dye that labels nuclei (blue). Neurons were identified as cells immunostained with antibodies against MAP2 (green). GFAP was used as a marker for astrocytes (red). Nuclei labeled with DAPI that were not associated with GFAP or MAP2 staining were categorized as unstained cells (arrow). This image was selected to contain both a GFAP-positive and an unstained cell; at this magnification most fields did not contain either. The table summarizes the quantification for the 3 cultures used for isolation of the RNA.
Expression of mRNAs Encoding Cytoskeletal Proteins and Cell-Type Specific Markers
Given the limited source material available from primary neuronal cultures, only a PCR-based strategy was feasible for simultaneous quantification of the expression of dozens of kinesin genes. Real-time PCR is the best method available for quantifying expression of both low- and high-abundance transcripts in the same sample. We used this approach to assay the expression of 38 kinesin transcripts as well as transcripts encoding several cyotskeletal proteins and cell-type specific markers. We assayed the expression of these messages in five sets of samples (details about replicate number and statistical measures of reproducibility are described in Methods). As a starting point, we examined kinesin expression in hippocampal tissue dissected from E16 mouse hippocampal tissue. This sample represents the starting material used to prepare hippocampal cell cultures. At this stage of development, nearly all pyramidal neurons in the hippocampus have completed cell division, but generation of neurons in the adjoining dentate gyrus has not yet begun (Banker and Cowan 1977). Then we assayed mRNA expression in hippocampal cultures at three time points, after 4, 7, and 14 DIV. At 4 DIV, hippocampal neurons have established axons, but dendritic development has barely begun. A 7 DIV, cells are undergoing rapid dendritic growth and branching and active synaptogenesis; by 14 DIV these developmental processes begin to plateau. Finally, we examined expression levels in adult hippocampal tissue, which contains a heterogeneous cell population, including mature neurons and a variety of glial cell types.
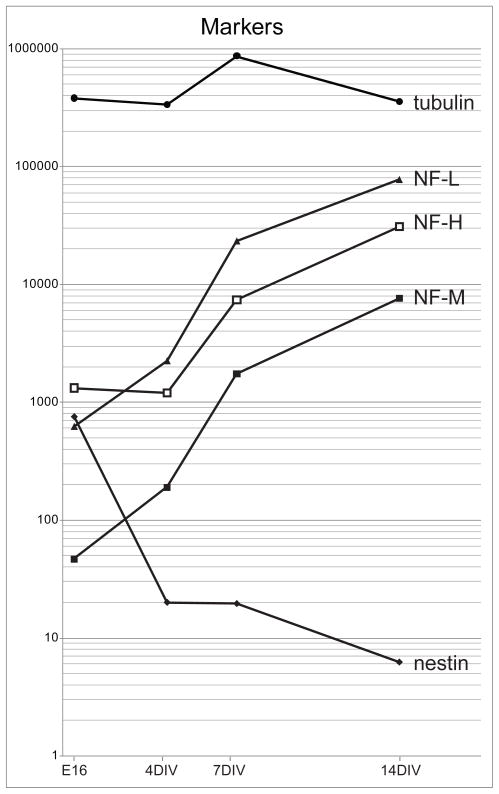
Before considering the data on kinesin expression, it is instructive to examine the expression patterns of mRNAs encoding neuronal cytoskeletal proteins, whose developmental expression has been well characterized (Figure 2). Not surprisingly, messages encoding β-tubulin subunits were the most abundant of all transcripts assayed, with a cycle threshold only a few cycles different from the 18S ribosomal RNA standard. The expression of tubulin messages did not change significantly during development, relative to 18S rRNA. This was surprising, given that microtubules are the major cytoskeletal elements in axons and dendrites, which undergo a massive increase in length during development. Since tubulin mRNA was already highly abundant in E16 hippocampus, it is likely that subsequent increases in tubulin message expression are matched by the increased abundance of the protein biosynthetic machinery, including 18S RNA. In contrast to tubulin mRNA, messages for the three neurofilament subunits increased markedly during development in culture (from 20- to 150-fold for the different subunits). This result agrees with previous results based on immunostaining, which show that neurofilaments become abundant only at a relatively late stage of development in hippocampal cultures (Benson et al. 1996). Conversely, transcripts for the intermediate filament protein nestin decreased markedly during development in culture, consistent with previous reports that nestin is expressed in neuroepithelial progenitor cells and postmitotic neuroblasts, but its expression declines rapidly as neurons differentiate (Dahlstrand et al. 1995).
Figure 2.
The Expression of Cytoskeletal Protein Transcripts during the Development of Hippocampal Neurons in Culture. This figures illustrates the levels of expression of mRNAs encoding cytoskeletal proteins in embryonic day 16 hippocampus and in hippocampal cultures a 4, 7, and 14 days in vitro. β-tubulin subunit mRNA was expressed at extremely high levels throughout development. In contrast, neurofilament transcripts increased dramatically during development in culture. The expression of nestin mRNA declined markedly. Values show transcript abundance relative to that of KIF5B in 14 DIV cultures, which was arbitrarily set at 10,000. We first calculated the difference in cycle threshold (CT) between each kinesin and KIF5B (ΔCT = CT KIFX − CT KIF5B), then determined relative abundance by evaluating the expression 2−ΔCT × 10,000.
