Abstract
Lipid-protein interactions are rarely characterized at a structural molecular level due to technical difficulties; however, the biological significance of understanding the mechanism of these interactions is outstanding. In this report, we provide mechanistic insight into the inhibitory complex formation of the lipid mediator sphingosylphosphorylcholine with calmodulin, the most central and ubiquitous regulator protein in calcium signaling. We applied crystallographic, thermodynamic, kinetic, and spectroscopic approaches using purified bovine calmodulin and bovine cerebral microsomal fraction to arrive at our conclusions. Here we present 1) a 1.6-Å resolution crystal structure of their complex, in which the sphingolipid occupies the conventional hydrophobic binding site on calmodulin; 2) a peculiar stoichiometry-dependent binding process: at low or high protein-to-lipid ratio calmodulin binds lipid micelles or a few lipid molecules in a compact globular conformation, respectively, and 3) evidence that the sphingolipid displaces calmodulin from its targets on cerebral microsomes. We have ascertained the specificity of the interaction using structurally related lipids as controls. Our observations reveal the structural basis of selective calmodulin inhibition by the sphingolipid. On the basis of the crystallographic and biophysical characterization of the calmodulin–sphingosylphosphorylcholine interaction, we propose a novel lipid-protein binding model, which might be applicable to other interactions as well.—Kovacs, E., Harmat, V., Tóth, J., Vértessy, B. G., Módos, K., Kardos, J., Liliom, K. Structure and mechanism of calmodulin binding to a signaling sphingolipid reveal new aspects of lipid-protein interactions.
Keywords: X-ray crystallography, isothermal titration calorimetry, stopped flow, lipid binding, calcium signaling
Calmodulin (CaM), the ubiquitous Ca2+ sensor of eukaryotic cells, is involved in a wide variety of signaling events, regulating numerous proteins, including kinases, phosphatases, ion channels, and pumps (1). CaM is a small (148 residues), acidic (pI=4.1), evolutionarily conserved protein, comprising homologous N- and C-terminal globular domains connected by a flexible central linker. Both domains possess a pair of α-helical Ca2+-binding EF-hand motifs (2). Association with Ca2+ induces the exposure of hydrophobic patches on CaM that allow binding of target proteins (3); in these complexes, CaM adopts a compact globular structure wrapped around its targets (4, 5). Key modulation of CaM function via Ca2+ binding was described decades ago. So far, no other second messenger has been shown to directly regulate CaM activity. The pharmacologic inhibitors of CaM are all synthetic aromatic molecules, of which trifluoperazine (TFP) is the most well characterized (6–8). There are reports on natural compounds presenting anti-CaM activity (9, 10), but these substances can only be found in certain species; they are not universal to eukaryotic or even mammalian cells.
Previously, we have demonstrated that sphingosylphosphorylcholine (SPC), a putative lipid second messenger (11, 12), can selectively bind to CaM and inhibit the protein's activity on target enzymes calcineurin and phosphodiesterase, while structurally similar lysophospholipid mediators, such as sphingosine-1-phosphate (S1P), lysophosphatidylcholine (LPC), and lysophosphatidic acid (LPA), showed no effect (13). This finding proposes a novel type of endogenous regulation for CaM and also suggests an intracellular target for SPC, the newly recognized sphingolipid mediator. Information on SPC metabolism is rather scarce compared to the intensively studied S1P and ceramide, yet SPC has been detected under several physiological and pathological conditions (14–17).
Here, we give an in-depth structural and mechanistic characterization of CaM binding to SPC. We believe that our results largely contribute to the understanding of lipid-protein interactions and that the inhibitory CaM-SPC complex might be of particular importance in cellular signaling.
MATERIALS AND METHODS
Preparation of CaM and SPC solutions
CaM was purified from bovine brain according to Gopalakrishna and Anderson (18), using phenyl-Sepharose affinity chromatography as the main purification step. d-erythro-sphingosylphosphorylcholine was purchased from Avanti Polar Lipids (Alabaster, AL, USA) and was delivered from a 10 mM methanolic stock solution.
X-ray crystallography
CaM and SPC were cocrystallized using the sitting drop method at room temperature. Protein solution (2 μl) at 1 mM concentration was mixed with an equal volume of precipitant solution and 10 mM SPC solution in methanol, as well as 1.5 μl 0.5% LM agarose gel. The precipitant solution was 50 mM sodium cacodylate adjusted to pH 5.5 and containing 10 mM CaCl2, 10 mM MgCl2, and 28% (w/v) polyethylene glycol 8000. Microseeding was carried out 24 h after setting up crystallization. The cryosolution contained 10% glycerol.
Data were collected at the ID 23-1 beamline of the European Synchrotron Radiation Facility. Wavelength of the beam was 0.97620 Å, and temperature was set to 100 K. Data were processed using Mosflm (19) and scaled using the SCALA program (20). The crystals are isostructural with structures of CaM complexed with various small molecules (TFP, arylalkylamine).
The structure was solved by molecular replacement using the program MOLREP (21) of the Collaborative Computational Project 4 (22). The N- and C-terminal CaM domains of PDB entry 1LIN were used as search models. Refinement was carried out with the REFMAC5 program, using TLS refinement and restrained maximum likelihood refinement (23). One TLS domain was defined for both CaM domains and one for the SPC molecules. Hydrogen atoms were generated in the riding positions. Model building was carried out using the Coot program (24).
The final model contains CaM residues 3–78 and 80–146, with 12 disordered side chains and 3 side chains in alternate positions, as well as 4 calcium ions, 4 SPC molecule fragments, and 79 water molecules. The stereochemistry of the structure was assessed with PROCHECK (25), 94.4 and 5.6% of the CaM residues are in the most favored and additional allowed regions of the Ramachandran plot, respectively. Data collection and refinement statistics are shown in Table 1. The atomic coordinates and structure factors were deposited in the PDB with accession code 3IF7.
Table 1.
Data collection and refinement statistics for the Ca2+ CaM/SPC complex
| Parameter | Ca2+CaM/SPC |
|---|---|
| Data collection | |
| Space group | P 3221 |
| Cell dimensions | |
| a, b, c (Å) | 39.67, 39.67, 170.34 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 29.393–1.60 (1.69–1.60) |
| Rmergea | 0.105 (0.446) |
| I/σI | 12.3 (4.1) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 8.6 (7.9) |
| Refinement | |
| Resolution (Å) | 29.09–1.60 (1.64–1.60) |
| Reflections | 20,426 (1554) |
| Rwork/Rfree | 0.195/0.238 (0.200/0.252) |
| Atoms | |
| Protein | 1096 |
| Ligand/ion | 80 SPC/4Ca2+ |
| Water | 79 |
| B factors (Å2) (net/residual)b | |
| Protein | 32.168/26.664 |
| Ligand | 71.108/48.393 |
| Ion | 21.570/18.778 |
| Water | 34.547 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.025 |
| Bond angles (°) | 2.085 |
Values in parentheses are for highest-resolution shell.
A high-resolution and a low-resolution data set were collected from the same crystal and were merged.
Net B factors are sum of the residual and TLS contributions.
