Abstract
We investigated whether apurinic/apyrimidinic (AP/abasic) sites were more frequent in regions of DNA replication in cells and whether their number increased during oxidative stress. DNA fiber spreading and fluorescent immunostaining were used to detect areas of DNA replication and sites of AP lesions in extended DNA fibers. The distribution of AP sites was determined in DNA fibers from vertebrate cells maintained under normal culture conditions or stressed with exogenous H2O2. AP lesions per unit length were enumerated in bulk DNA or at replication sites. The background density of AP sites in DNA fibers was 5.4 AP sites/106 nt, while newly replicated DNA contained 12.9 AP sites/106 nt. In cells exposed to 20 μM H2O2, AP sites in newly replicated DNA increased to 20.8/106 nt. Determinations of AP site density in bulk DNA by fiber analysis or standard slot blot assays agreed to within 10%. Our findings show that the fiber assay not only accurately determines the frequency of AP sites but also shows their distribution. They also reveal that there is increased susceptibility to oxidative damage in DNA regions undergoing replication, which may explain the previously observed clustering of AP sites.—Chastain II, P. D., Nakamura, J., Rao, S., Chu, H., Ibrahim, J. G., Swenberg, J. A., Kaufman, D. G. Abasic sites preferentially form at regions undergoing DNA replication.
Keywords: DNA damage, apurinic/apyrimidinic sites, oxidative stress, hydrogen peroxide, DNA fiber immunostaining
Reactive oxygen species (ROS) are a class of reactive ions and free radicals generated within cells by oxidative reactions both as products of endogenous metabolism and in response to environmental exposures. Inside the cell, ROS are generated in a variety of ways, as byproducts of energy production in mitochondria, as part of an antimicrobial or antiviral response, and in detoxification reactions carried out by the cytochrome P-450 system. Environmental factors such as chlorinated compounds, radiation, metal ions, barbiturates, phorbol esters, some peroxisome proliferating compounds, and ultraviolet light can also induce the formation of ROS inside the cell. Once formed, ROS can react with macromolecules and lipids. In DNA they create several distinct oxidative DNA damage products: 8-hydroxyguanine (8-oxoG) and apurinic/apyrimidinic sites (AP/abasic sites) are the damage products most studied (1, 2). The base excision repair (BER) pathway repairs these DNA base lesions (in addition to the lesions generated by alkylation and deamination). BER includes two major processes, the single-nucleotide (SN)-BER and long-patch (LP)-BER pathways, distinguished by their repair patch size and the enzymes they require. In addition to the formation of AP sites during BER, AP sites form through spontaneous depurinations and depyrimidinations reactions (3, 4).
Normally, the cell's antioxidant defense mechanisms are able to eliminate most of the ROS that are formed and minimize the formation of ROS-induced AP sites. When cells cannot efficiently eliminate ROS, they suffer the consequences of oxidative stress, including increased ROS-induced damage of DNA. The excessive production of ROS and subsequent oxidative stress and cellular damage has been linked to the pathogenesis of many age-related and chronic diseases. These include ischemia/reperfusion injuries (5, 6), Alzheimer's disease (7–9), amyotrophic lateral sclerosis (ALS) (10), Parkinson's disease (11–14), atherosclerosis (15–18), cataract formation (19–22), macular degeneration (23, 24), the aging process (25–28), and some cancers (29–32).
The role of DNA damage induced by oxidative stress was investigated previously using DNA fiber analysis to determine the global distribution of AP sites in animal organs and in cells grown in culture (2). In the previous studies, calf thymus and HeLa cell DNA fibers were visualized by electron microscopy and most of the DNA fibers (average size 20 kb) were found to contain only one or a few AP sites. However, in 3% of the fibers, 10 to 50 AP sites (with an average of 15) were detected per fiber and were spaced ∼250 bp from one another. Therefore a 20-kb fiber that, on the basis of global measurements, should contain on average only 0.3 AP sites, had as many as 12 AP sites closely spaced in a 3-kb length of DNA. The groups of oxidative DNA damage in close proximity were called “AP clusters.” Clusters of endogenous oxidative DNA damage have been found by others in hematopoietic stem and progenitor cells, human skin in three dimensional reconstructions, cultured skin cells, and human tumor tissue (33–36). Clusters of oxidative DNA damage have also been found under hypoxic conditions (37). Since the spacing of AP sites so close together is not typical, we concluded that some special features or configuration of the target DNA might contribute to their formation (2). The presence of AP site clustering may be an indicator of, for example, an exceptionally exposed state of chromatin that may increase its vulnerability to chemical attack. Open chromatin typically is found in regions undergoing transcription or DNA replication. In this paper, we explored whether oxidative damage preferentially occurs in areas undergoing replication. Our data indicate that not all areas of the genome are equally sensitive to DNA damage, especially to AP site clustering. DNA at sites undergoing replication after cells were exposed to H2O2 was more vulnerable to AP site formation than genomic regions that had replicated just prior to exposure, or to the bulk DNA that was not undergoing replication at that time.