It is also instructive to compare the abundance of messages encoding cell type-specific proteins in mature hippocampal cultures (14 DIV) with adult hippocampal tissue (Supplemental Table 3). Hippocampal cultures contain few non-neuronal cells whereas hippocampal tissue contains the full complement of astrocytes, oligodendrocytes, microglia and vascular cells characteristic of mature neural tissue. As expected, mRNAs that are preferentially expressed in neurons (such as those encoding neurofilament proteins) were slightly more abundant in samples from hippocampal cultures than from hippocampal tissue. mRNAs that are preferentially expressed in oligodendroglia, such as those encoding myelin basic protein and proteolipid protein, were far more abundant in samples from hippocampal tissue than from hippocampal cultures. Surprisingly, the level of glial fibrillary acidic protein (GFAP) message was similar in 14 DIV cultures and hippocampal tissue. In the cultures, neurons outnumber astrocytes by 30 to 1, whereas astrocytes outnumber neurons in hippocampal tissue. It may be that astrocytes in culture express some features of the reactive phenotype (Ahlemeyer et al. 2003; Sergent-Tanguy et al. 2006), including comparatively high GFAP expression. GFAP is upregulated by more than 80-fold in reactive astrocytes, as compared with the non-reactive astrocytes present in normal tissue (Burbach et al. 2004).
Expression patterns of mRNAs encoding kinesins that function in organelle transport and mitosis
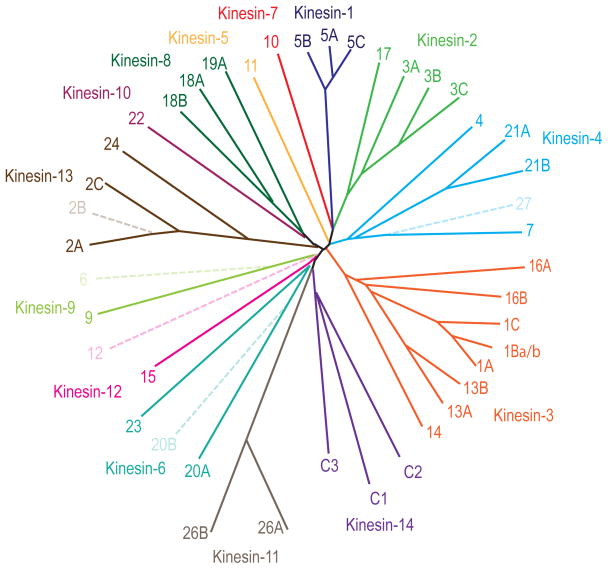
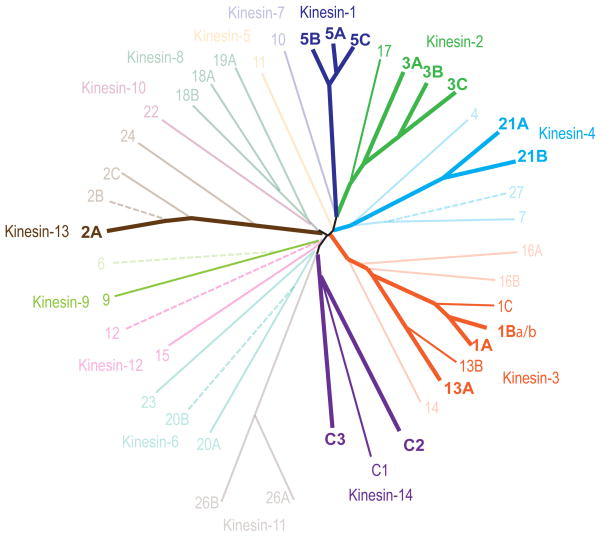
We reliably detected the expression of 35 different kinesin transcripts from 14 different subfamilies in the embryonic hippocampus, the adult hippocampus, or in both (Figure 3). The level of expression of these kinesin transcripts varied widely. The most abundant kinesins were expressed at levels only 10 to 50-fold less than beta-tubulin, one of the most highly abundant of all neuronal proteins; other kinesins were four orders of magnitude less abundant.
Figure 3.
Dendrogram of the Kinesin Genes in the Mouse Genome. The genes can be grouped into 14 families based on the comparison of their conserved motor domains. Solid lines represent the kinesin genes reliably detected in embryonic or adult hippocampus in this study. The dendrogram was generated using web-based clustalW and drawtree (http://evolution.genetics.washington.edu/phylip/software.html).