Isothermal titration calorimetry
Thermodynamics of the interaction of Ca2+CaM and apoCaM with SPC was examined using a VP-ITC instrument (MicroCal, Piscataway, NJ, USA). Five- to 10-μl aliquots of the protein at a concentration of 300 or 75 μM in a buffer containing 10 mM HEPES, pH 7.4; 100 mM KCl; and either 5 mM CaCl2 or 1 mM EGTA was injected into the ITC cell containing 200 μM SPC in the same buffer at 25°C. A time interval of 10 minutes was applied between injections. The analysis of titration curves and the energetics calculations were done using the Origin software provided by MicroCal.
Dynamic light scattering
Size distributions of SPC-CaM solutions were assessed by an ALV goniometer with a Melles Griot diode-pumped solid-state laser at 457.5 nm wavelength (type: 58 BLD 301). The intensity of the scattered light was measured at 90° at room temperature. The autocorrelation function was calculated using an IBM PC-based data acquisition system developed in the Institute of Biophysics and Radiation Biology, Semmelweis University. The most probable particle distribution was estimated using the maximum entropy method that produces normalized distribution (26). The hydrodynamic radii of the particles were calculated based on the Einstein-Stokes equation. Size distributions calculated in this manner bear a se of <5%. Micelle size distribution is essentially independent of total SPC concentration (27); we therefore optimized SPC concentrations for the best signal-to-noise ratios.
ANS fluorescence
Micelle integrity was monitored using the fluorescent hydrophobic probe 8-anilinonapthalene-1-sulfonic acid (ANS; Fluka 10417; Sigma-Aldrich, St. Louis, MO, USA) (28) on a Jobin Yvon Fluoromax-3 spectrofluorometer (Jobin Yvon Horiba, Longiumeau, France) at 25°C. 5 μM ANS in a solution of 200 μM SPC; 10 mM HEPES, pH 7.4; 100 mM KCl; and either 5 mM CaCl2 or 1 mM EGTA was titrated with CaM in a concentration range of 0.1–20 μM in case of apoCaM and 0.1–40 μM in case of Ca2+CaM. The fluorophore was excited at 388 nm; emission was monitored from 400 to 600 nm. The excitation slit was adjusted to 2 nm, the emission slit was adjusted to 5 nm, and the maximum intensities were read.
Stopped-flow measurements
Dansyl-CaM was prepared according to Kovacs et al. (13). Fluorescence time courses were recorded at 25°C in buffers 10 mM HEPES, pH 7.4; 100 mM KCl; and either 5 mM CaCl2 or 1 mM EGTA, using an SX-20 (Applied Photophysics, Surrey, UK) stopped-flow apparatus having 2 ms dead time. Dansyl fluorescence was excited at 340 nm, and emission was selected with a 455-nm long-pass filter. At least 5 individual curves were collected and averaged for each data point. Time courses were analyzed using the curve-fitting software provided with the stopped-flow apparatus or by Origin 7 (OriginLab Corp., Northampton, MA, USA). The amplitude vs. concentration curves were fitted with the following quadratic equation (derived in ref. 29):
where [AB] is the concentration of the complex, s is y at x = 0, Amp is amplitude, c is the concentration of the constant component, and K is the dissociation constant.
Measurement of CaM binding to its cerebral microsomal targets
Cerebral microsomes were isolated from bovine brain by differential centrifugation based on the procedure of Volpe et al. (30). The interaction between microsomal proteins and CaM was monitored using the fluorescence of dansyl-labeled CaM in a Jobin Yvon Fluoromax-3 spectrofluorometer at 25°C in 10 mM HEPES, pH 7.4; 100 mM KCl; and 1 mM CaCl2. Bandwidths were set to 5 nm. Dansyl was excited at 340 nm; emission was monitored from 400 to 600 nm. Microsomal protein and dansyl-CaM concentrations were 0.5 mg/ml and 0.2 μM, respectively. Each spectrum was corrected for corresponding lipid, protein, and buffer effects by subtracting a matching buffer scan.
RESULTS
Crystal structure of the Ca2+CaM/SPC complex
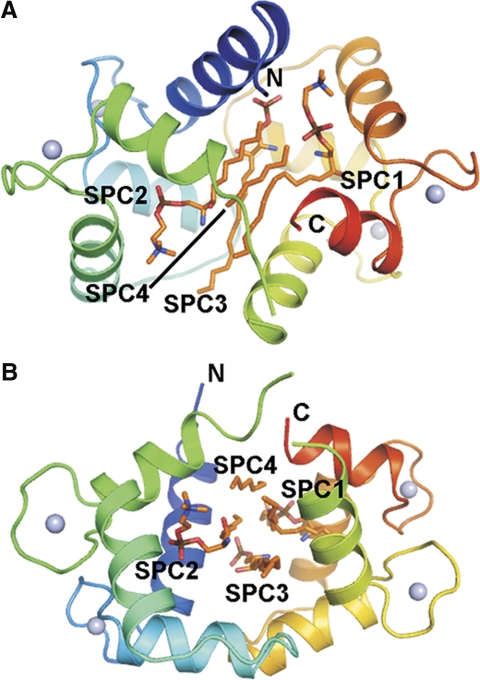
The crystal structure of the Ca2+CaM/SPC complex was refined to 1.6 Å resolution (Fig. 1). The protein is well defined by electron density, except for the central linker region and the ends of the protein chain known to be highly flexible and often disordered in CaM structures (4–6, 31). The protein is in a collapsed conformation with a binding channel of mainly hydrophobic character formed between its two domains. The hydrophobic pockets known to be adaptable to the bound ligand are open.
Figure 1.
Overall structure of the Ca2+CaM/SPC complex. A) Side view shows SPC1 and SPC2 bound in quasi-symmetry-related positions occupying the hydrophobic pockets of CaM. B) Top view shows the 4 SPC molecules bound in nearly parallel orientations. Note that not all parts of the SPC molecules can be identified in the electron density map (see text for further details). Figure was prepared using PyMol (DeLano Scientific LLC, South San Francisco, CA, USA).
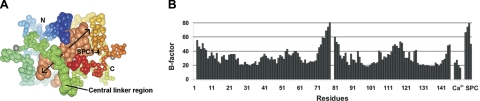
Four SPC molecules could be identified in the calculated electron density maps (Supplemental Fig. S1). Their most rigid parts are the long extended alkyl chains bound in nearly parallel positions. The binding channel between the two calmodulin domains is filled by these four chains. Parts of SPC molecules farther from the center of the complex show higher mobility with their B-factor values similar to the central linker region of the protein (Fig. 2). The alkyl chains of the sphingosyl moiety spread outside the binding channel (∼17 Å long), and the overhangs are disordered. The phosphocholine moieties could be built in electron density maps fully for two SPC molecules and partially for the third one; however, they show significantly lower electron density than the alkyl chains. Two of the SPC molecules are bent near their phosphate moieties and fill the hydrophobic pockets of CaM, while their positively charged choline groups are pointing toward the acidic regions of CaM, forming electrostatic interactions. The strongest electrostatic interactions of these charged choline groups are established with Glu11 and Glu127 (2.9–3 Å), as well as with Glu54 and Glu123 (5.4 Å). The low occupancy of the phosphocholine moieties suggests that they are partially disordered in the crystal and possibly flexible in solution. This results in a nondirected binding of the phosphocholine headgroup and in a possible mobility of the SPC molecule along its axis within the binding channel.