MATERIALS AND METHODS
Cell lines and culture condition
Avian DT40 cells (38, 39) were grown at 39.5°C, the normal avian body temperature, in a humidified atmosphere supplemented with 5% CO2 as a suspension in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Sigma, St. Louis, MO, USA), 1% chicken serum (Sigma and Invitrogen), and containing 100 μg/ml penicillin and 100 μg/ml streptomycin (Invitrogen).
Detection of endogenous AP sites by slot-blot analysis
DNA was isolated from DT40 cells in normal culture conditions or experiencing oxidative stress and processed for slot-blot analysis, as described previously (2).
DNA labeling and fiber spreading
The detection of areas undergoing replication in isolated DNA fibers was originally performed by Bensimon (40, 41) and later modified by Jackson and Pombo (42) to generate DNA fibers directly from lysed cells instead of using purified DNA. The DNA fiber extension methodology used in this paper is a modified version of the protocol initially described by Merrick et al. (43), which is a modification of Jackson and Pombo's method. Briefly, cells growing in culture were first labeled for 10 min in medium with 100 μM iododeoxyurine (IdU), and then centrifuged to remove the medium containing IdU. Cells were resuspended in unlabeled medium and exposed to H2O2 for 10 min. H2O2 exposure was terminated by the addition of catalase (3 U/ml) for 10 min then centrifuged, and thereafter, the cells were resuspended in medium with 50 μM chlorodeoxyuridine (CldU) for 20 min to provide a second DNA label. After exposure to the second halogenated nucleotide, the cells were harvested by centrifugation and resuspended in ice-cold PBS at ∼200 cells/μl.
For the preparation of the DNA fiber spreads on slides, 2 μl of cell suspension were spread on a Silane-Prep slide (Sigma; S4651), close and parallel to the label. The sample was allowed to evaporate until almost, but not completely dry and then overlaid with 10 μl of spreading buffer (0.5% SDS in 200 mM Tris-HCl, pH 7.4, and 50 mM EDTA). After ∼10 min, the slide was tilted at ∼15° to allow the cell lysate to slowly move down the slide, and the resulting DNA spreads were air dried, fixed in 3:1 methanol/acetic acid for 2 min, air-dried overnight, then stored at −20°C for at least 24 h.
For the detection of IdU and CldU within the DNA fiber spreads, the slides were treated with 2.5 M HCl for 30 min, washed several times in PBS, and blocked in 3% BSA in PBS for 60 min. The slides were incubated at room temperature with the antibodies indicated below, rinsed 3 times in PBS, and incubated for 30 min in blocking buffer between each of the following incubations: 1) 1 h in 1:500 rat anti-bromodeoxyuridine (detects CldU) (OBT0030; Accurate, Westbury, NY, USA) plus 1:500 mouse anti-bromodeoxyuridine (detects IdU) (Becton Dickinson, Franklin Lakes, NJ, USA); 2) 30 min in 1:500 AlexaFluor 488-conjugated chicken anti-rat (Molecular Probes, Carlsbad, CA, USA) plus 1:500 AlexaFluor 594-conjugated rabbit anti-mouse; and 3) 30 min in 1:500 AlexaFluor 488-conjugated goat anti-chicken plus AlexaFluor 594-conjugated goat anti-rabbit. In addition, prior to the blocking step between the first and second antibody incubations, the slides were placed for 15 min in a stringency buffer containing 10 mM Tris HCl (pH 7.4), 400 mM NaCl, 0.2% Tween-20, and 0.2% Nonidet P40 (Nonidet P-40) to remove any nonspecifically bound primary antibodies. The slides were rinsed 3 times in PBS and mounted in antifade (UNC Microscopy Core). Microscopy was carried out using an Olympus FV500 confocal microscope (Olympus, Tokyo, Japan) in sequential scanning mode.
Fluorescence visualization of AP sites
We tried 3 approaches for the labeling and visualization of AP sites in the DNA fiber spreads. Approach 1: biotin-tagged aldehyde reactive probe (ARP), which reacts with the ring-open form of AP sites to generate a biotin-tagged AP site, was added to the cells 1 h before DNA fibers were prepared. Approach 2: DNA fibers were prepared first, and then the AP sites were reactively tagged with ARP. Approach 3: same as approach 2, but using a fluorescent form of ARP called F-ARP. Using confocal microscopy, either the fluorescently tagged AP sites in the fibers could be directly visualized, or the biotin-tagged AP sites could be detected using fluorescent antibodies against biotin. DNA was labeled with a DNA dye (YOYO-1; Invitrogen) which provides a bright green signal when it associates with DNA. Regardless of the method used, we found that the distribution of AP sites in the DNA fiber spreads was equivalent. Since all three approaches seemed to give similar results and approach 2 required the least time and cost for reagents, we chose to use that methodology for our analyses.
