Abstract
Previous transgenic-reporter and targeted-deletion studies indicate that the subset-specific expression of CD8αβ heterodimers is controlled by multiple enhancer activities, since no silencer elements had been found within the locus. We have identified such a silencer as L2a, a previously characterized ~220 bp nuclear matrix associating region (MAR) located ~4.5 kb upstream of CD8α. L2a transgenes driven by the E8I enhancer showed no reporter expression in thymic subsets or T cells in splenic, inguinal and mesenteric lymph node peripheral T cells. Deletion of L2a resulted in significant reporter de-repression, even in the CD4+CD8+ double positive (DP) thymocyte population. L2a contains binding sites for two MAR-interacting proteins, SATB1 and CDP. We found that that binding of these factors was markedly influenced by the content and spacing of L2a sub-motifs (L and S) and that SATB1 binds preferentially to the L motif both in vitro and in vivo. A small fraction of the transgenic CD8+ single positive (SP) thymocytes and peripheral CD8+ T cells bypassed L2a-silencing to give rise to variegated expression of the transgenic reporter. Crossing the L2a-containing transgene onto a SATB1 knockdown background enhanced variegated expression, suggesting that SATB1 is critical in overcoming L2a-silenced transcription.
Keywords: CD8, transcriptional silencing, transgenic mice, SATB1
Introduction
The mechanism underlying CD4 and CD8 T cell lineage commitment has been the subject of intense investigation over many years. Identification of cis-acting regulatory elements and factors controlling CD4 and CD8 gene transcription has been critical to the understanding of CD4/CD8 lineage differentiation. DNAse I hypersensitivity site (DH) measurements and transgenic reporter assays of the ~80 kb region spanning the murine genomic CD8α and CD8β locus identified four clusters (CI-CIV) of DH sites (Fig. 1A;(Hostert et al., 1997a). Subsequently four enhancers (E8i–E8iv, Fig. 1A), which overlap DH sites between CD8α and CD8β, were shown to be involved in CD8 gene expression in CD8αβ+ T cells (Ellmeier et al., 1997; Ellmeier et al., 1998; Ellmeier et al., 2002; Garefalaki et al., 2002; Hostert et al., 1998; Hostert et al., 1997b; Kioussis and Ellmeier, 2002). Each enhancer is CD8 lineage-specific and is active at a defined T cell developmental stage.
Figure 1. The L2a element.
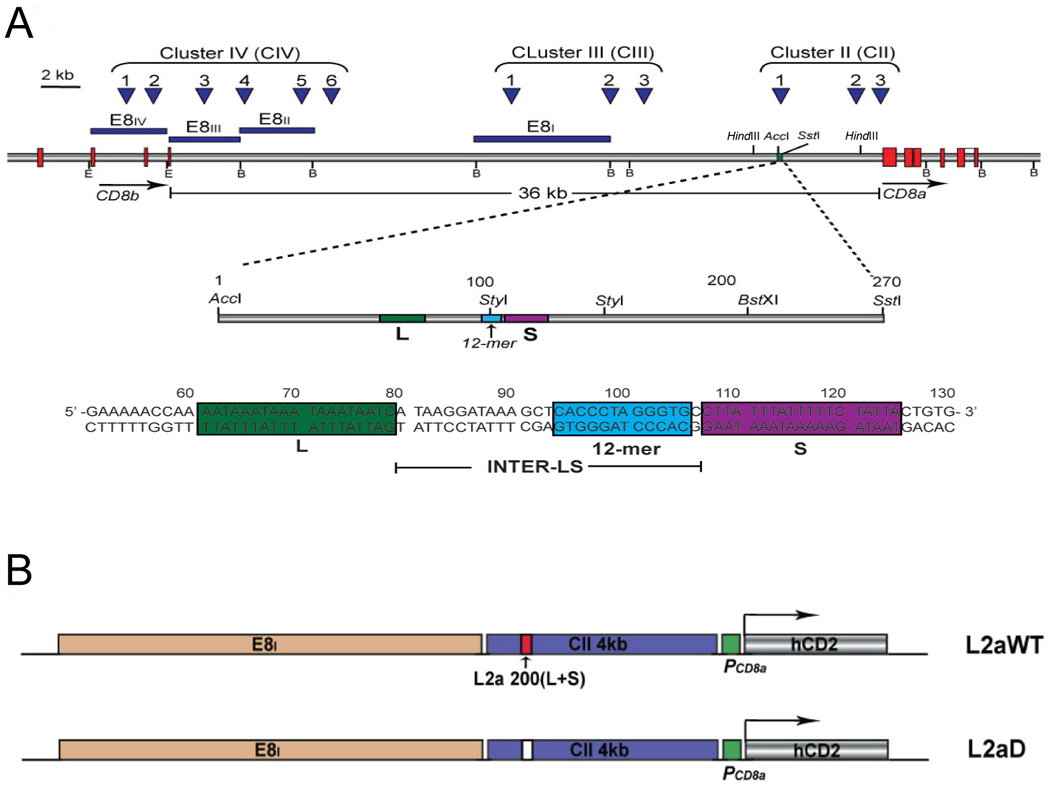
(A) Schematic of the murine CD8 locus and intergenic region with expanded details of L2a. Positions of established exons (red boxes), enhancer regions (blue lines) and DNAse 1 hypersensitivity sites (blue arrows) shown to be critical for CD8 transcriptional regulation are indicated. A third cluster of two hypersensitive sites (Cluster I, not shown) resides 3’ to CD8a but is yet to be implicated in its regulation. The L2a element (green) is located ~4.5 kb upstream of the mouse CD8a gene. The 270 bp AccI/SstI fragment spanning L2a is expanded to show the L, S, and INTER-LS regions identified in footprinting studies. A 12 bp palindromic sequence is found located at the end of the INTER-LS region close to the S region. (B) Schematic of the transgenic constructs. Both wildtype (WT) and L2a deleted (L2aD) constructs were derived from a human CD2 reporter gene construct (Sawada et al., 1994) and a CD8 intergenic construct (Tg-a) modified by Ellmeier et al. (1997). For L2aWT, a 4.3 kb HindIII/HindIII DH Cluster II fragment containing L2a was introduced upstream of the CD8α promoter, and a 7.6 kb BamHI/BamHI fragment containing the E8I enhancer was subcloned upstream of DH cluster II fragment. Components of the constructs are highlighted. The L2aD deletion mutant was generated by deletion of a 200 bp AccI/BstXI fragment spanning L2a. This fragment served as the WT200 L+S probe in EMSA experiments.