Figure 2.
Comparison of the mobility of SPC molecules and CaM. A) Total atomic B factors of the protein backbone atoms (in rainbow colors) and SPC atoms (light orange) represented as thermal ellipsoids. Arrows show B factor growing along SPC chains. Most mobile regions of the complex are the central linker region of CaM (residues 75–81) and SPCs. Figure was drawn using CCP4mg (39). B) B-factor plot of the structure.
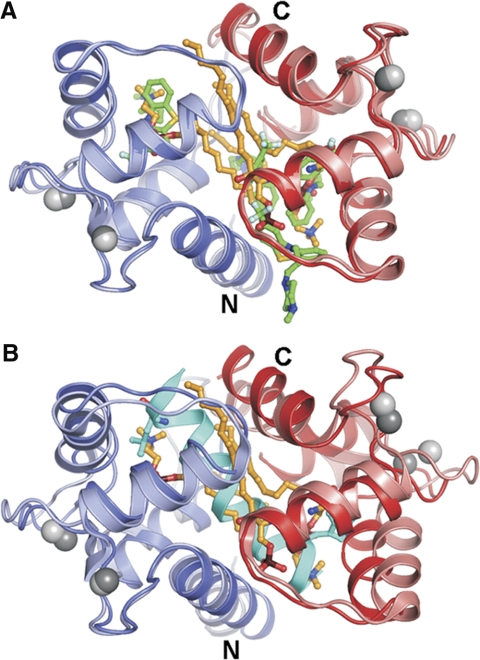
Most importantly, the overall conformation of CaM is well defined in our crystal structure and resembles its complexes with the small molecules TFP (8) and the arylalkylamine derivative AAA (6) and many of its peptide complexes (e.g., refs. 4, 5) (Fig. 3). Accordingly, residues involved in SPC binding could be also mapped to the small molecule inhibitor and target peptide binding sites (Supplemental Fig. S2). In the Ca2+CaM/SPC complex, CaM is in a collapsed overall conformation with SPC molecules bound between the two CaM domains. The fact that the lipid occupies the same binding pocket as the target peptides of CaM explains competitive inhibition witnessed in the calcineurin functional assay (13) and in peptide-binding assays (29).
Figure 3.
Conformation of Ca2+CaM in the Ca2+CaM/SPC structure is collapsed, similarly to Ca2+CaM complexes with small molecules and most of its target peptides. A) Structural alignment with the Ca2+CaM/4TFP complex (PDB accession code 1LIN). N- and C-terminal CaM domains of the Ca2+CaM/SPC complex are shown in blue and red, respectively. Compared structure is shown in lighter colors. Carbon atoms of the SPC and TFP molecules are shown in orange and green, respectively. B) Structural alignment with the Ca2+CaM/myosin light-chain kinase target peptide complex (PDB accession code 2K0F). Target peptide is shown in cyan, with its residues anchoring in the hydrophobic pockets of CaM shown as sticks. Figure was prepared using PyMol (DeLano Scientific).
Isothermal titration calorimetry provides clues for a 2-step binding model
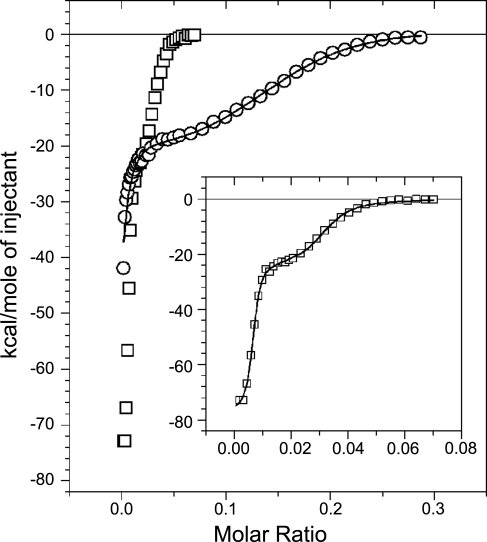
During preliminary studies on the interaction between SPC and CaM (13), we have found that in order to efficiently bind to the protein, the lipid has to be clustered, that is, in vitro, it has to form micelles. We have determined the CMC of SPC to be 33 ± 2 μM (13) and estimated a micelle to consist of ∼50–150 monomers, based on geometric calculations. As a consequence of this mode of binding, the characterization of the interaction requires special treatment. Hence, instead of applying the conventional isothermal titration calorimetry (ITC) experimental setup of ligand injection, we injected the protein into the cell containing 200 μM SPC. By keeping the lipid concentration constant, well above its critical micelle concentration, we avoided the heat effects caused by the change in the micellar state of the lipid.
In contrast to traditional CaM inhibitors, which bind only to Ca2+CaM (8, 32), SPC binds both in the presence and absence of Ca2+ (13). Therefore, we carried out experiments with both Ca2+CaM and apoCaM (Fig. 4). Parameters obtained from the fitting process are presented in Table 2. ΔH, ΔS, and Kd values correspond to the binding of one CaM molecule to one apparent binding site. Both titrations generated two clearly distinguishable binding processes. The binding of apoCaM to SPC was very similar to Ca2+CaM binding to the lipid, only it took place in a narrower concentration range, that is, with different stoichiometry (see inset in Fig. 4). In case of apoCaM, we observed an additional endothermic heat reaction, which gradually decreased during the whole experiment and overlapped with the two binding reactions (Supplemental Fig. S3). We interpreted this process as micelle rearrangement, the effect of which could be removed by subtraction for the whole concentration range, yielding an undistorted double-binding curve for data evaluation. Parameters given for the first process of the Ca2+CaM titration should be taken as approximate values (Table 2), because the beginning of the reaction could not be resolved. In general, the observed stoichiometry range for the SPC–CaM interaction is similar to what we and others have found previously for other lipid-protein interactions (33, 34).
Figure 4.
Calorimetric traces of titrating 200 μM SPC with Ca2+CaM (○) or apoCaM (□). Inset depicts the titration with apoCaM on a more appropriate scale. Solid lines show the fit by a 2-site binding model provided by the Origin for ITC software, yielding parameters listed in Table 2.
Table 2.
Thermodynamic parameters of the CaM–SPC interaction obtained from ITC measurements
| First process |
Second process |
|||||||
|---|---|---|---|---|---|---|---|---|
| n1 | Kd1 (nM) | ΔH1 (kcal/mol) | −TΔS1 (kcal/mol) | n2 | Kd2 (μM) | ΔH2 (kcal/mol) | −TΔS2 (kcal/mol) | |
| Ca2+CaM | 100–200a | 61 ± 10 | −56 ± 1 | +46 | 7 ± 0.15 | 2.1 ± 0.2 | −22.9 ± 0.5 | +15 |
| ApoCaM | 164 ± 5 | 2.4 ± 0.4 | −77 ± 1 | +65 | 40 ± 1 | 0.22 ± 0.03 | −24.7 ± 0.6 | +15.5 |
n1 and n2 are the numbers of SPC molecules bound by one CaM molecule for the given binding process. ΔH, ΔS, and Kd are values corresponding to the binding of one CaM molecule to one apparent binding site. −TΔS = R · T · ln Kd − ΔH. Reliability of the fittings is shown by the error values. Each experiment was repeated 3 times. Deviation between the individual measurements was found to be <5%.