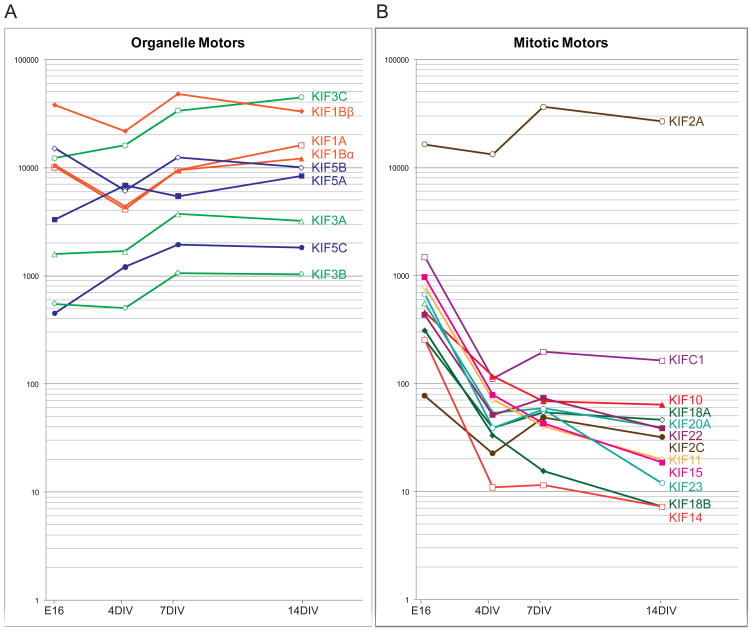
We first examined the developmental patterns of expression of several kinesins whose functions have been well established. Figure 4A shows the relative abundance of the members of the Kinesin-1 (KIF5A, 5B, and 5C), Kinesin-2 (KIF3A, 3B, and 3C), and Kinesin-3 (KIF1A, 1Bα, and 1Bβ) families that are known to serve as organelle motors in long-range axonal transport. All were highly expressed in embryonic and adult hippocampal tissue, and in hippocampal cultures at all stages of development. In 14 DIV cultures, the expression of these kinesins ranged from about 10-fold less than KIF5B to about 5-fold above KIF5B. There were no statistically significant increases or decreases in the expression of these messages during development in cell culture or in adult versus embryonic hippocampal tissue (Supplementary Figure S1; see methods for description of statistical analysis).
Figure 4.
The Expression Patterns of Kinesins Known to Participate in Organelle Transport and Mitosis. A illustrates the relative abundance of messages encoding 9 members of the Kinesin-1, -2, and -3 families that have well-established roles in mediating rapid axonal transport of membranous organelles. All are expressed at high levels in embryonic hippocampus and their expression did not change significantly during neuronal development in culture. B shows the expression patterns of mRNAs encoding 11 kinesin that play well-established roles in mitosis. All but one were expressed at moderate levels in embryonic hippocampus and at distinctly lower levels in hippocampal cultures. One mitotic kinesin, the Kinesin-13 KIF2A, was expressed at high levels in embryonic hippocampal tissue and its expression remained high throughout neuronal development in culture. Values show abundance relative to the level of KIF5B mRNA in 14 DIV cultures, calculated as in Figure 2.
Next we examined the expression patterns of 12 kinesins known to play a role in mitosis, based on knock-down studies in human and Drosophila cell lines (Zhu et al. 2005). Most of the mitotic kinesins exhibited a characteristic pattern of expression that was distinctly different from that of the known organelle motors. Nearly all mitotic kinesins were expressed at moderate levels in E16 hippocampal tissue, averaging about 10-fold less than the kinesins that mediate organelle transport (compare Figure 4B with 4A). In hippocampal cultures, the level of expression of mitotic kinesins was far lower, roughly 100-fold less than the known organelle motors. The mitotic motors were also expressed at very low levels in adult hippocampal tissue. These expression patterns parallel the extent of cell division occurring at each time point. The E16 hippocampus contains a significant population of mitotic cells that gives rise to dentate neurons and glia, whereas hippocampal cultures consist almost exclusively of postmitotic neurons. KIF2A, a member of the Kinesin-13 family, exhibited a strikingly different pattern of expression. It was expressed at high levels in E16 hippocampal tissue, higher even than KIF5B, and its expression remained high in adult hippocampal tissue and throughout neuronal development in culture. KIF2A causes depolymerization of microtubules at their plus ends, which contributes to chromosome movements during anaphase. Evidence from mice lacking KIF2A indicates that this kinesin also regulates the microtubule cytoskeleton in post-mitotic neurons (Homma et al. 2003); its pattern of expression in hippocampal cultures is consistent with this role. The expression of the minus-end directed kinesin KIFC1 also differed somewhat from most mitotic motors. KIFC1 was expressed at moderate levels in E16 hippocampus, but its level of expression in mature cultures was somewhat higher than other mitotic kinesins.