Studies on the cis-regulatory elements within these enhancers revealed a complex network of multiple lineage-specific elements. E8I, which overlaps CIII-1,2 (DH cluster III sites 1 and 2), was shown to regulate expression of the CD8a gene in mature CD8+ T cells from spleen and inguinal lymph nodes and in CD8αα homodimer-expressing intraepithelial lymphocytes (IELs). Inactive in double positive (DP) thymocytes, E8I becomes functional after positive selection (Ellmeier et al., 1998; Hostert et al., 1998). Enhancer E8II (CIV-4, 5) directs reporter expression in DP and CD8SP thymocytes as well as in mature peripheral CD8+ T cells. E8III (CIV-3) is only active in DP thymocytes, whereas E8IV (CIV-1,2) directs reporter gene expression in both CD8+ T cells and a subset of CD4+ T cells (Ellmeier et al., 1998). These enhancers (II-IV) are active in thymus-derived T cells, but not in CD8αα+ IELs.
Targeted disruption of E8I (CIII-1,2) in vivo had no effect on CD8α and CD8β expression in thymus-derived T cells, but CD8αα expression in IELs was eliminated, suggesting that other cis-acting elements could compensate for the loss of E8I in thymus-derived T cells (Ellmeier et al., 1998; Hostert et al., 1998). Combined deletion of E8I and E8II resulted in variegated expression of CD8SP and DP thymocytes as well as in reduced CD8 expression on mature CD8+ T cells (Ellmeier et al., 2002). Further studies demonstrated that E8I and E8II deletion led to impaired chromatin remodeling during T cell development (Bilic et al., 2006). Combined targeted deletion of E8II and E8III did not significantly alter expression levels of CD8α and CD8β in thymocytes or T cells (Feik et al., 2005). DH cluster II was inactive in transgenic reporter analysis, but as observed in E8I-E8II double deletion mice, cluster II deletion led to impaired CD8 SP frequencies with a small % of variegated expression observed in peripheral CD8+ T cells (Garefalaki et al., 2002).
The L2a element is located ~4.5 kb upstream of the CD8a gene (Fig. 1A, lower section) where it resides within the first DH site of DH cluster II (CII-1), a short distance upstream of two GATA-3 binding sites (Landry et al., 1993). L2a was defined by in vitro nuclear matrix binding assays as a nuclear matrix attachment region (MAR) (Banan et al., 1997). MARs frequently reside in close proximity to promoters and enhancers and have been shown to be important transcriptional regulators during chromatin remodeling (Hart and Laemmli, 1998). In stable transfection studies, inclusion within constructs of a 900 bp fragment that spanned L2a significantly reduced the frequency of CD8+ T cells, suggesting that L2a may negatively regulate CD8 transcription (Lee et al., 1994). This interpretation is consistent with the observations (cited above) that deletion of the DH cluster II/CII-1,2 region (which spans L2a) results in variegated CD8 expression (Garefalaki et al., 2002) and that inclusion of a fragment spanning DH cluster II enables E8I to activate reporter gene expression in DP thymocytes (Hostert et al., 1998).
Two MAR-binding proteins, SATB1 (Special AT-rich Binding protein 1) and CDP (CCAAT displacement protein), bind L2a within AT-rich regions (L and S) separated by a DH-containing region, INTER-LS (Banan et al., 1997; Lee et al., 1994) (Fig. 1A). SATB1 is predominantly expressed in the thymus and has been shown to bind to numerous MARs, including those within the IgH and TCRβ enhancers (Scheuermann and Garrard, 1999), and to associate with chromatin remodeling complexes (Yasui et al., 2002). SATB1 has been shown to be critical for expression of the MHC Class I locus and the TH2 cytokine locus (Cai et al., 2006; Galande et al., 2007; Kumar et al., 2007). Targeted disruption of SATB1 blocked thymocyte development primarily at the DP stage, suggesting an essential role for SATB1in the development and maturation of CD+ 8 SP T cells (Alvarez et al., 2000; Sinclair et al., 2001). Recent studies showed that SATB1 can promote breast tumor growth and metastasis by re-programming chromatin organization and transcriptional profiles (Han et al., 2008). CDP is ubiquitously expressed and binds to MARs and regulatory regions of many genes of immunological interest. CDP knockout mice display neonatal lethality (Luong et al., 2002) with both intrinsic and extrinsic defects in lymphoid development (Sinclair et al., 2001; Zhu et al., 2004).