Image processing and calculation
Images were processed using ImageJ software (http://rsbweb.nih.gov/ij/) and the MBF ImageJ for Microscopy (http://www.macbiophotonics.ca/imagej/) collection of plug-ins. To determine the number of AP sites within a given field of DNA fibers, the average fluorescence intensity per nucleotide was determined as follows: 1) a section of one of the fibers was erased, and the total intensity of the image was recalculated; 2) the intensity of the erased fiber was determined by subtracting the new total fluorescent intensity from the previous total intensity; 3) the number of nucleotides in the erased DNA fiber was determined by measuring the erased fiber length in micrometers and then multiplying that value by 6000 nt/μm (i.e., 3000 bp/μm × 2) (44); and 4) the average fluorescent intensity per nucleotide was obtained by dividing the fluorescent intensity of the DNA fiber by its length expressed in nucleotides. For each image, we determined the fluorescent intensity of at least 5 different fibers located in different areas of the image. This allowed us to calculate the average fluorescence intensity per nucleotide for that image. To obtain the total amount of DNA (expressed in nucleotides) in a given image, the total fluorescence intensity of the image was divided by the average intensity per nucleotide for that image.
The intensity of the fiber F was calculated as follows:
where Ib = total green fluorescence intensity of the image before the fiber is subtracted and Ia = intensity after the fiber is subtracted.
The average fluorescent intensity of each nucleotide was calculated as follows:
where n = 5 (number of fibers measured per image), F = intensity of the fiber, 6000 = number of nucleotides per micrometer of DNA, and L = length of fiber.
The total number of nucleotides in the image Tnt was calculated as follows:
We also devised a method to determine the total number of AP sites (labeled by red fluorescence) on well-defined DNA fibers in the same images. We used the ImageJ program to subtract background red fluorescence and focus the quantitative analysis on AP sites that met empirically determined criteria. We examined an image and compiled a distribution of the sizes and intensities of the entire red fluorescent signal. We determined empirically that a true single AP site had an area of 1 (as defined by the ImageJ software) and intensity between 45 and 80 intensity units (3). Since we were interested only in red signal associated with AP sites overlapping with clearly identifiable green DNA fibers, we used the colocalization function of ImageJ to identify red signal that colocalized with green DNA fibers. To exclude from analysis any red signal that was not associated with AP sites (i.e., red signal below the intensity of 45), we evaluated different settings for the lower limit threshold for red fluorescence. We found that the red fluorescence signal decreased as the threshold was increased, up to 50 intensity units. Subsequent small increases of the threshold did not reduce the number of apparent AP sites detected and only slightly reduced the overall red fluorescence signal. On the basis of these observations, we set the lower limit threshold for red signal at 50 (anything above 50 was determined to be an AP site signal and anything below was not). The red signals that colocalized with green DNA fibers were counted using the particle counter function of ImageJ. Red signal with an area equivalent to twice the signal of 1 AP site was counted as 2 AP sites, 3 times the signal as 3 AP sites, etc.
To determine the number of nucleotides that had been replicated (i.e., incorporated IdU or CldU) in normal culture conditions, the total amount of fluorescence from both IdU and CldU was measured. The number of AP sites colocalized in areas where DNA had replicated was assessed using the colocalization function provided by Image J. In this series of experiments, AP sites were marked by blue signal, and those that colocalized with areas undergoing replication were counted using the particle counter function of Image J. To determine the amount of replicating DNA (nt), the total amount of red and green fluorescence (IdU and CldU, respectively) was determined and then divided by fluorescence intensity per nucleotide, as described above.
The intensity of the red track R was calculated as follows:
where Trb = total red fluorescence intensity of the image before the fiber is subtracted and Tra = intensity after the fiber is subtracted.
The average fluorescent intensity of each nucleotide was calculated as follows:
where n = 5 (number of fibers measured per image), R = intensity of the red track, 6000 = number of nucleotides per micrometer of DNA, and L = length of fiber.
The total number of red nucleotides in the image Tr_nt was calculated as follows:
The intensity of the green track G was calculated as follows:
where Tgb = total green fluorescence intensity of the image before the fiber is subtracted and Tga = intensity after the fiber is subtracted.
The average fluorescent intensity of each nucleotide was calculated as follows:
where n = 5 (number of fibers measured per image), G = intensity of the green track, 6000 = number of nucleotides per micrometer of DNA, and L = length of fiber.