Expression patterns of other kinesin mRNAs
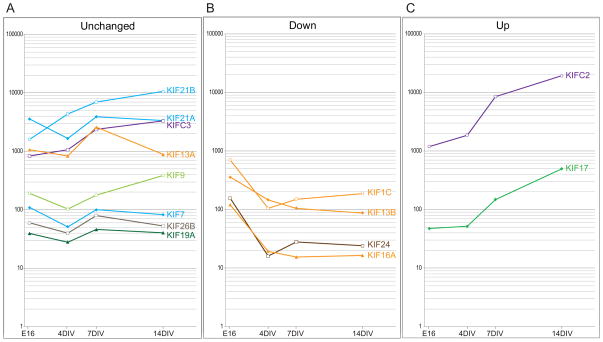
In addition to the kinesins described above, which have been the subject of extensive study, we detected the expression of 14 other kinesins in cultured hippocampal neurons. Their pattern of expression is illustrated in Figure 5. Of these kinesins, the expression of 8 transcripts did not change significantly during development (Fig. 5A). These kinesins largely fell into two groups, based on their levels of expression. Four members of the Kinesin-3 (KIF13A), Kinesin-4 (KIF21A and 21B), and Kinesin-14 (KIFC3) families were expressed at high levels throughout development in hippocampal cultures, comparable to the kinesins known to mediate axonal transport. Transcripts for these four kinesin were also expressed at high levels in embryonic and adult hippocampal tissue. A Kinesin-4 (KIF7), Kinesin-8 (19A), and Kinesin–11 (KIF26B) were expressed at far lower levels, 100–500 times less than KIF5B. The Kinesin-9 family member KIF9 was expressed at a level intermediate between these two groups.
Figure 5.
Changes in the Relative Abundance of mRNAs for Kinesin Family Members during Development of Hippocampal Neurons in Culture. A illustrates the levels of mRNA encoding 8 kinesins whose expression did not change significantly during development in culture. About half were expressed at levels comparable to known organelle motors. B illustrates the expression patterns of the two kinesins whose expression increased significantly during development in culture (p < 0.05). C shows the expression patterns of 4 kinesins whose expression declined significantly during development in culture (p < 0.05). Two declined to very low levels, comparable to known mitotic kinesins. The data show abundance relative to the level of expression of KIF5B mRNA in 14 DIV cultures, calculated as in Figure 2.
The expression of four kinesins decreased significantly during development (Fig. 5B). Again these fell into two classes. Transcripts for a Kinesin-3 (KIF16A) and a Kinesin-13 (KIF24) were expressed at low levels in E16 hippocampal tissue and at even lower levels during development in culture and in adult hippocampal tissue. Thus their pattern of expression was similar to that of the known mitotic motors. KIF1C and KIF13B, both members of the Kinesin-3 family, were expressed at moderate levels in E16 hippocampal tissue. Their expression declined somewhat during development in culture, but remained significantly higher than the levels of mitotic kinesins.
Only two kinesin genes increased significantly during development (Fig. 5C). One of these, the Kinesin-14 family member KIFC2, was already expressed at high levels in E16 hippocampal tissue and its expression increased still further during development in culture and in vivo, reaching levels higher than KIF5B. The Kinesin-2 family member KIF17 was expressed at low levels in E16 hippocampal tissue, but its expression increased about 10-fold during development in culture and in vivo.
Kinesin expression in hippocampal cell cultures versus hippocampal tissue
Finally, we compared kinesin expression in mature hippocampal cultures, which are highly enriched in neurons, with expression in adult hippocampal tissue, which contains a full complement of glial cells as well as mature neurons (based on the data summarized in Supplementary Table 3). Transcripts for the great majority of kinesins were expressed at comparable levels in 14 DIV hippocampal cultures and in adult hippocampal tissue, but there were four exceptions. Some members of the Kinesin-3 (KIF1C and KIF13B) and Kinesin-4 (KIF21A) families were expressed at 4–7 fold higher levels in adult hippocampal tissue, which suggests they may be preferentially expressed in glial cells. Only one kinesin, the Kinesin-4 family member KIF21B, was expressed at notably higher levels (17 fold) in neuronal cultures than in hippocampal tissue.
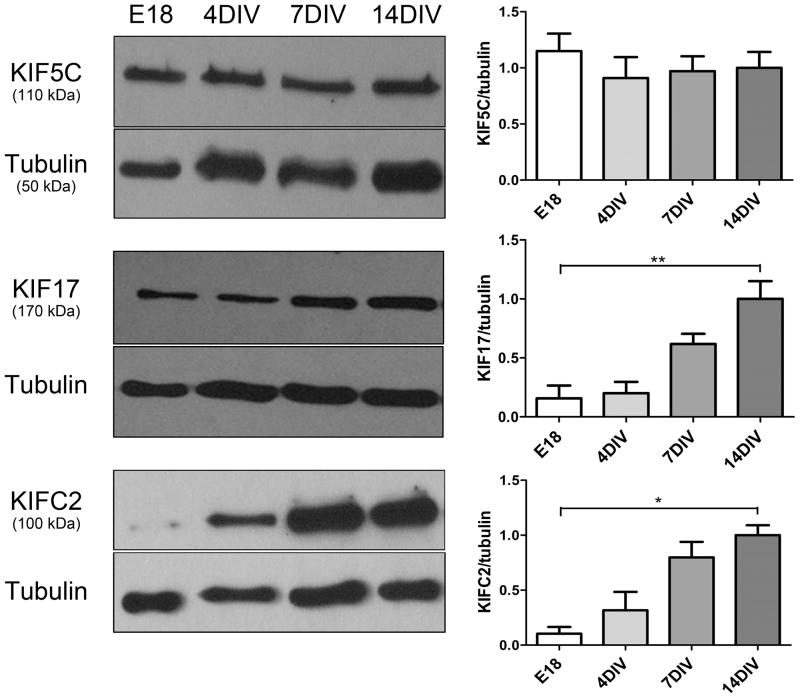
Kinesin protein levels reflect mRNA expression levels
Protein levels do not always parallel mRNA expression levels due to differences in protein half-life or stability. Thus we sought to determine if the developmental increases in kinesin mRNA expression that we detected by RT-PCR were paralleled by an uprergulation in the level of protein expression. Immunoblotting revealed an increase in levels of KIF17 and KIFC2 in developing hippocampal cultures (Figure 6). KIF17 protein levels increased approximately fivefold and KIFC2 protein levels increased approximately ten-fold between E18 and 14DIV. Both of these changes parallel the increases in the expression of their mRNAs (Figure 5C). In contrast, there was no change in the level of KIF5C protein during these stages of development, again paralleling unchanging expression of KIF5C mRNA. These data indicate that changes in message levels for developmentally regulated kinesins are also reflected at the level of protein expression.