Here we utilized a transgenic approach to test whether L2a is the element within DH cluster II responsible for modifying the function of the E8I enhancer. Our results identify L2a as a silencer for regulating CD8 expression. They further implicate SATB1 as a positive transactivator whose expression contributes to reversing the L2a-mediated silenced state.
2. Materials and Methods
2.1 Generation of transgenic mice
Wildtype (L2aWT) transgene construction was performed as described previously (Ellmeier et al., 1998). The human (h) CD2 reporter gene was derived from a mouse CD4 reporter gene construct produced by Sawada and coworkers (Sawada et al., 1994). hCD2 contains the mouse CD4 exon I, a portion of intron I lacking the CD4 silencer, and the untranslated portion of exon II (a CD4 splicing module) fused to a human CD2 cDNA (Sayre et al., 1987) with the SV40 polyadenylation site. It was modified by Ellmeier et al. (1998) to contain a polylinker and a PCR-amplified mouse CD8a promoter (Nakauchi et al., 1987). Mutant (L2aD) was created by a deleting the 210 bp AccI-BstX1 fragment spanning L2a from L2aWT. The ~18 kB inserts were excised from the vector using Not I, purified, and microinjected into the male pronucleus of C57BL/6 zygotes as previously described (Nie et al., 2005). The C57BL/6 zygotes were chosen to generate transgenic mice to obtain an inbred background. After the microinjection of transgene constructs, the zygotes that survived injection were cultured overnight and those that developed to 2-cell embryos were transferred into the oviducts of 0.5-dpc pseudopregnant female C57BL/6 mice.
2.2 Generation of transgenic mice with reduced levels of SATB1 protein
L2aWT, L2aD and L2aR transgenic mice were crossed to mice termed SATB1 “reduced” (Nie et al., 2005) in which SATB1 expression was reduced specifically in T cells by homozygous anti-sense inhibition (strain designated as As+/+) as measured by quantitative western analysis in peripheral T cells and in thymocytes. In addition, to further reduce SATB1 in T cells, one SATB1 allele in As+/+ was disrupted by homologous recombination (strain designation As+/+SATB1+/−) (Nie et al., 2005). Backcrosses of L2aWT and L2aD mice with As+/+ mice produced L2aWT+As+/+ and L2aD+As+/+ mice. These mice were then bred to As+/+SATB1+/− such that ~25% of the offspring resulted in the most highly reduced (~32% of wildtype) L2aWT+As+/+SATB1+/− and L2aD+As+/+SATB1+/− genotypes. Southern blots of tail DNA were used to identify positive founders that developed from injected zygotes using a probe from the CD4 splicing module of the hCD2 portion of the transgenes (Sawada et al., 1994). Founders were bred to C57BL/6 to obtain transgenic progeny for subsequent analyses. All animals analyzed were between 4 and 8 wk old following euthanasia according to IACUC guidelines.
2.3 Isolation of lymphocytes from mouse thymus, lymph nodes and spleen
Thymus, inguinal lymph nodes and spleen were removed from euthanized mice and placed into 60mm dishes containing HBSS (Sigma) buffer. Tissues were passed through a 70 micron nylon cell strainer (BD Biosciences) to prepare single cell suspensions. To remove red blood cells, isolated cells were incubated in RBC lysis buffer (0.5M NH4Cl, 0.15M Tris-HCl [pH7.65]) for 5 minutes at room temperature. Cells were washed with HBSS and ready for desired treatment and analysis.
2.4 Isolation of intestinal IELs
IELs were isolated by a modified method based on a procedure previously described (Ellmeier et al., 1997). The small intestine was removed from euthanized mice and washed with RPMI medium. The small intestine was turned inside-out over a glass tubing and incubated in 30ml of RPMI for 45 minutes at 37°C with low speed rotation to release the IELs. The released IELs was passed through a 70 micron cell strainer to filter out debris. Cells were centrifuged (1,000 rpm, room temperature) and resuspended in appropriate volume of RPMI medium. Cells were then purified with Ficoll-Pague Plus (Amersham) centrifugation (2,000 rpm, 30 min, room temperature), and washed with HBSS buffer.
2.5 Flow cytometric analyses
Isolated cells were washed with HBSS (Sigma) buffer twice at 1,000 rpm, 4°C, and resuspended in Hanks buffer (HBSS with 2% FBS and 0.1% sodium azide) on ice. Cells were counted and ~1×106 cells were used for subsequent staining. After incubation on ice with Fc-block for 15 min, cells were stained with the desired antibodies for 45 min. Flow cytometric analysis was carried out with monoclonal antibodies (BD Pharmingen) directed at the following cell surface molecules: hCD4 (RM4–5), CD8α (53-6.7), CD8β (53-5.8) and human CD2. Following two washes with Hanks buffer and one wash with HBSS, cells were fixed in 1% paraformaldehyde and analyzed immediately. Cells requiring secondary antibody staining were incubated on ice with the appropriate reagent for 30 min after the wash steps of the first staining. Cells were then washed and analyzed on a BD FACScalibur using CellQuest Pro software. Accurate CD8α transcriptional reporting by hCD2 staining has been established in previous studies (Ellmeier et al., 1997; Ellmeier et al., 1998; Ellmeier et al., 2002; Garefalaki et al., 2002; Hostert et al., 1998; Hostert et al., 1997b; Kioussis and Ellmeier, 2002) and was confirmed in several of our transgenic lines by semi-quantitative RT-PCR (data not shown).