The total number of green nucleotides in the image Tg_nt was calculated as follows:
Statistical analysis
To estimate the effect of H2O2 on AP site formation in DNA fibers globally and areas undergoing replication, a Poisson regression was used to model the distribution of AP sites. A Wald test was used to determine the statistical significance of the H2O2 effect. All statistical analyses were done using SAS 9.2 (SAS Institute, Cary, NC, USA).
RESULTS AND DISCUSSION
Average number of AP sites in DNA fiber spreads
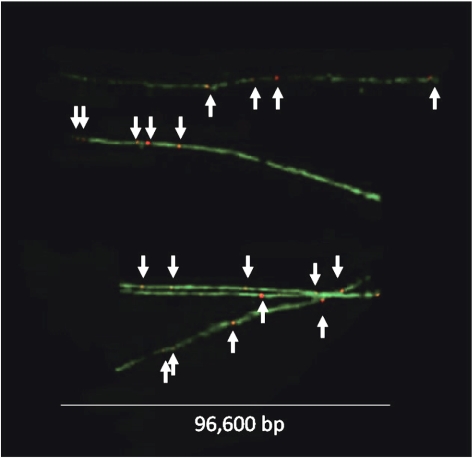
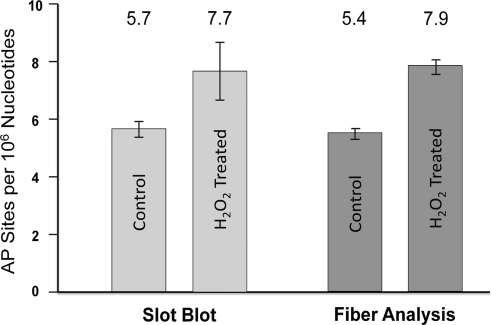
To quantify the number of AP sites per 106 nt in the YOYO-labeled DNA fiber spreads, we determined the total amount of DNA (nt) in a given image by dividing the total green fluorescence intensity of the image by the average intensity per nucleotide for that image, as outlined in Materials and Methods. We then determined the total number of AP sites located in well-defined DNA fibers in the image by counting the number of AP sites that colocalized to those fibers (see Materials and Methods). Figure 1 presents a composite image of a number of isolated DNA fibers that were observed in our samples. Using this approach, we analyzed >109 nt of DNA and determined that our sample of DNA from cells growing in normal culture conditions contained a basal value of 5.4 AP sites/106 nt, while slot-blot analysis gave a value of 5.7 AP sites/106 nt (Fig. 2). The number of AP sites detected in DT40 cells in this study is similar to what was found previously in HeLa cells and calf thymus DNA using a different methodology (2). When we analyzed 7 × 108 nt in DNA fiber spreads from cells that were exposed to 20 μM H2O2, we found that the number of AP sites increased to 7.9/106 nt, also consistent with the 7.7 AP sites/106 nt found by slot blot analysis (Fig. 2). The observed difference in AP sites in the absence and presence of H2O2 (5.4 vs. 7.9 AP sites/106 nt, respectively) is statistically significant with P < 0.0001, as determined using the Poisson regression model. It was interesting to note that there was considerably less variance in the values of AP sites per 106 nt obtained using fiber analysis than there was using the slot blot technique. Nonetheless, in view of the far greater effort involved in making these measurements of AP sites per 106 nt by fiber analysis, the slot blot technique remains the logical choice for routine assessment of the quantity of AP sites.
Figure 1.
Detection of AP sites in DNA fiber spreads. Composite image of DNA fibers stained with YOYO-1 green fluorescent dye. AP sites (white arrows) were tagged with biotin using ARP (see Materials and Methods) and detected with a red fluorescent anti-biotin antibody. Scale bar = 96.6 kbp.
Figure 2.
Number of AP sites in DNA from cells under typical tissue culture conditions or exposed to 20 μM H2O2. Number of AP sites per 106 nt as determined by slot-blot (left) and fiber-spread analysis (right) for cells under normal culture conditions and after exposure to 20 μM hydrogen peroxide. Average values are listed at top. Slot blot average was determined from 3 independent experiments; fiber analysis values were determined from analysis of 6 different slides.
AP sites in areas of DNA undergoing replication
In this study we observed that many AP sites occurred in clusters in both untreated and H2O2-treated cells, confirming the observations previously made in HeLa cells and calf thymus DNA. Furthermore, clustering appeared to be more common in the H2O2-treated cells (Fig. 1) as it had in the previous study. As noted earlier, we hypothesized that AP site formation in DNA might result from a higher propensity for ROS to attack chromatin that has an unusually open or exposed state, such as is found in genomic regions undergoing transcription or DNA replication. Recently, others and we have reported the capability to identify where DNA replication is occurring in extended DNA fibers (42, 43, 45). By incorporating two thymidine analogs in short sequential pulses, the direction of DNA replication can be determined and replication structures such as origins and termination site can be identified (42–46). These new techniques for analysis of DNA replication, when combined with our demonstration of AP sites using fluorescent probes, allow us to examine the formation of AP sites with regard to areas of DNA replication and to address very important questions about how replication is affected by oxidative stress.