Figure 6.
The expression of kinesin motor proteins KIF5C, KIF17 and KIFC2 during the development of rat hippocampal neurons in culture. Shown is the level of protein expression of KIF5C, KIF17, and KIFC2 motor proteins in embryonic day 18 hippocampus and hippocampal cultures at 4, 7, and 14 DIV. KIF17 and KIFC2 protein expression increased significantly in developing hippocampal neurons, paralleling the increase in the expression of their mRNAs. There was no change in the levels of KIF5C protein. The kinesin motor protein densitometry values were calculated as a ratio to the tubulin loading control and were normalized to the level expressed in 14 DIV cultures, which were arbitrarily taken as 1. The values represent the mean ± SEM from at least three independent experiments. *, p < 0.05, **, p < 0.01, statistically significant differences vs. 14 DIV.
Discussion
The principal goal of this study was to determine whether the great diversity of kinesin expression in the brain reflects differences in the set of kinesins expressed by different types of neurons or the diverse roles played by different kinesins within a single neuron. Our results show that a homogeneous population of cultured pyramidal neurons expresses a great diversity of kinesins, as summarized in Figure 7. In all, we detected moderate to high levels of 20 different kinesin transcripts in mature hippocampal cultures. Of these, the great majority probably function as organelle motors. We detected 15 transcripts from the Kinesin-1, -2, -3 and -4 families. Nine of these are known to mediate long-range organelle transport and the remainder may likewise function as organelle motors. Thus it seems likely that the diversity of kinesin expression largely reflects the diversity of cargoes that undergo microtubule-based transport in neurons and the need to transport cargoes to different subcellular domains. Cultured hippocampal neurons also express high levels of KIF2A, a Kinesin-13 that depolymerizes microtubules, and four other kinesins, including a Kinesin-9 and 3 minus-end directed Kinesin-14s. The function of the latter kinesins is not known.
Figure 7.
Kinesin genes expressed in mature hippocampal neurons. Transcripts derived from 19 genes of 7 different kinesin families were detected at high (thick solid lines) or moderate (thin solid lines) levels in 14 DIV cultures. Both known splice variants of the Kinesin-3 family member KIF1B, KIF1Bα and KIF1Bβ, were detected at high levels.
Plus-end Directed Kinesins that Mediate Organelle Transport
Strong evidence suggests that members of the Kinesin-1, -2, and -3 families participate in long-range anterograde organelle transport in axons and dendrites. Our assay detected high expression levels of each of these kinesins in embryonic hippocampal tissue and immature hippocampal cultures, and robust expression was maintained throughout neuronal development and at maturity. These kinesins include all of the Kinesin-1 family members, KIF5A, 5B, and 5C; the Kinesin-2 family members KIF3A, 3B, and 3C; and the Kinesin-3 family members KIF1A, 1Bα, and 1Bβ.
The Kinesin-1s were the first axonal transport motors to be identified and are known to play a role in the transport of many different cargoes, including mitochondria (Tanaka et al. 1998) and vesicles containing proteins destined for the axonal plasma membrane, such as presenilin-1, amyloid precursor protein, and the apolipoprotein E receptor 2 (Kamal et al. 2000; Kamal et al. 2001; Verhey et al. 2001). The Kinesin-3 family members KIF1A and the two splice variants of KIF1B are thought to mediate axonal transport of synaptic vesicle precursors; a second isoform of KIF1B, KIF1Bβ, may contribute to the transport of mitochondria (Nangaku et al. 1994; Okada et al. 1995; Yonekawa et al. 1998; Zhao et al. 2001; Zhang and Sperry 2004). Kinesin-1 and Kinesin-3 family members have also been linked to the trafficking of dendritic membrane proteins, such as AMPA receptors (Setou et al. 2002) and EphB2 receptors (Hoogenraad et al. 2005), as well mRNA granules (Kanai et al. 2004). Finally, Kinesin-1 family members may also mediate the slow axonal transport of neurofilaments (Shah and Cleveland 2002; Xia et al. 2003). The Kinesin-2 family members KIF3A, 3B, and 3C all accumulate at the proximal side of peripheral nerve ligatures (Kondo et al. 1994; Yamazaki et al. 1995; Yang and Goldstein 1998), indicating that they undergo rapid axonal transport, but less is known about the cargoes they carry (Takeda et al. 2000).