The fitting provided acceptable results in this n1 range; Kd1 and ΔH1 values are calculated for n1 = 200.
The stoichiometry of the two binding processes enabled us to envisage the following scenarios. The first, stronger binding, for which the fitting process yielded a stoichiometry of 100–200 SPC monomers/protein and a Kd of 61 ± 10 nM for Ca2+CaM, and a stoichiometry of 164 ± 5 SPC monomers/protein and a Kd of 2.4 ± 0.4 nM for apoCaM, may be interpreted as the binding of the protein to available surfaces on micelles. The second binding process is weaker, yielding a stoichiometry of 7 ± 0.15 SPC monomers/protein and a Kd of 2.1 ± 0.2 μM for Ca2+CaM and a stoichiometry of 40 ± 1 SPC monomers/protein and a Kd of 0.22 ± 0.03 μM for apoCaM, and could result from additional protein molecules binding to the micelles. However, this scenario is quite unlikely in the case of Ca2+CaM, where the estimated number of SPC monomers per protein molecule is 7 at the end of the second ITC phase. This would mean that ∼30 CaM molecules bind to one micelle, which is hardly possible considering that the size of the protein and the micelle are comparable (see dynamic light-scattering results for evidence). More likely, the binding of further CaMs could result in the disintegration of micelles by the protein. Ca2+CaM could then isomerize in a collapsed conformation wrapped around only a few SPC monomers, as we see it in our crystal structure. ApoCaM, on the other hand, follows a reaction with the stoichiometry of ∼40 SPC monomers/protein. This stoichiometry would allow the binding of several apoCaM molecules to one micelle or, alternatively, the disintegration of micelles into smaller “parts” stabilized by the protein.
Considering the enthalpy and entropy values (Table 2), we can conclude that both reactions are entropically largely unfavorable. A study of Brokx et al. (35) on target peptide binding by CaM revealed that binding can be either entropically or enthalpically driven, but even in the second case, it is only slightly unfavorable entropically with the highest −TΔS value of approximately +5 kcal/mol. In contrast, our titrations yielded −TΔS values of +46 and +65 kcal/mol for the first reaction and +15 and +15.5 kcal/mol for the second reaction, in case of Ca2+CaM and apoCaM, respectively. Accordingly, our ΔH values are larger compared to the results of Brokx et al. (35) who obtained ΔH values between approximately −15 and +15 kcal/mol in contrast to our −56 ± 1 and −77 ± 1 kcal/mol for the first reaction and −22.9 ± 0.5 and −24.7 ± 0.6 kcal/mol for the second reaction for Ca2+CaM and apoCaM, respectively. On the other hand, ΔG (around −10 kcal/mol) and Kd values (in the nanomolar range) are similar for interactions of CaM with target peptides and SPC. The large differences in the thermodynamic properties between SPC–CaM and single ligand–CaM interactions could be explained by the different nature of the two interactions. In our experiments, the initial state of SPC is micellar, not monomeric, and instead of one ligand, a CaM molecule embraces several SPC molecules on binding.
Dynamic light scattering affirms that CaM first binds to SPC micelles, which are then disintegrated by the protein
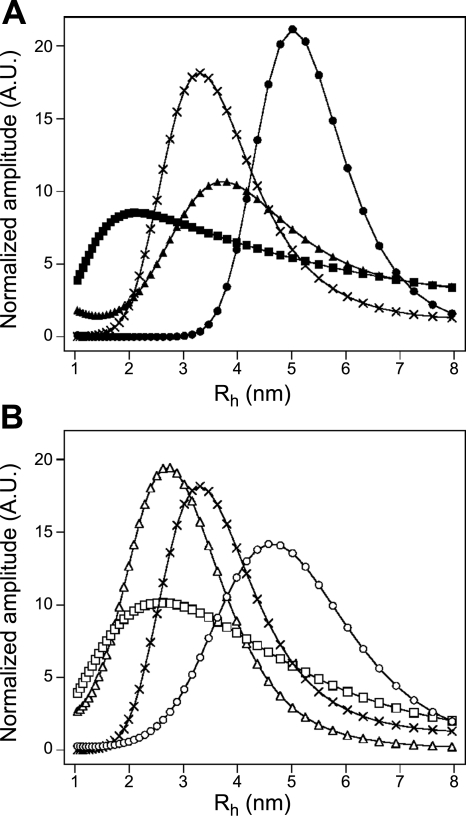
To ascertain the size distribution profile of SPC–CaM mixtures of varying stoichiometries, dynamic light scattering (DLS) measurements were carried out (Fig. 5). It is worth noting that the intensity dimension of the DLS data is not informative, since it is greatly influenced by the larger contaminating particles in the solution that vary from sample to sample. To compare the size distributions of the species present, curves were normalized to their area. The Rh values of the peaks correspond to the average hydrodynamic radii of the particles, and are, therefore, the relevant values characterizing the size of a given particle. Samples of 40 μM Ca2+CaM and 40 μM apoCaM both gave homogenous size distributions of 3.3 nm and 2.6 nm hydrodynamic radii, respectively, in accordance with preceding reports (36). A sample of pure SPC micelles revealed that micelles have an average hydrodynamic radius of 3.1 nm. In mixtures of 200 μM SPC with 8 μM Ca2+CaM or 3 μM apoCaM (concentrations near the end of the first ITC phase), an increase in particle size could be observed compared to SPC-free Ca2+CaM and apoCaM (Fig. 5); 200 μM SPC with 8 μM Ca2+CaM yielded particles with an average hydrodynamic radius of 5 nm, while 200 μM SPC with 3 μM apoCaM yielded particles with an average hydrodynamic radius of 4.5 nm. At concentrations corresponding to the end of the second ITC phase, 200 μM SPC with 40 μM Ca2+CaM or 12 μM apoCaM, the size distribution profile decreased substantially in agreement with the hypothesis that the large protein–micelle particles are disintegrating. The size distribution became less homogenous, a small amount of the large complexes could still be observed. In case of Ca2+CaM, the resulting particles are significantly smaller than Ca2+CaM alone, which indicates that the protein adopts a more compact conformation. Together with the 7 SPC monomers/protein stoichiometry estimated from the ITC fitting, this suggests that the ensuing Ca2+CaM/SPC complex is in the typical collapsed conformation seen in Ca2+CaM/inhibitor and Ca2+CaM/target peptide complexes.
Figure 5.
Dynamic light-scattering profiles of different CaM-SPC solutions in case of Ca2+CaM (A) and apoCaM (B). Symbols represent 500 μM SPC (×), 40 μM Ca2+CaM (▴), 40 μM apoCaM (▵), 200 μM SPC with 8 μM Ca2+CaM (●), 200 μM SPC with 3 μM apoCaM (○) (plateau of the first ITC phase), 200 μM SPC with 40 μM Ca2+CaM (■), and 200 μM SPC with 12 μM apoCaM (□) (plateau of the second ITC phase). Note that curves are normalized to their area, and their amplitudes are not informative. Rh values corresponding to the peaks describe the average size distribution of the population.