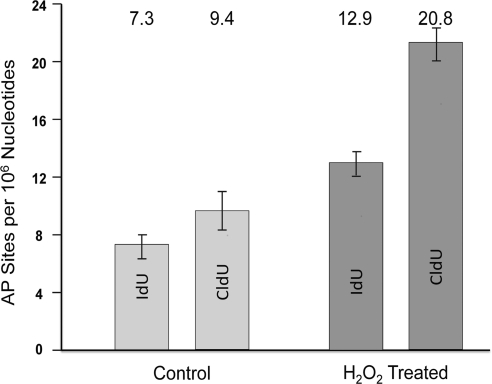
To determine the number of AP sites per unit length of DNA in areas undergoing replication, cells were first pulsed with IdU (a nucleotide precursor), and then pulsed with CldU (a different precursor). To determine the effects of oxidative stress, cells were exposed to H2O2 between the two pulses, while control cells were treated similarly but without inclusion of H2O2. Fluorescently labeled AP sites were readily detected and could be quantified with respect to the fluorescent tracks of IdU and CldU-labeled DNA (see Materials and Methods). In the control cells not exposed to H2O2, we found that the number of AP sites in areas that were labeled with IdU was 7.3 AP sites/106 nt, while in areas incorporating CldU that replicated later, the number was 9.4 AP sites/106 nt (Fig. 3). Both of these values were higher than the overall amount of AP site formation throughout the genome, which was found to be 5.4 AP sites/106 nt (Fig. 2); these differences are statistically significant with P < 0.0001, as determined by the Poisson regression model. This result indicates that AP sites are 1.5- to 2-fold (50 to 100%) more likely to be present in areas where DNA replication is in progress. It does not, however, distinguish whether more AP sites are formed in these regions or whether they accumulate there because they are not removed as efficiently from these areas as from other genomic sites.
Figure 3.
Number of AP sites in areas undergoing replication. Comparison between average number of AP sites found in replicating DNA fibers before and after exposure to 20 μM H2O2. In these experiments, only DNA fibers with replicating DNA were quantified. Average values are listed at top. Fiber analysis values were determined from analysis of 3 slides.
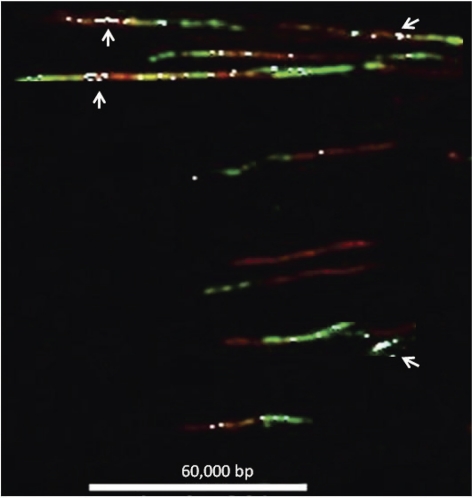
Finally we examined AP site formation in regions undergoing DNA replication in cells that had been further stressed by exposure to H2O2. Exposure to H2O2 occurred during the interval between the first pulse labeling of replicating DNA with IdU and the start of the second pulse labeling of replicating DNA with CldU. While the number of AP sites per 106 nt found globally in DNA exposed to H2O2 was 7.9, in the IdU tracks, this increased to 12.9, and in the CldU tracks, it increased to 20.8. Similar to the results shown for AP sites in replicating regions where H2O2 was not added, AP sites per 106 nt in replicating regions exposed to H2O2 are increased by 50 to 150%. These differences in AP sites per 106 nt observed at replication sites are statistically significant with P < 0.0001, as determined by the Poisson regression model. The greater increase in the formation of AP sites in areas undergoing replication indicates sites of replication are particularly vulnerable to the formation of AP sites by ROS-induced oxidative stress. AP sites were distributed as single and multiple events in the areas undergoing replication. Clustering of AP sites was detected in areas of the genome undergoing replication (Fig. 4), particularly in areas of transition between red and green label, that is, in areas replicated during exposure to hydrogen peroxide (Fig. 4). However, not all transitional areas had clusters. Occasionally, we detected small stretches with only green label (CldU, second pulse) in which AP sites were clustered heavily (one example is illustrated in Fig. 4). These areas represent origins of replication that were activated during or after exposure to hydrogen peroxide and began replicating in the presence of the second (green) pulse. We interpret this observation as indicating that some replication origins are extraordinarily sensitive to the effects of oxidative stress (AP site formation). The observation that there is an increased density of AP sites in regions of DNA fibers replicated during or after exposure to H2O2 suggests that open regions in the chromatin that form at or ahead of active replication forks are preferential targets for oxidative damage. These results are consistent with earlier observations that regions of DNA that were replicated while they were exposed to benzo(a)pyrene-diol-epoxide were more extensively adducted than nearby unreplicated regions near the replication fork (47).