Of the remaining plus-end directed kinesins that we assayed, three showed the same pattern of expression as kinesins that mediate organelle transport–all were expressed at high levels at E16 and remained at this level throughout development. This group includes a Kinesin-3 family member, KIF13A, and two closely related members of the Kinesin-4 family, KIF21A and 21B. Previous reports indicate that KIF13A is involved in intracellular trafficking of AP1 and mannose 6-phosphate receptor (Nakagawa et al. 2000). Mutations in KIF21A cause congenital fibrosis of extraocular muscles (Tiab et al. 2004; Chan et al. 2007), which is believed to reflect a failure in development of the axons that innervate these muscles and could be explained by a dysfunction in axonal transport of proteins required for nerve-muscle interactions. KIF21B is thought to be enriched in dendrites (Marszalek et al. 1999), although its expression pattern resembles kinesins involved in axonal transport. Comparatively little is known about the cargoes carried by these Kinesin-4 family members.
Four transcripts encoding plus-end directed kinesins were expressed at moderate levels in mature hippocampal cultures, less than kinesins known to participate in organelle transport but distinctly higher than mitotic motors. Based solely on expression level, such kinesins might be candidates to mediate transport of cargoes that are not abundant or that are transported within specific compartments within the cell. Included in this group are two Kinesin-3 family members, KIF13B and KIF1C, whose expression declined significantly during development in culture, the Kinesin-2 family member KIF17 whose expression increased significantly, and the Kinesin-9 family member, KIF9, whose expression was moderate throughout development. KIF13B mediates the transport of vesicles enriched in PIP3 in developing axons (Horiguchi et al. 2006), and regulates membrane trafficking events required for myelination by Schwann cells (Bolis et al. 2009). The expression of KIF13B is about 5-fold higher in adult hippocampal tissue, which contains a significant population of oligodendroglia, than in mature hippocampal neuronal cultures; this result suggests that KIF13B might also be involved in CNS myelination (Bolis et al. 2009). KIF1C cargoes have not yet been identified (Nakajima et al. 2002) and the function of KIF9 is unknown.
KIF17 is the only plus-end kinesin whose expression increased during development, from very low levels prior to 4 DIV to moderate levels at 14 DIV. The timing of its increase corresponds to the period of peak dendritic growth and branching. KIF17 has been implicated in the transport of several membrane proteins that are localized to dendrites, but excluded from the axon (Setou et al. 2000; Chu et al. 2006; Kayadjanian et al. 2007). The pattern of expression of KIF17 is consistent with a role in the transport organelles principally within the dendritic domain. KIF17 also plays a role in ciliary transport. Cilia are present in cultured hippocampal neurons (Berbari et al., 2007) and the requirement for ciliary transport may also increase during development, reflecting a need for increased KIF17 expression.
Plus-end Directed Kinesins that Regulate Microtubule Organization
Comprehensive RNAi-based screens in Drosophila (Goshima and Vale 2003) and human cell lines (Zhu et al. 2005) have identified the kinesins that are essential for mitosis (see Fig. 4B). During mitosis, these kinesins regulate the parallel or antiparallel alignment of microtubules, induce microtubule sliding, and control microtubule depolymerization. Do these kinesins also regulate microtubule organization in postmitotic cells? With one exception, all of the plus-end directed kinesins identified in these screens exhibited the expression pattern one might expect for kinesins that function exclusively during mitosis. They were expressed at moderate levels in E16 hippocampal tissue, then dropped to far lower levels in hippocampal cultures and adult hippocampal tissue. Two other kinesins of unknown function, KIF16A and KIF24, displayed the same pattern of development as the known mitotic motors.
The expression pattern of KIF2A was distinctly different than the other mitotic kinesins. This Kinesin-13 family member was expressed at high levels in the embryonic hippocampus and its expression remained high in hippocampal cultures and in adult hippocampal tissue. During mitosis, KIF2A mediates the depolymerization of spindle microtubules. KIF2A-null mice exhibit aberrant axonal branching, characterized by an overabundance of axon collaterals, suggesting that this kinesin also regulates microtubule organization in postmitotic neurons (Homma et al. 2003). Our results show that KIF2A is maintained at high levels throughout the life of the neuron. Two other mitotic kinesins, KIF23 (CHO1) and KIF11 (Eg5), which regulate the antiparallel organization of spindle microtubules during mitosis, have been hypothesized to mediate the establishment of minus-end out microtubules in dendrites (Ferhat et al. 1998a; Ferhat et al. 1998b; Kaech and Banker 2006). Our results indicate that KIF11 and KIF23 are expressed in embryonic hippocampus at levels comparable to other mitotic kinesins and their expression drops to very low levels in hippocampal cultures, like those kinesins thought to function exclusively in mitosis.