ANS fluorescence reveals the disruption of SPC micelles by CaM
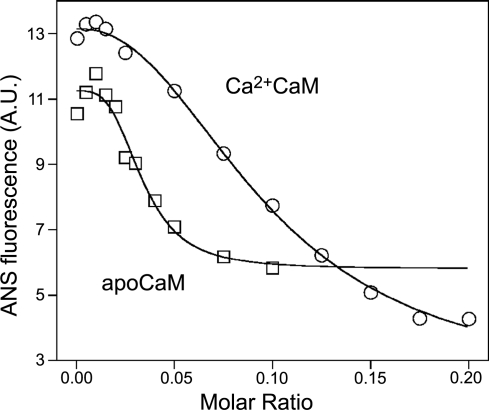
To further verify the proposed model, we monitored micelle integrity during titration with CaM. Under conditions identical to that of ITC experiments, we followed the fluorescence of the hydrophobic probe, ANS. As the intensity of ANS fluorescence increases greatly when incorporated into lipid micelles (28), it is well suited to certify the presence of micelles in a solution. The change in ANS fluorescence in the presence of 200 μM SPC on incremental addition of Ca2+CaM or apoCaM is depicted in Fig. 6. It is clearly demonstrated that increasing protein concentration disrupts micelles; moreover, their disruption occurs in the same range of molar ratios (0.02–0.2 for Ca2+CaM and 0.01–0.05 for apoCaM) as the second phase of the ITC measurements. This observation along with the results of the DLS measurements strongly argues for the explanation that the second phase witnessed in ITC experiments is a micelle disintegration process.
Figure 6.
Monitoring micelle integrity with ANS fluorescence. 200 μM SPC was titrated with Ca2+CaM (○) or apoCaM (□) similarly to ITC measurements, only monitoring the fluorescence of ANS instead of released heat. Disruption of micelles occurs in the same range of molar ratios as the second phase of ITC measurements.
Kinetic properties of the 2-step binding process
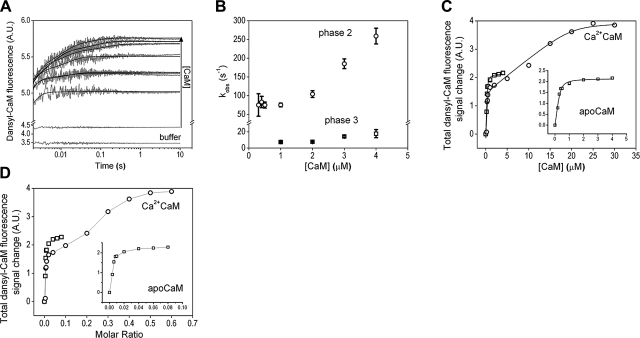
We previously determined several kinetic parameters of the CaM–SPC interaction but only at relatively low CaM-to-SPC ratios (29). In the current study, we aimed at characterizing the processes seen in the ITC experiments and implicated by the obtained Ca2+CaM/SPC crystal structure. We carried out rapid mixing experiments with both apo and Ca2+-saturated dansyl-CaM binding to SPC micelles (Fig. 7). The reaction of Ca2+CaM with SPC at the applied concentrations was too fast to be measured; the entire signal change was lost in the dead time of the stopped-flow apparatus (2 ms). Therefore, we could analyze the final fluorescence levels but not the kinetics of the fluorescence change. In the case of apoCaM, however, we could obtain transients on binding to 50 μM SPC (Fig. 7A). Similarly to the changes previously observed in equilibrium measurements (13), we obtained a large fluorescence increase on binding. The intensity increase partitioned into three kinetically distinguishable phases. The first, fast phase can only be inferred from the shift in the initial observed fluorescence compared to the reaction with buffer as the time course is lost in the dead time. This fast, initial fluorescence increase has the largest amplitude of all phases and most likely represents the collision complex of a dansyl-CaM molecule with a micelle. The second phase displayed concentration dependence only above 1 μM CaM concentration (Fig. 7B). Below 1 μM CaM, an apparently first-order process having a rate constant of 77 ± 4 s−1 is observed. The concentration-dependent part of the curve can be fitted with a linear function with a second-order rate constant of 79 ± 10 μM−1s−1. The third kinetic phase appears also above 1 μM CaM concentration and has a rate constant of 12 ± 4 s−1.
Figure 7.
Kinetic analysis of the CaM–SPC interaction. A) Time courses of 50 μM SPC binding to 0, 0.2, 0.4, 0.5, 1, 2, 3, or 4 μM apoCaM, as reported by dansyl-CaM fluorescence in a stopped-flow experiment. Smooth lines through the data represent the best exponential fits (double exponentials to the 0.4–0.8 μM, and triple exponentials to the 1–4 μM CaM data). Note the logarithmic time scale and y axis break applied for better viewing. B) Concentration dependence of the observed rate constants of the two slower phases extracted from time courses shown (A). The first kinetic phase was lost in the dead time of the stopped-flow apparatus. C) Concentration dependence of the total signal amplitudes observed in time courses recorded on mixing 50 μM SPC with various concentrations of apo (□) or Ca2+ (○) CaM. A quadratic fit to the saturating apo CaM curve yielded a Kd of 0.05 ± 0.03 μM and an apparent concentration of 0.36 μM for SPC, implying 140 monomers/micelle. The Ca2+CaM curve was fitted with a double quadratic equation and yielded Kd,1 = 0.02 ± 0.07 μM and an apparent SPC concentration of 0.44 μM (114 monomers/micelle), Kd,2 = 0.43 ± 0.80 μM and an apparent SPC concentration of 18 μM (3 monomers/interacting species). D) Fluorescence change data from C is calculated in molar ratios to show a graph comparable to the ITC results.
The concentration dependence of the total fluorescence change (Fig. 7C, inset) could be fitted with a quadratic function yielding an apparent Kd = 0.05 ± 0.03 μM. The apparent concentration of the interacting species from the quadratic fit is 0.36 μM. Given that the SPC monomer concentration was 50 μM, the interacting micelle should consist of 140 monomers, a number close to the one estimated from the ITC experiments (Table 2). Considering the monophasic nature of the saturation curve and that the obtained Kd and monomers/micelle ratio values are close to the ones observed in the first ITC binding phase, we presume that the dansyl fluorescence predominantly reports on this first process, and only a small-amplitude fraction reports on the second one. Therefore, the equilibrium constant of the two processes cannot be resolved, and the apparent Kd lies in between the relevant ones of the ITC measurement (Table 2). On the other hand, plotting the signal change in function of the molar ratios as done when displaying ITC data (Fig. 7D, inset), it is apparent that the stoichiometry dependence of the second phase and the appearance of the third phase coincide with the appearance of the second ITC process at molar ratio = 0.02. This may indicate that we can observe the binding of additional CaMs to the CaM-micelle complex (concentration-dependent second phase) and a subsequent conformational change coupled to it (third kinetic phase). These events are much slower than the formation of the initial complex likely involving only one CaM binding to a micelle.