Figure 4.
Detection of AP sites in areas undergoing replication in DNA fiber spreads. Composite image of multiple DNA fibers containing AP sites and areas undergoing replication. Fiber spreads were prepared from cells that were pulsed with IdU (red fluorescence) for 10 min, exposed to H2O2, and then pulsed with CldU (green fluorescence) for 20 min. IdU and CldU were identified as described in Materials and Methods. AP sites were tagged with biotin using ARP, and biotin was identified using a blue fluorescent antibody. For ease of viewing, the blue signal corresponding to AP sites was electronically changed to white. Scale bar = 60 kbp.
We applied our ability to detect AP sites in replicating DNA to determine also whether replication forks prematurely terminate when they reach AP sites, or they are able to bypass the damage and continue to replicate the DNA template. As shown in Fig. 4, AP sites can clearly be detected in the CldU tracks. Thus, it appears that replication forks are able to advance past these lesions even though they were not yet repaired, and the process proceeds rapidly since we can see multiple AP sites in tracks generated by a 20-min labeling with CldU.
AP site clustering was once thought to occur only as a result of ionizing radiation (48–50). However, recent research suggests that clustering may be a normal occurrence within cells (34–36), most likely due to endogenous ROS, and maybe more prevalent in tumors (33). The occurrence of these clusters in the genome of normal cells leads us to believe that there may be regions within the genome with increased vulnerability to ROS damage, such as regions undergoing replication. Our current analysis supports this assertion, as DNA that is in the process of replicating acquires 50 to 150% more AP sites than DNA that is not replicating, or replicated just prior to H2O2 exposure, even though the type of oxidative damage induced in areas undergoing replication is similar to what is found in bulk DNA [i.e., there are regions without any damage, regions with a single AP site and those that contain many AP sites (clusters)]. We also detected clusters of AP sites in some regions between adjoined IdU and CldU tracks (Fig. 4A, top), and also in newly initiated origins, indicating that some areas of the genome undergoing replication may constitute preferential targets for AP site formation and cluster formation. An uneven distribution of AP sites (i.e., clustering) may imply that the detrimental effects of ROS in the development of disease may not simply be due to the total number of AP sites present, but to how AP sites are distributed in the genome during replication.
The new technology presented here makes it possible to analyze a large number of genomic DNA regions during metabolically important stages, such as replication (as shown in this article) and transcription. This technology can be applied to detecting virtually every type of DNA damage, being limited only by the availability of antibodies to damage sites. We were able to confirm our prediction that replicating DNA is more vulnerable to the attack of ROS (2), as shown here by the increased level of AP site formation in regions labeled during DNA replication. In the future, this analysis could be performed with even more specificity by determining the genomic location of sites of replication that show enhanced vulnerability. This could be accomplished by coupling this technology for detection of DNA damage in DNA fibers with fluorescent in situ hybridization (FISH) to localize the damage sites in selected genomic regions that are identified by hybridization of fluorescent genomic probes.
Acknowledgments
This work was supported by U.S. National Institutes of Health grants R01-CA084493 and R21-CA125337 to D.G.K.; J.N. and J.A.S. were supported by grants P30-ES10126 and P42-ES05948; P.D.C. was also supported by grants T32-ES07017 and P42-ES05948. The authors have no conflicts of interest or financial disclosures to report.