2.7 Cell sorting
Thymic, lymph node and splenic tissues were teased to a single-cell suspension through a 70 µm cell strainer (BD Labware) in tissue medium. Cells were centrifuged at 485g for 5 min and resuspended in tissue medium. Cells were labeled with anti-CD4 and CD8 antibodies and separated on magnetically labeled MicroBeads (Miltenyi Biotec GmbH) as previously described (Nie et al., 2008). Retained fractions were eluted and used immediately for culture and subsequent studies. Thymocyte populations were also sorted by flow cytometry on a BD FACS Aria to >95% purity based on CD4/CD8 expression off a CD3 gate (Nie et al., 2005).
2.8 Preparation of nuclear extracts
Nuclear extracts were prepared as described by Dignam et al. (Dignam et al., 1983). All steps were performed at 4°C or on ice. Approximately 2×108 cells were harvested and washed in PBS. The cells were resuspended in 5 ml buffer A (10mM HEPES [pH7.9], 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, and protease inhibitor cocktail (Roche) and incubated for 10 min. The cells were pelleted at 1,000g for 10 min and resuspended in 2ml of buffer A. Cells were lysed with an homogenizer (10 strokes, B pestle), and the nuclei were pelleted at 1,000g for 10 min. The nuclei were washed once with 2 ml of buffer A and then resuspended in 0.5–1.0 ml of buffer C (20mM HEPES [pH7.9], 25% glycerol, 0.42M NaCl, 1.5mM MgCl2, 0.2mM EDTA, 0.5mM DTT, and protease inhibitor cocktail), and homogenized (20 strokes, A pestle). The sample was magnetically stirred for 30 min and then pelleted at 15,000g for 30 min. The supernatant was recovered and dialyzed against buffer D (20mM HEPES [pH 7.9], 20% glycerol, 0.1M KCl, 0.2mM EDTA, 0.5mM DTT, and protease inhibitor cocktail) for 3 hr. After dialysis, nuclear extracts were centrifuged at 15,000g for 20 min, and supernatants were quick frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined by Bradford assay.
2.9 Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts (2–5µg) were mixed with poly-(dI-dC)(2µg) in binding buffer (20mM HEPES [pH 7.9], 20% glycerol, 0.1M KCl, 0.2mM EDTA, 10mM DTT, and protease inhibitor cocktail). Binding reactions were performed in 25µl total volume at room temperature for 5 min. After 20 min incubation with end-labeled probe (0.2 ng), samples were electrophoresed at 120V for ~3 hr through a 4% polyacrylamide gel (29:1) in 1xTBE buffer (90mM Tris-HCl [pH 8.0], 90mM boric acid, and 2mM EDTA). Gels were dried for 1 hr and autoradiographed for 4 hr, using phosphoimage screens, or overnight, using films with intensifying screens at −80°C. For competition assays, unlabeled competitor DNA fragments were added to the reaction before the addition of radiolabeled probes. For antibody inhibition assays, 2µl of appropriate dilutions of polyclonal anti-SATB1 (Banan et al., 1997) and anti-CDP (Wang et al., 1999) were added to the reaction before the addition of radiolabeled probes. Immunodepletions were carried out as previously described (Banan et al., 1997) under conditions and criteria detailed in the legend to Figure 3.
Figure 3. Differential recognition and binding modalities of SATB1 and CDP for L2a.