The concentration and stoichiometry dependence of the total fluorescence change on Ca2+CaM binding to SPC can be seen in Fig. 7C, D, respectively. The smooth line through the data in Fig. 7C represents the best double quadratic fit yielding Kd,1 = 0.02 ± 0.07 μM and Kd,2 = 0.43 ± 0.80 μM. The large fitting errors restrain us to resolve only the order of magnitude of the two Kds, not their precise values. Nevertheless, two distinct processes can clearly be observed with a significant difference between the two dissociation constants. The apparent SPC concentrations from the quadratic fits (0.44 and 18 μM) imply that the interacting entities consist of ∼114 and ∼3 monomers in the first and second process, respectively. Relative Kds and the monomer numbers of the CaM-interacting partners are close to the ones calculated from the ITC experiments, suggesting that we observed the same events in both experimental setups. Comparing Fig. 7D with Fig. 4, it is even more striking that the same processes are observed using two utterly different techniques. Furthermore, the 3 SPC/Ca2+CaM ratio determined in solution is very close to the stoichiometry of the complex resolved in our crystal (4 SPC/Ca2+CaM) and inferred from the ITC experiments (7 SPC/Ca2+CaM). This implies that our observations in the crystal and in the solution phase apply to the same phenomenon and consistently describe a uniform system.
SPC disrupts the interaction between CaM and CaM-binding proteins of cerebral microsomes
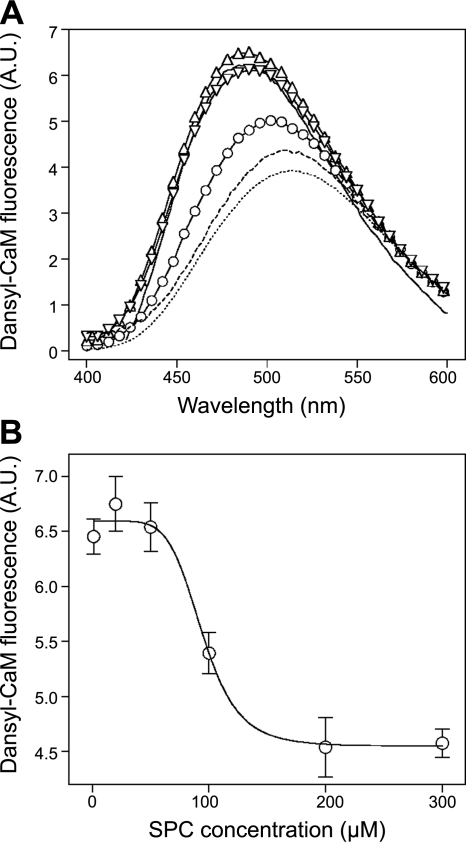
To assess the possible functional consequences of the binding mechanism described here in detail, we explored the effect of SPC on CaM bound to its targets on a cerebral microsomal fraction mimicking the intracellular environment. We used dansyl-CaM to monitor the state of CaM in the preparation, because the fluorescence of dansyl-CaM increases when bound to target proteins compared to the free or SPC-bound form (29, 37). Figure 8 reveals that in the presence of microsomes, dansyl-CaM exhibits increased fluorescence, while the addition of SPC suppresses intensity to the level of dansyl-CaM bound to SPC alone. These results indicate that the addition of SPC to cerebral microsomes displaces CaM from its targets. The displacement is selective to SPC as other structurally related lysophospholipids, such as LPC or LPA, did not show the effect (Fig. 8A). The dose response for SPC yielded an IC50 of 94.4 ± 2.2 μM (Fig. 8B). This value is ∼5-fold higher than we would expect from peptide binding assays (29) and also higher than one would estimate cellular SPC levels to be. However, we should take into account the significant amount of endogenous CaM (and other possible SPC-binding proteins) in such cellular fractions, and that all CaM, endogenous and dansyl-labeled, should equally be displaced from its targets in order to get a complete signal from the dansyl fluorophore. We should also keep in mind that CaM regulation by SPC is most probably a highly localized one, and therefore, it does not require the inhibition of a large amount of CaM-target interactions globally, as in our experimental setup.
Figure 8.
Disruption of complexes between CaM and its targets on cerebral microsomes by SPC. A) Spectra of 0.2 μM Ca2+-saturated dansyl-CaM alone (dotted trace), with 100 μM SPC (broken trace), with 0.5 mg/ml microsomal protein (solid trace), with 0.5 mg/ml microsomal protein and 100 μM SPC (○), with 0.5 mg/ml microsomal protein and 100 μM LPC (▵) and with 0.5 mg/ml microsomal protein and 100 μM LPA (▿). Spectra are representatives of three separate measurements. B) Concentration dependence of the complex dissociating ability of SPC. Ca2+-saturated dansyl-CaM and microsomal protein concentrations were 0.2 μM and 0.5 mg/ml, respectively. Fitting a sigmoidal dose-response function yielded an IC50 value of 94.4 ± 2.2 μM. Data points depict means ± se (n=3).
DISCUSSION
Previously, we have shown that the putative lipid second messenger SPC is capable of selectively inhibiting CaM in vitro (13). This finding is of particular importance, as it proposes an intracellular target site for SPC and also suggests an utterly novel type of in vivo regulation for the well-known Ca2+ sensor protein CaM. The so far characterized CaM inhibitors are all synthetic (6–8) or occur only in specific species (9, 10), while SPC is presumably present in most eukaryotic cells as an intermediate of sphingolipid metabolism (11, 12). Moreover, we have demonstrated that SPC can bind to CaM efficiently only if it is clustered, which form is manifested in micelles in vitro, raising several intriguing questions about lipid-protein interactions. It is for the above reasons that we examined the CaM–SPC interaction using several independent biochemical and biophysical methods that allow evaluation of distinct features of the binding process. On the basis of our results, we propose a complex and consistent model of CaM binding to SPC both in the presence and absence of Ca2+.
As Ca2+CaM is the functionally more important and better-characterized species, we present a detailed model on the Ca2+CaM–SPC interaction (Fig. 9). Both ITC and stopped-flow measurements revealed a strong binding of Ca2+CaM to SPC (Kd of 60 nM and 20 nM, respectively) with a stoichiometry, suggesting that the protein binds to micelles (values of 100–200 and 114 SPC/Ca2+CaM, respectively). DLS measurements verified this concept as the size of this species was determined to be significantly larger than SPC micelles or CaM alone. The driving force of this interaction is probably an electrostatic attraction between the positively charged choline groups of SPC and the negatively charged residues of CaM. This notion is aided by the fact that SPC micelles are supposedly perceived as positively charged spheres with their hydrophobic carbon chains buried inside, and also that this phase was shown to be entirely enthalpically driven in ITC experiments.
Figure 9.