Minus end-directed kinesins
The Kinesin-14 family members KIFC2 and KIFC3 were among the most highly expressed kinesins in hippocampal cultures and both increased in expression as neurons matured (although the increase in KIFC3 expression did not reach statistical significance). KIFC1 was also expressed at moderate levels in mature neurons. The function of these minus-end directed kinesins in neurons is not understood. KIFC1 is important for spindle formation and function during mitosis (Zhu et al., 2005; Hentrich and Surrey, 2010) and also serves as a minus-end directed motor for organelle transport in some non-neuronal cells (Zhang and Sperry 2004; Nath et al. 2007). KIFC2 and KIFC3 have also been thought of as organelle motors (Hanlon et al. 1997; Saito et al. 1997), but knockouts of KIFC2 or KIFC3, or simultaneous knockouts of both, exhibited no detectable phenotype (Yang et al. 2001b; Yang et al. 2001a). Thus the role of the Kinesin-14s remains paradoxical.
Concluding comments
Most of the kinesin genes identified in the mouse genome are expressed in the brain at one or another stage of development (Miki et al., 2003 and others). We now show that a single, homogeneous population of cultured neurons expresses a great diversity of kinesins. Of the 19 kinesins expressed in mature hippocampal neurons, three are minus-end motors, one regulates microtubule organization, and the remainder likely mediate plus-end directed organelle transport. This diversity of kinesin expression must reflect the diversity of cargoes that are transported in neurons and the requirement to convey these cargoes to different cellular domains. In order to elucidate the role these diverse kinesins play in neuronal trafficking and the maintenance of neuronal polarity, it will be essential to identify the cargoes that each kinesin carries.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 MH66179 and R21AG19562 (to GB) and National Science and Engineering Research Council of Canada grant #327100–06 (to MS). Our thanks to Drs. J. Petersen, C-F Huang, and M. Leroux for their helpful comments on this manuscript.
References
- Ahlemeyer B, Kolker S, Zhu Y, Hoffmann GF, Krieglstein J. Cytosine arabinofuranoside-induced activation of astrocytes increases the susceptibility of neurons to glutamate due to the release of soluble factors. Neurochemistry International. 2003;42 (7):567–581. doi: 10.1016/s0197-0186(02)00164-x. [DOI] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85(21):8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Research. 1977;126:397–425. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- Benson DL, Mandell JW, Shaw G, Banker G. Compartmentation of alpha-internexin and neurofilament triplet proteins in cultured hippocampal neurons. J Neurocytol. 1996;25(3):181–196. doi: 10.1007/BF02284795. [DOI] [PubMed] [Google Scholar]
- Benson DL, Watkins FH, Steward O, Banker G. Characterization of GABAergic neurons in hippocampal cell cultures. J Neurocytol. 1994;23(5):279–295. doi: 10.1007/BF01188497. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Bishop GA, Askwith CC, Lewis JS, Mykytyn K. Hippocampal neurons possess primary cilia in culture. J Neurosci Res. 2007 Apr;85(5):1095–100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- Bolis A, Coviello S, Visigalli I, Taveggia C, Bachi A, Chishti AH, Hanada T, Quattrini A, Previtali SC, Biffi A, et al. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci. 2009;29(27):8858–8870. doi: 10.1523/JNEUROSCI.1423-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BW, Russel K. MULTI: identifying important results from multiple stastistical tests (ver. 2) 1996 URL: http://biostatistics.mdanderson.org/SoftwareDownload.
- Burbach GJ, Dehn D, Del Turco D, Staufenbiel M, Deller T. Laser microdissection reveals regional and cellular differences in GFAP mRNA upregulation following brain injury, axonal denervation, and amyloid plaque deposition. Glia. 2004;48(1):76–84. doi: 10.1002/glia.20057. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol. 2005;34(3):597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Chan WM, Andrews C, Dragan L, Fredrick D, Armstrong L, Lyons C, Geraghty MT, Hunter DG, Yazdani A, Traboulsi EI, et al. Three novel mutations in KIF21A highlight the importance of the third coiled-coil stalk domain in the etiology of CFEOM1. BMC Genet. 2007;8:26. doi: 10.1186/1471-2156-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PJ, Rivera JF, Arnold DB. A role for Kif17 in transport of Kv4.2. J Biol Chem. 2006;281(1):365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- Conforti L, Buckmaster EA, Tarlton A, Brown MC, Lyon MF, Perry VH, Coleman MP. The major brain isoform of kif1b lacks the putative mitochondria-binding domain. Mamm Genome. 1999;10(6):617–622. doi: 10.1007/s003359901056. [DOI] [PubMed] [Google Scholar]
- Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res. 1995;84(1):109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- Ferhat L, Cook C, Chauviere M, Harper M, Kress M, Lyons GE, Baas PW. Expression of the mitotic motor protein Eg5 in postmitotic neurons: implications for neuronal development. J Neurosci. 1998a;18(19):7822–7835. doi: 10.1523/JNEUROSCI.18-19-07822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhat L, Kuriyama R, Lyons GE, Micales B, Baas PW. Expression of the mitotic motor protein CHO1/MKLP1 in postmitotic neurons. Eur J Neurosci. 1998b;10(4):1383–1393. doi: 10.1046/j.1460-9568.1998.00159.x. [DOI] [PubMed] [Google Scholar]