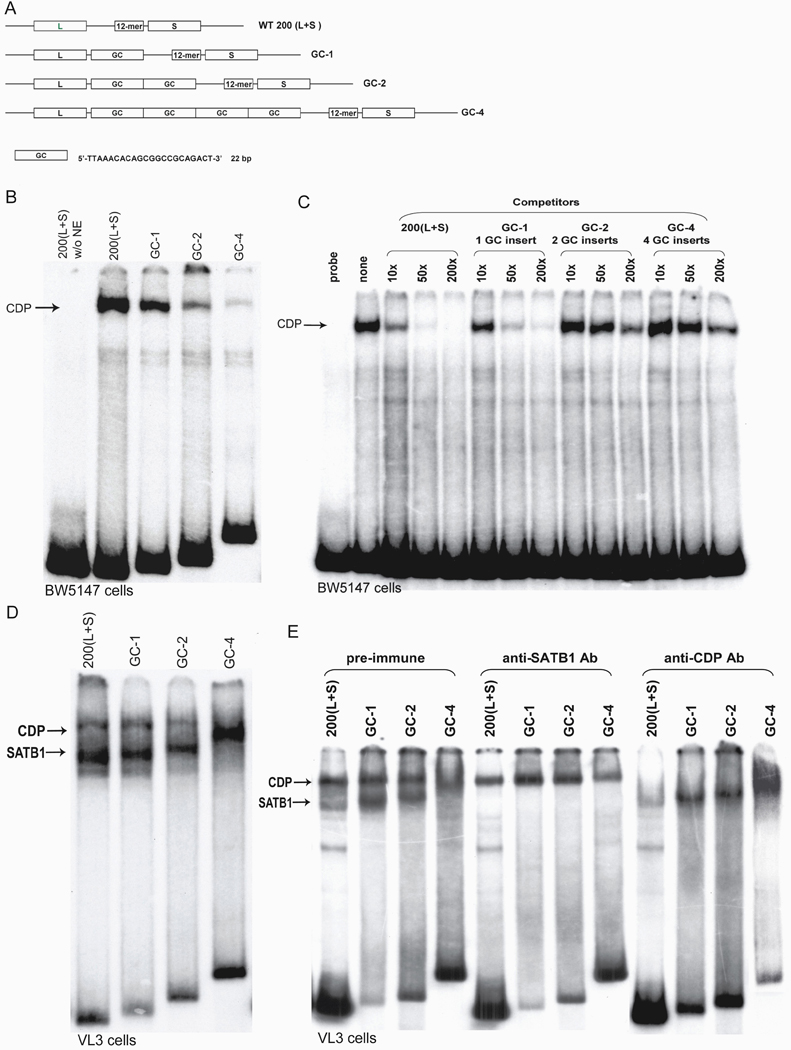
(A) Schematic of the wildtype L2a element, WT200 (L+S), and mutants with elongated INTER-LS regions. The insert sequence (GC) is shown below the construct schematics. GC-1, 2 and 4 contained one insert, two inserts and four spacer inserts, respectively, separating the L and S sites. (B) Weakened binding of CDP to L2a mutants with elongated GC inserts between the L and S sites. EMSA analysis was performed with BW5147 nuclear extract, radiolabeled 200(L+S) probe and mutated probes. The arrow indicates CDP complexes. (C) Probes with increased distance between the L and sites are unable to compete for CDP binding. Competition EMSAs were performed with BW5147 nuclear extract, WT200(L+S) probe, and unlabeled mutated GC-1, GC-2, GC-4 competitors. 10–200 molar excess of competitors were added after the addition of radiolabeled probe. (D) Supershifted pattern of SATB1 binding to mutant probes. EMSA was performed with VL3 nuclear extract, radiolabeled 200(L+S), and mutated 200(L+S) GC probes. (E) CDP and SATB1 binding patterns in the absence of the other protein. Antibodies to SATB1 or CDP used in EMSA supershift assays in (D) were added to VL3 nuclear extracts followed by addition of radiolabeled WT200(L+S) and mutated GC probes. The arrows indicate SATB1 or CDP complexes. No shifted complexes were observed for probe alone lanes or antibody plus probe only lanes (data not shown). Please note that the ratios of the SATB1/CDP bands are higher in the absence of pre-immune sera (compare lanes of Fig. 3D to equivalent lanes in Fig 3E, pre-immune). Since pre-immune sera does not react with either of the purified proteins or with their DNA complexes, we suspect that selective degradation or non-specific interactions with other proteins in the nuclear extracts accounts for these differences.
2.10 Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (Sheldon et al., 2001) with minor modifications. Total thymocytes or magnetically sorted DP and CD8SP thymocytes were fixed in PBS containing 1% formaldehyde at 37 °C for 10 min, followed by further incubation at 4 °C for 2 hr. Nuclei were isolated and digested with micrococcal nuclease at 37 °C for 10 min. Soluble material containing predominantly mono-nucleosome and bi-nucleosome (>90%) was isolated by centrifugation and diluted with buffer containing 0.01% SDS, 1% Triton X-100, 1.5 mM EDTA, 15 mM Tris, pH 8.0, 15 mM NaCl, 5 mM sodium butyrate, 10 mM NaF, and 1X protease inhibitor cocktail (Roche). Nucleosomes were pre-cleared first by incubation with protein G beads alone, then mouse IgG1 and protein G beads. Pre-cleared samples were divided and incubated overnight with anti-SATB1 mouse monoclonal antibody (Transduction Labs) or mouse IgG1. Agarose-conjugated immune complexes were washed, eluted and then digested with proteinase K, followed by phenol/chloroform extraction and ethanol precipitation with glycogen, and the precipitate was resuspended in 50 µl of TE buffer. Five ng of DNA was added to a PCR reaction using either primers that amplify the entire L2a region (forward: CATCATGCTGGCAATGAACT; reverse: GGGAAGCAGTGAGCAACTTC) or primers specific for the L sub-region (forward: AAGTGAGGTCCAGGACAGTC; reverse: GACCAGCATGAGTGCTTCTG). Twenty five cycles of PCR were performed (TaqDNA polymerase kit; Invitrogen Life Technologies) in which each cycle consisted of 30 s of denaturing at 94°C, 1 min of annealing at 59°C, 90 s of elongation at 72°C, and 10 min at 72°C. PCR products were resolved on 5% acrylamide gels and stained during electrophoresis by ethidium bromide.