REFERENCES
- 1.Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J. (2003) Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 [DOI] [PubMed] [Google Scholar]
- 2.Chastain P. D., 2nd, Nakamura J., Swenberg J., Kaufman D. (2006) Nonrandom AP site distribution in highly proliferative cells. FASEB J. 20, 2612–2614 [DOI] [PubMed] [Google Scholar]
- 3.Liu Y., Kao H. I., Bambara R. A. (2004) Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 4.Nakamura J., Swenberg J. A. (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 59, 2522–2526 [PubMed] [Google Scholar]
- 5.Gill R., Tsung A., Billiar T. (2010) Linking oxidative stress to inflammation: Toll-like receptors. Free Radic. Biol. Med. 48, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misra M. K., Sarwat M., Bhakuni P., Tuteja R., Tuteja N. (2009) Oxidative stress and ischemic myocardial syndromes. Med. Sci. Monit. 15, RA209–RA219 [PubMed] [Google Scholar]
- 7.Bozner P., Grishko V., LeDoux S. P., Wilson G. L., Chyan Y. C., Pappolla M. A. (1997) The amyloid beta protein induces oxidative damage of mitochondrial DNA. J. Neuropathol. Exp. Neurol. 56, 1356–1362 [DOI] [PubMed] [Google Scholar]
- 8.Multhaup G., Ruppert T., Schlicksupp A., Hesse L., Beher D., Masters C. L., Beyreuther K. (1997) Reactive oxygen species and Alzheimer's disease. Biochem. Pharmacol. 54, 533–539 [DOI] [PubMed] [Google Scholar]
- 9.Scapagnini G., Caruso C., Vasto S., Pascale A., Romeo L., D'Agata V., Intrieri M., Sapere N., Li Volti G. (2010) Genetic risk factors and candidate biomarkers for Alzheimer's disease. Front. Biosci. (Schol. Ed.) 2, 616–622 [DOI] [PubMed] [Google Scholar]
- 10.Barber S. C., Shaw P. J. (2010) Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic. Biol. Med. 48, 629–641 [DOI] [PubMed] [Google Scholar]
- 11.Mukherjee S. K., Adams J. D., Jr. (1997) The effects of aging and neurodegeneration on apoptosis-associated DNA fragmentation and the benefits of nicotinamide. Mol. Chem. Neuropathol. 32, 59–74 [DOI] [PubMed] [Google Scholar]
- 12.Radunovic A., Porto W. G., Zeman S., Leigh P. N. (1997) Increased mitochondrial superoxide dismutase activity in Parkinson's disease but not amyotrophic lateral sclerosis motor cortex. Neurosci. Lett. 239, 105–108 [DOI] [PubMed] [Google Scholar]
- 13.Miller R. L., James-Kracke M., Sun G. Y., Sun A. Y. (2009) Oxidative and inflammatory pathways in Parkinson's disease. Neurochem. Res. 34, 55–65 [DOI] [PubMed] [Google Scholar]
- 14.Halliwell B. (1992) Reactive oxygen species and the central nervous system. J. Neurochem. 59, 1609–1623 [DOI] [PubMed] [Google Scholar]
- 15.Alexander R. W. (1998) Atherosclerosis as disease of redox-sensitive genes. Trans. Am. Clin. Climatol. Assoc. 109, 129–145; discussion 145–126 [PMC free article] [PubMed] [Google Scholar]
- 16.Fiorillo C., Oliviero C., Rizzuti G., Nediani C., Pacini A., Nassi P. (1998) Oxidative stress and antioxidant defenses in renal patients receiving regular haemodialysis. Clin. Chem. Lab. Med. 36, 149–153 [DOI] [PubMed] [Google Scholar]
- 17.Singh U., Jialal I. (2006) Oxidative stress and atherosclerosis. Pathophysiology 13, 129–142 [DOI] [PubMed] [Google Scholar]
- 18.De Rosa S., Cirillo P., Paglia A., Sasso L., Di Palma V., Chiariello M. (2010) Reactive oxygen species and antioxidants in the pathophysiology of cardiovascular disease: does the actual knowledge justify a clinical approach? Curr. Vasc. Pharmacol. 8, 259–275 [DOI] [PubMed] [Google Scholar]
- 19.Tissie G., Guillermet V., Latour E., Coquelet C., Bonne C. (1988) Oxidative stress and lens opacity: an overall approach to screening anticataractous drugs. Ophthalmic Res. 20, 27–30 [DOI] [PubMed] [Google Scholar]
- 20.Varma S. D., Devamanoharan P. S., Morris S. M. (1995) Prevention of cataracts by nutritional and metabolic antioxidants. Crit. Rev. Food Sci. Nutr. 35, 111–129 [DOI] [PubMed] [Google Scholar]
- 21.Varma S. D., Devamanoharan P. S. (1995) Peroxide damage to rat lens in vitro: protective effect of dehydroascorbate. J. Ocul. Pharmacol. Ther. 11, 543–551 [DOI] [PubMed] [Google Scholar]
- 22.Berthoud V. M., Beyer E. C. (2009) Oxidative stress, lens gap junctions, and cataracts. Antioxid. Redox Signal. 11, 339–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolas M. G., Fujiki K., Murayama K., Suzuki M. T., Shindo N., Hotta Y., Iwata F., Fujimura T., Yoshikawa Y., Cho F., Kanai A. (1996) Studies on the mechanism of early onset macular degeneration in cynomolgus monkeys. II. Suppression of metallothionein synthesis in the retina in oxidative stress. Exp. Eye Res. 62, 399–408 [DOI] [PubMed] [Google Scholar]
- 24.Plafker S. M. (2010) Oxidative stress and the ubiquitin proteolytic system in age-related macular degeneration. Adv. Exp. Med. Biol. 664, 447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadtman E. R., Berlett B. S. (1998) Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab. Rev. 30, 225–243 [DOI] [PubMed] [Google Scholar]