- Goldstein LSB, Yang Z. Microtubule-Based Transport Systems in Neurons: The Roles of Kinesins and Dyneins. Annual Review of Neuroscience. 2000;23(1):39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Gong TW, Winnicki RS, Kohrman DC, Lomax MI. A novel mouse kinesin of the UNC-104/KIF1 subfamily encoded by the Kif1b gene. Gene. 1999;239(1):117–127. doi: 10.1016/s0378-1119(99)00370-4. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162(6):1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon DW, Yang Z, Goldstein LS. Characterization of KIFC2, a neuronal kinesin superfamily member in mouse. Neuron. 1997;18(3):439–451. doi: 10.1016/s0896-6273(00)81244-1. [DOI] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010 May 3;189(3):465–80. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci. 2006;26 (27):7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004a;301(1):50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr Opin Neurobiol. 2004b;14(5):564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. 2005;6(3):201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114(2):229–239. doi: 10.1016/s0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8 (7):906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174(3):425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MR, Wang K, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278(2):175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Kinesin-mediated axonal transport of a membrane compartment containing beta-secretase and presenilin-1 requires APP. Nature. 2001;414(6864):643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28(2):449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kayadjanian N, Lee HS, Pina-Crespo J, Heinemann SF. Localization of glutamate receptors to distal dendrites depends on subunit composition and the kinesin motor protein KIF17. Mol Cell Neurosci. 2007;34(2):219–230. doi: 10.1016/j.mcn.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125(5):1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167(1):19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol. 1999;145(3):469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15(9):467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Hirokawa N. Kinesin superfamily proteins (KIFs) in the mouse transcriptome. Genome Res. 2003;13(6B):1455–1465. doi: 10.1101/gr.984503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98(13):7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Milligan RA. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J Cell Sci. 2006;119(Pt 19):3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103(4):569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Takei Y, Tanaka Y, Nakagawa T, Nakata T, Noda Y, Setou M, Hirokawa N. Molecular motor KIF1C is not essential for mouse survival and motor-dependent retrograde Golgi apparatus-to-endoplasmic reticulum transport. Mol Cell Biol. 2002;22(3):866–873. doi: 10.1128/MCB.22.3.866-873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79(7):1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Nath S, Bananis E, Sarkar S, Stockert RJ, Sperry AO, Murray JW, Wolkoff AW. Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol Biol Cell. 2007;18(5):1839–1849. doi: 10.1091/mbc.E06-06-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81(5):769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar S. NLME: linear and non-linear mixed effect models. R package version 3.1–23 2002 [Google Scholar]
- Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N. KIFC2 is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron. 1997;18(3):425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- Schweder T, Spjøtvoll E. Plots of P-values to evaluate many tests simultaneously. Biometrika. 1982;69(3):493–502. [Google Scholar]
- Sergent-Tanguy S, Michel DC, Neveu I, Naveilhan P. Long-lasting coexpression of nestin and glial fibrillary acidic protein in primary cultures of astroglial cells with a major participation of nestin(+)/GFAP(−) cells in cell proliferation. J Neurosci Res. 2006;83(8):1515–1524. doi: 10.1002/jnr.20846. [DOI] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288(5472):1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417(6884):83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Slow axonal transport: fast motors in the slow lane. Curr Opin Cell Biol. 2002;14(1):58–62. doi: 10.1016/s0955-0674(01)00294-0. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends in Cell Biology. 2009;19(7):306–16. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Takeda S, Yamazaki H, Seog DH, Kanai Y, Terada S, Hirokawa N. Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J Cell Biol. 2000;148(6):1255–1265. doi: 10.1083/jcb.148.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93(7):1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Tiab L, d’Alleves Manzi V, Borruat FX, Munier F, Schorderet D. Mutation analysis of KIF21A in congenital fibrosis of the extraocular muscles (CFEOM) patients. Ophthalmic Genet. 2004;25(4):241–246. doi: 10.1080/13816810490902828. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152(5):959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17(1):82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Xia CH, Roberts EA, Her LS, Liu X, Williams DS, Cleveland DW, Goldstein LS. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol. 2003;161(1):55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130(6):1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Goldstein LS. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol Biol Cell. 1998;9(2):249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Roberts EA, Goldstein LS. Functional analysis of mouse C-terminal kinesin motor KifC2. Mol Cell Biol. 2001a;21(7):2463–2466. doi: 10.1128/MCB.21.7.2463-2466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Roberts EA, Goldstein LS. Functional analysis of mouse kinesin motor Kif3C. Mol Cell Biol. 2001b;21(16):5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141(2):431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sperry AO. Comparative analysis of two C-terminal kinesin motor proteins: KIFC1 and KIFC5A. Cell Motil Cytoskeleton. 2004;58(4):213–230. doi: 10.1002/cm.20008. [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105(5):587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16(7):3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.