2.11 DNase I footprinting
The DNase I footprinting of EMSA-isolated bands was performed as described (Banan et al., 1997). Following addition of optimized concentrations of DNase I (Ambion), samples were were loaded onto a 4% polyacrylamide gel (29:1) and electrophoresed at 120V for ~3 hr. The retarded bands and free probes were excised, eluted in 0.2M NaCl-TE, phenol:chloroform (2:1) extracted and ethanol precipitated with 100µg/ml yeast tRNA carrier. The dried DNA samples were resuspended in 0.1M NaOH:formamide (1:2 v/v), 0.1% xylene cyanol, 0.1% bromophenol blue) and then electrophoresed through an 8% sequencing gel. The markers were produced by PCR using a DNA sequencing kit (Promega).
2.12 Cell lines
The murine T cell lines VL3.B4 (provided by Dr. I. Weissman, Stanford University) and BW5147 were grown in DMEM media supplemented with 10% fetal bovine serum (FBS).
3. Results
3.1 Construction and initial expression analyses of wild-type and L2a-lacking transgenes
An hCD2 reporter gene previously demonstrated to accurately report CD8α expression (Owen et al., 1988) was utilized for our transgenic constructs (Fig. 1A). In addition to the wild type L2a-containing construct (L2aWT), a 210 bp AccI-BstX1 fragment was deleted from the 4 kb DH cluster II fragment to generate a construct lacking L2a (termed L2aD; Fig. 1B). Seven independent L2aWT and 5 independent L2aD mutant strains with copy numbers varying from 2 to >10 (Table 1, Suppl. Fig. 1) were constructed in C57BL/6 mice. We observed no correlation of copy number with reporter expression (Table 1), nor did we observe significant variation of reporter expression within any of the transgenic lines after 2–3 generations (data not shown). Data were consistent among all of the independent lines with the exception of L2aWT1 and L2aD3 (Table 1) in which aberrant expression likely resulted from positional effects. No significant alterations in T cell development were observed in any of the transgenic strains (representative analyses in Fig. 2A–C, left panels and full data sets in Suppl. Figs. 2–4).
Table 1.
Summary of L2a transgenic analyses.
| Transgene |
L2aWT |
L2aD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Line | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 |
| Copy Number | 2 | >10 | 4 | >10 | >10 | >10 | >10 | >10 | 2 | 1 | >10 | >10 |
| CD8SP (%hCD2/MFI) | 80.4/69 | 1.5/47 | 0 | 5.4/42 | 2.5/49 | 1.1/91 | 0 | 85.1/46 | 23.7/26 | 0 | 25.7/89 | 74.6/63 |
| DP (%hCD2/MFI) | 80.3/58 | 0 | 0 | 0 | 0 | 0 | 0 | 79.8/93 | 66.4/29 | 0 | 25.8/45 | 41.2/33 |
| Conclusion in Thymus | Active | Sil/Var | inactive | Sil/Var | Sil/Var | Sil/Var | inactive | Active | Active | inactive | Active | Active |
| CD8+ (%hCD2/MFI) | 99.3/69 | 1.6/107 | 0 | 8.9/61 | 7.1/82 | 2.6/129 | 0.37/33 | 99.8/186 | 16.2/25.7 | 0 | 16.1/93 | 99.6/96 |
| Conclusion in Lymph Node | Active | Sil/Var | inactive | Sil/Var | Sil/Var | Sil/Var | Sil/Var | Active | Active | inactive | Active | Active |
| CD8αβ (%hCD2/MFI) | 74.2/38 | N.A. | 0 | 4.0/43 | 5.5/50 | N.A. | N.A. | 83.1/45 | 8.0/26 | 0 | N.A. | 90.3/52 |
| CD8αα (%hCD2/MFI) | 89.1/54 | N.A. | 0 | 1.5/56 | 0.46/96 | N.A. | N.A. | 95.4/112 | 14.1/25 | 0 | N.A. | 92.3/81 |
| Conclusion in IEL | Active | N.A. | inactive | Sil/Var | Sil/Var | N.A. | N.A. | Active | Active | inactive | N.A. | Active |
The copy number, percentages and MFIs of hCD2 positive (>1.0%) cells within CD8SP, DP and mature CD8 subsets are shown. For each transgenic line, at least 3 mice were tested, and no significant variation of reporter expression was observed in any transgenic line after 2–3 generations. Sil/Var: Silenced/Variegated. N.A, not analyzed.
Figure 2. L2a silences CD8 reporter expression on thymic, lymph node and intraepithelial (IEL) T cells.