- 26.Beckman K. B., Ames B. N. (1998) The free radical theory of aging matures. Physiol. Rev. 78, 547–581 [DOI] [PubMed] [Google Scholar]
- 27.Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., Wohlgemuth S. E., Hofer T., Seo A. Y., Sullivan R., Jobling W. A., Morrow J. D., Van Remmen H., Sedivy J. M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T. A. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484 [DOI] [PubMed] [Google Scholar]
- 28.Finkel T., Holbrook N. J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 29.Vuillaume M. (1987) Reduced oxygen species, mutation, induction and cancer initiation. Mutat. Res. 186, 43–72 [DOI] [PubMed] [Google Scholar]
- 30.DeWeese T. L., Shipman J. M., Larrier N. A., Buckley N. M., Kidd L. R., Groopman J. D., Cutler R. G., te Riele H., Nelson W. G. (1998) Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc. Natl. Acad. Sci. U. S. A. 95, 11915–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halliwell B. (2007) Oxidative stress and cancer: have we moved forward? Biochem. J. 401, 1–11 [DOI] [PubMed] [Google Scholar]
- 32.Ralph S. J., Rodriguez-Enriquez S., Neuzil J., Saavedra E., Moreno-Sanchez R. (2010) The causes of cancer revisited: “mitochondrial malignancy” and ROS-induced oncogenic transformation - why mitochondria are targets for cancer therapy. Mol. Aspects Med. 31, 145–170 [DOI] [PubMed] [Google Scholar]
- 33.Nowsheen S., Wukovich R. L., Aziz K., Kalogerinis P. T., Richardson C. C., Panayiotidis M. I., Bonner W. M., Sedelnikova O. A., Georgakilas A. G. (2009) Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat. Res. 674, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett P., Ishchenko A. A., Laval J., Paap B., Sutherland B. M. (2008) Endogenous DNA damage clusters in human hematopoietic stem and progenitor cells. Free Radic. Biol. Med. 45, 1352–1359 [DOI] [PubMed] [Google Scholar]
- 35.Bennett P. V., Cuomo N. L., Paul S., Tafrov S. T., Sutherland B. M. (2005) Endogenous DNA damage clusters in human skin, 3-D model, and cultured skin cells. Free Radic. Biol. Med. 39, 832–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland B. M., Bennett P. V., Cintron N. S., Guida P., Laval J. (2003) Low levels of endogenous oxidative damage cluster levels in unirradiated viral and human DNAs. Free Radic. Biol. Med. 35, 495–503 [DOI] [PubMed] [Google Scholar]
- 37.Pastukh V., Ruchko M., Gorodnya O., Wilson G. L., Gillespie M. N. (2007) Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic. Biol. Med. 43, 1616–1626 [DOI] [PubMed] [Google Scholar]
- 38.Buerstedde J. M., Takeda S. (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67, 179–188 [DOI] [PubMed] [Google Scholar]
- 39.Matsuzaki Y., Adachi N., Koyama H. (2002) Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 30, 3273–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrick J., Bensimon A. (1999) Single molecule analysis of DNA replication. Biochimie (Paris) 81, 859–871 [DOI] [PubMed] [Google Scholar]
- 41.Lebofsky R., Bensimon A. (2003) Single DNA molecule analysis: applications of molecular combing. Brief Funct. Genomic. Proteomic. 1, 385–396 [DOI] [PubMed] [Google Scholar]
- 42.Jackson D. A., Pombo A. (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrick C. J., Jackson D., Diffley J. F. (2004) Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279, 20067–20075 [DOI] [PubMed] [Google Scholar]
- 44.Frum R. A., Chastain P. D., 2nd, Qu P., Cohen S. M., Kaufman D. G. (2008) DNA replication in early S phase pauses near newly activated origins. Cell Cycle 7, 1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chastain P. D., 2nd, Heffernan T. P., Nevis K. R., Lin L., Kaufmann W. K., Kaufman D. G., Cordeiro-Stone M. (2006) Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle 5, 2160–2167 [DOI] [PubMed] [Google Scholar]
- 46.Frum R. A., Khondker Z. S., Kaufman D. G. (2009) Temporal differences in DNA replication during the S phase using single fiber analysis of normal human fibroblasts and glioblastoma T98G cells. Cell Cycle 8, 3133–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paules R. S., Cordeiro-Stone M., Mass M. J., Poirier M. C., Yuspa S. H., Kaufman D. G. (1988) Benzoαpyrene diol epoxide I binds to DNA at replication forks. Proc. Natl. Acad. Sci. U. S. A. 85, 2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang N., Galick H., Wallace S. S. (2004) Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair (Amst.) 3, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 49.Tian K., McTigue M., de los Santos C. (2002) Sorting the consequences of ionizing radiation: processing of 8-oxoguanine/abasic site lesions. DNA Repair (Amst.) 1, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 50.Georgakilas A. G., Bennett P. V., Sutherland B. M. (2002) High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acids Res. 30, 2800–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]