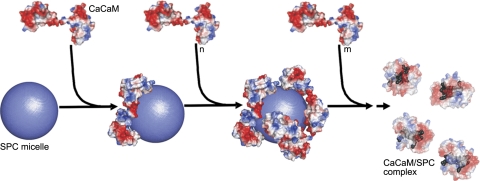
Stoichiometry-dependent binding model for the Ca2+CaM–SPC interaction. The SPC micelle is represented as a sphere with a radius of 30 Å. Molecular surfaces are colored according to surface charge distribution (red, negative; blue, positive). First process (steps 1 and 2): Ca2+CaM molecules bind to the positively charged micelle. Electrostatic interaction with the acidic CaM regions is important in this phase. Second process (step 3): Saturation of the micelle surface with protein molecules will eventually result in the disintegration of the micelle and Ca2+CaM adopting a collapsed conformation (transparent view) embracing several SPC monomers (gray). In this phase, hydrophobic interactions play an important role in complex formation. The exact mechanism by which the protein disintegrates the micelle is unknown, as depicted by the dashed arrow. Crystal structures of Ca2+CaM (PDB structure 1UP5) and Ca2+CaM/SPC (PDB structure 3IF7) were used for modeling the complexes. Note that while the Ca2+CaM–SPC micelle complex is only a model, the structure of the resulting Ca2+CaM/SPC complex has been determined by X-ray crystallography. The figure was prepared using PyMol (DeLano Scientific).
ITC and stopped-flow measurements also demonstrated that a second process takes place during binding of Ca2+CaM to SPC micelles, which occurs when micelles become limiting. This second process results in a complex with an estimated stoichiometry of 3–7 SPC monomers/protein. As our crystal structure of the Ca2+CaM/SPC complex revealed that Ca2+CaM can adopt a collapsed conformation wrapped around a few SPC molecules (4 in our structure), we hypothesized that the species resulting from the second process is identical to the one that we see in the crystal structure. This concept was supported by DLS experiments since the size of this species was significantly smaller than micelles or the protein alone. Furthermore, fluorescence measurements with ANS clearly demonstrated that micelles are disrupted during the second process. This species strikingly resembles the collapsed conformation of CaM, which the protein adopts on binding to target peptides or synthetic inhibitors (Fig. 3). Intriguingly, the peptide binding site of CaM is now occupied by several lipid molecules. Since the exact positions of the SPC headgroups were ambiguous to some extent in the crystal structure, hydrophobic interactions between the aliphatic chain of the lipid and the hydrophobic residues of the protein are more likely to contribute to the stabilization of this complex.
We envision that the surface of SPC micelles are coated by CaM, until there is not enough free space for the negatively charged protein to bind to the positive surface provided by the choline headgroups of SPC without sterically hindering each other (Fig. 9). As micelles are dynamic structures, we propose that they fluctuate during the competition of CaM molecules for their positive surface, which leads to the appearance of the collapsed protein conformation engulfing several lipid monomers. CaM molecules embracing several SPC monomers continuously leave the micelle-protein complex, thus gradually resulting in disintegration of the micelle. It is important to emphasize that this scenario only pictures one possible way the two species (CaM bound to micelles and the collapsed conformation) might arise, other possibilities also exist. What we have demonstrated unambiguously is the stoichiometry-dependent appearance of the two species, both of which are inhibitory. The lipid shields the target peptide binding sites in both CaM conformations, resulting in an inhibitory effect in enzyme activity (13) and peptide-binding assays (29).
The binding of apoCaM resembles that of Ca2+CaM, as it can be described by similar two processes, but with a difference in the complex arising in the second phase. The apoCaM-SPC complex can be characterized by a stoichiometry of ∼40 SPC monomers/protein and is larger than the one appearing in the presence of Ca2+. Presumably, this phenomenon can be explained by the fact that apoCaM is unable to adopt a collapsed conformation with a hydrophobic binding channel surrounded by acidic residues as Ca2+CaM can. The fact that apoCaM can bind to SPC makes SPC unique among CaM inhibitors since the classical CaM inhibitors are unable to bind to apoCaM (8, 32). This special behavior might be explained by the fact that the binding of the protein to the micelle is almost entirely driven by electrostatic forces, as the hydrophobic chains of the lipid are buried within the micelle. Hence, the open hydrophobic patches of the protein are not required in the initial stage of binding. In contrast, traditional aromatic inhibitors bind as monomers, offering mostly the possibility of hydrophobic interactions with CaM. The unique property of SPC—that it binds to both apo and Ca2+CaM—raises the possibility of an entirely new regulation of proteins that constitutively bind CaM (38).
We have demonstrated that SPC is capable of displacing CaM from its targets on cerebral microsomes, indicating that the interaction that we have structurally extensively characterized might have important implications in cellular signaling. A membrane surface enriched in SPC might serve as an in vivo equivalent for the SPC surface presented by micelles in vitro. It is important to note that the precise affinity and stoichiometry values determined here characterize the protein-micelle system and should not be directly applied to cellular conditions. What our model reveals about the in vivo lipid-protein interaction is the following: 1) CaM interacts with SPC clusters at high (nanomolar) affinity; 2) once the SPC-CaM complex forms, it can adopt different conformations depending on several factors, most likely on the curvature and size of the lipid cluster and the local abundance of CaM. The fact that the interaction can manifest itself in both mixed lipid (13, 29) and protein (measurements on cerebral microsomes) environments points to the plausibility of the same phenomenon to occur under cellular conditions.
In summary, we present a structural model for the binding of CaM to the lipid mediator SPC. At low protein-to-lipid ratios, CaM binds to intact micelles, whereas at high protein-to-lipid ratios micelles are disintegrated by the protein, eventuating in a compact globular conformation of Ca2+CaM, also visualized in our crystal structure. Both the micelle-bound and the collapsed conformation are inhibitory, as the lipid shields the binding sites for target peptides, potentially having important consequences in cell signaling. Moreover, this unique 2-step binding model describes a novel and peculiar biochemical mechanism, which reveals how versatile a lipid-protein interaction can be.
Supplementary Material
Acknowledgments
The authors thank the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, and Juan Sanchez Weatherby for assistance in using beamline ID23-1. The authors thank Dr. Ferenc Tölgyesi and the Institute of Biophysics and Radiation Biology, Semmelweis University, for providing the instrumentation for calorimetry experiments.
This work was supported by the Hungarian Scientific Research Fund OTKA (grants 61501 to K.L.; 67800, 67937, and 72973 to V.H.; 68464 and 81950 to J.K.; 72008 to J.T.; and 68229 and 78646 to B.G.V.), the U.S. National Institutes of Health (grant 1R01TW008130-01 to J.T.), the Howard Hughes Medical Institutes (grant 55000342 to B.G.V.), and the EU (FP6 SPINE2c LSHG-CT-2006-031220 to B.G.V.). J.T. is a recipient of the Bolyai Fellowship.