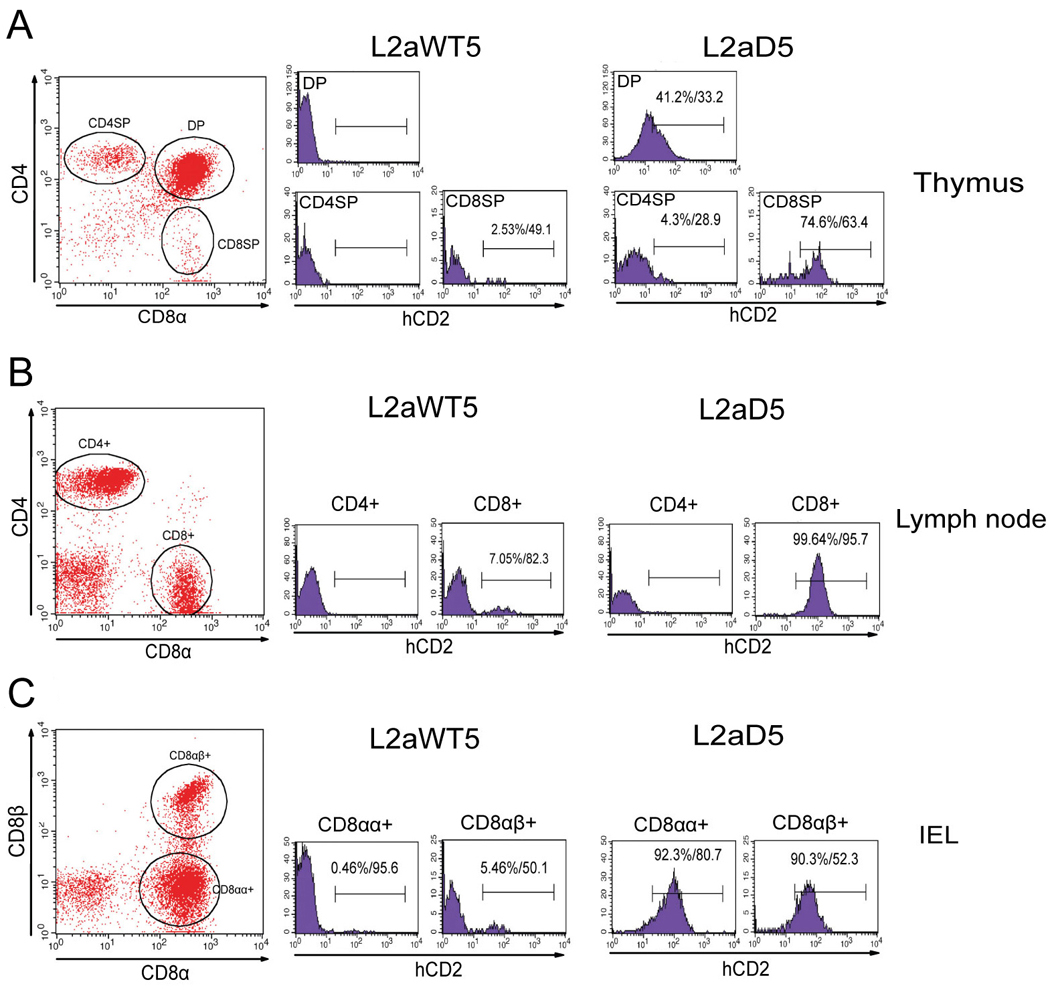
Representative fluorescent activated cytometric analyses of T cells isolated from one (L2aWT5) of seven independent L2aWT transgenic mice and one (L2aD5) of five independent L2aD transgenic lines. Transgenic thymic (A) and lymph node (B) were stained with anti-CD4, anti-CD8α and anti-hCD2. Transgenic intestinal IELs (C) were stained with anti-CD8α, anti-CD8β and anti-hCD2. Expression of the hCD2 reporter gene on gated populations (circled in red) were analyzed and results are represented in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets are shown.
Figure 4. SATB1 binds preferentially to L2a in vitro and in vivo.
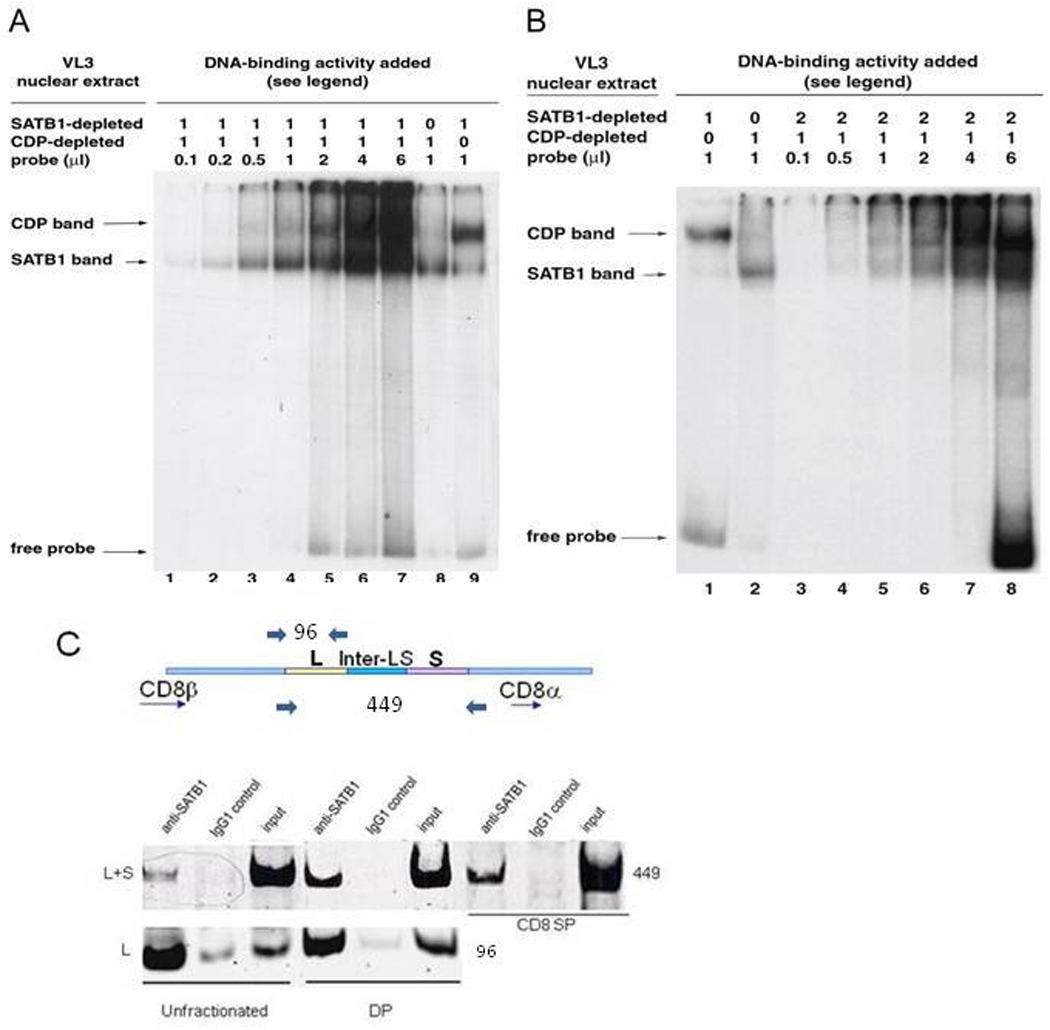
(A) In vitro EMSA binding competition by equivalent amounts of CDP and SATB1. Since purified preparations of endogenous, post-translationally modified SATB1 and CDP were unavailable, competition experiments were carried with VL3 nuclear extracts that had been immunodepleted of either protein. An equivalent amount of CDP or SATB1 was defined as the quantity in SATB1-depleted or CDP-depleted VL3 extract required to shift 90% of 1 ng of the wildtype L2a (L200 L+S) probe. Lanes 1–7 in (A) and Lanes 3–8 in (B) contain SATB1- depleted and CDP-depleted VL3 extract in amounts equivalent to CDP and SATB1 protein present singly in lanes 8 and 9 in (A), and Lanes 1 and 2 in (B). The small amount of SATB1- retarded band observed with the SATB1-depleted extract alone (A, Lane 9) reflects careful titration of the anti-SATB1 antibody used for depletion so that no free anti-SATB1 antibody remains behind to interact with SATB1 in the CDP-depleted extract following mixing. Similar precautions were taken to determine the amount of anti-CDP antibody used to deplete VL3 extract of CDP. (B) In vitro EMSA binding competition assays using CDP in 2-fold excess. Immunodepleted VL3 nuclear extract was tested essentially as described for (A) except that the CDP binding activity (SATB1-depleted extract, lanes 3–8) was in two-fold excess. Amounts of probe in (A) and (B) are as indicated above the figure. No shifted complexes were observed for probe alone lanes or antibody plus probe only lanes (data not shown). (C) In vivo recruitment of SATB1 to L2a chromatin in unfractionated, DP, and CD8SP thymocytes. Primer pairs