REFERENCES
- 1.Chin D., Means A. R. (2000) Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10, 322–328 [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyaya R., Meador W. E., Means A. R., Quiocho F. A. (1992) Calmodulin structure refined at 1.7 A resolution. J. Mol. Biol. 228, 1177–1192 [DOI] [PubMed] [Google Scholar]
- 3.LaPorte D. C., Wierman B. M., Storm D. R. (1980) Calcium-induced exposure of a hydrophobic surface on calmodulin. Biochemistry 19, 3814–3819 [DOI] [PubMed] [Google Scholar]
- 4.Meador W. E., Means A. R., Quiocho F. A. (1992) Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 5.Meador W. E., Means A. R., Quiocho F. A. (1993) Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science 262, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 6.Harmat V., Bocskei Z., Naray-Szabo G., Bata I., Csutor A. S., Hermecz I., Aranyi P., Szabo B., Liliom K., Vertessy B. G., Ovadi J. (2000) A new potent calmodulin antagonist with arylalkylamine structure: crystallographic, spectroscopic and functional studies. J. Mol. Biol. 297, 747–755 [DOI] [PubMed] [Google Scholar]
- 7.Osawa M., Swindells M. B., Tanikawa J., Tanaka T., Mase T., Furuya T., Ikura M. (1998) Solution structure of calmodulin-W-7 complex: the basis of diversity in molecular recognition. J. Mol. Biol. 276, 165–176 [DOI] [PubMed] [Google Scholar]
- 8.Vandonselaar M., Hickie R. A., Quail J. W., Delbaere L. T. (1994) Trifluoperazine-induced conformational change in Ca2+-calmodulin. Nat. Struct. Biol. 1, 795–801 [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Luis S., Perez-Vasquez A., Mata R. (2007) Natural products with calmodulin inhibitor properties. Phytochemistry 68, 1882–1903 [DOI] [PubMed] [Google Scholar]
- 10.Molnar A., Liliom K., Orosz F., Vertessy B. G., Ovadi J. (1995) Anti-calmodulin potency of indol alkaloids in in vitro systems. Eur. J. Pharmacol. 291, 73–82 [DOI] [PubMed] [Google Scholar]
- 11.Meyer zu Heringdorf D., Himmel H. M., Jakobs K. H. (2002) Sphingosylphosphorylcholine-biological functions and mechanisms of action. Biochim. Biophys. Acta 1582, 178–189 [DOI] [PubMed] [Google Scholar]
- 12.Nixon G. F., Mathieson F. A., Hunter I. (2008) The multi-functional role of sphingosylphosphorylcholine. Prog. Lipid. Res. 47, 62–75 [DOI] [PubMed] [Google Scholar]
- 13.Kovacs E., Liliom K. (2008) Sphingosylphosphorylcholine as a novel calmodulin inhibitor. Biochem. J. 410, 427–437 [DOI] [PubMed] [Google Scholar]
- 14.Betto R., Teresi A., Turcato F., Salviati G., Sabbadini R. A., Krown K., Glembotski C. C., Kindman L. A., Dettbarn C., Pereon Y., Yasui K., Palade P. T. (1997) Sphingosylphosphocholine modulates the ryanodine receptor/calcium-release channel of cardiac sarcoplasmic reticulum membranes. Biochem. J. 322, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liliom K., Sun G., Bunemann M., Virag T., Nusser N., Baker D. L., Wang D. A., Fabian M. J., Brandts B., Bender K., Eickel A., Malik K. U., Miller D. D., Desiderio D. M., Tigyi G., Pott L. (2001) Sphingosylphosphocholine is a naturally occurring lipid mediator in blood plasma: a possible role in regulating cardiac function via sphingolipid receptors. Biochem. J. 355, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Lafrasse C., Vanier M. T. (1999) Sphingosylphosphorylcholine in Niemann-Pick disease brain: accumulation in type A but not in type B. Neurochem. Res. 24, 199–205 [DOI] [PubMed] [Google Scholar]
- 17.Okamoto R., Arikawa J., Ishibashi M., Kawashima M., Takagi Y., Imokawa G. (2003) Sphingosylphosphorylcholine is upregulated in the stratum corneum of patients with atopic dermatitis. J. Lipid Res. 44, 93–102 [DOI] [PubMed] [Google Scholar]
- 18.Gopalakrishna R., Anderson W. B. (1982) Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem. Biophys. Res. Commun. 104, 830–836 [DOI] [PubMed] [Google Scholar]
- 19.Leslie A. G. W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsl. Protein Crystallogr. 26, 27–33 [Google Scholar]
- 20.Evans P. (2005) Scaling and assessment of data quality. Acta Crystallogr. D62, 72–82 [DOI] [PubMed] [Google Scholar]
- 21.Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 22.Collaborative Computational Project N. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R. A., MacArthur M., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 26.Bryan R. K. (1990) Solving oversampled data problems by maximum entropy. In Maximum Entropy and Bayesian Methods (Fougere P. F. ed) pp. 221–232, Kluwer Academic Publishers, The Netherlands [Google Scholar]
- 27.Yoshii N., Iwahashi K., Okazaki S.A molecular dynamics study of free energy of micelle formation for sodium dodecyl sulfate in water and its size distribution. J. Chem. Phys. 124, 184901, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Abuin E. B., Lissi E. A., Aspee A., Gonzalez F. D., Varas J. M. (1997) Fluorescence of 8-anilinonaphthalene-1-sulfonate and properties of sodium dodecyl sulfate micelles in water-urea mixtures. J. Colloid Interface Sci. 186, 332–338 [DOI] [PubMed] [Google Scholar]
- 29.Kovacs E., Toth J., Vertessy B. G., Liliom K. (2009) Dissociation of calmodulin-target peptide complexes by the lipid mediator sphingosylphosphorylcholine: implications in calcium signaling. J. Biol. Chem. 285, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpe P., Alderson-Lang B. H., Nickols G. A. (1990) Regulation of inositol 1,4,5-trisphosphate-induced Ca2+ release. I. Effect of Mg2+. Am. J. Physiol. 258, C1077–C1085 [DOI] [PubMed] [Google Scholar]
- 31.Persechini A., Kretsinger R. H. (1988) The central helix of calmodulin functions as a flexible tether. J. Biol. Chem. 263, 12175–12178 [PubMed] [Google Scholar]
- 32.Massom L., Lee H., Jarrett H. W. (1990) Trifluoperazine binding to porcine brain calmodulin and skeletal muscle troponin C. Biochemistry 29, 671–681 [DOI] [PubMed] [Google Scholar]
- 33.Pal-Gabor H., Gombos L., Micsonai A., Kovacs E., Petrik E., Kovacs J., Graf L., Fidy J., Naiki H., Goto Y., Liliom K., Kardos J. (2009) Mechanism of lysophosphatidic acid-induced amyloid fibril formation of beta(2)-microglobulin in vitro under physiological conditions. Biochemistry 48, 5689–5699 [DOI] [PubMed] [Google Scholar]
- 34.Andersen K. K., Oliveira C. L., Larsen K. L., Poulsen F. M., Callisen T. H., Westh P., Pedersen J. S., Otzen D. (2009) The role of decorated SDS micelles in sub-CMC protein denaturation and association. J. Mol. Biol. 391, 207–226 [DOI] [PubMed] [Google Scholar]
- 35.Brokx R. D., Lopez M. M., Vogel H. J., Makhatadze G. I. (2001) Energetics of target peptide binding by calmodulin reveals different modes of binding. J. Biol. Chem. 276, 14083–14091 [DOI] [PubMed] [Google Scholar]
- 36.Papish A. L., Tari L. W., Vogel H. J. (2002) Dynamic light scattering study of calmodulin-target peptide complexes. Biophys. J. 83, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucas J. L., Wang D., Sadee W. (2006) Calmodulin binding to peptides derived from the i3 loop of muscarinic receptors. Pharm. Res. 23, 647–653 [DOI] [PubMed] [Google Scholar]
- 38.Balshaw D. M., Yamaguchi N., Meissner G. (2002) Modulation of intracellular calcium-release channels by calmodulin. J. Membr. Biol. 185, 1–8 [DOI] [PubMed] [Google Scholar]
- 39.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.