indicated by arrows above the L2a schematic were designed (Materials and Methods) to amplify either the entire L2a region (L+S) or the sub-site (L), which was established by EMSA and footprinting experiments to be the binding region for SATB1 in vitro. The input lanes were loaded as 10% of IP lanes. Amplicon sizes are indicated on the schematic and to the right of the gel.
3.2 Transgenic expression of a CD8α reporter containing L2a is silenced in T cells
A representative analysis of thymocytes isolated from one of seven independent transgenic mice carrying the L2aWT transgene is shown in Fig. 2A (center panels). Six out of seven of our L2aWT transgenic mice showed very low to undetectable levels of hCD2 expression in thymus-derived DP and CD8SP or peripheral CD8+ T cells from inguinal LN (Figs. 2A and 2B, Table 1, Suppl. Figs. 2 and 3). Only one L2a-containing transgenic line, L2aWT1, expressed significant levels of hCD2 in CD8SP (80.4%) and DP (80.3%) cells (Table 1, Suppl. Figs. 2 and 3). This may result from a strongly enhancing position effect of transgene integration. Neither CD4SP thymocytes nor LN-derived CD4+ T cells expressed hCD2 on their surface, confirming the CD8+ T cell subset specificity of the transgenic E8I-driven construct (Figs. 2B and 2C, Suppl. Figs. 2 and 3).
3.3 Reporter expression in both thymocytes and mature T cells is observed in transgenic mice lacking the L2a element
Representative flow cytometric analyses of hCD2 expression on thymocytes and lymph nodes from one of five L2aD transgenic mice are shown in Figs. 2A and B (right panels) with full data sets shown in Suppl. Figs. 6 and 7, and summarized in Table 1. Significant CD8 reporter expression was observed in CD8SP and DP thymocytes and in LN CD8+ T cells in 4 of 5 L2a-deleted mice. Thus, L2a is the element responsible for silencing the reporter activity of our WT transgenes in the thymus and the periphery.
We also observed that four of five L2aD transgenics expressed detectable levels of hCD2 reporter activity in CD4SP thymocytes but not in peripheral CD4+ T cells (Fig. 2, Suppl. Figs. 6–8). These results suggest that in addition to its silencer function, L2a may contribute to the exclusive CD8 subset specificity of the E8I enhancer.
3.4 L2a represses CD8 reporter expression in CD8αα+ intraepithelial lymphocytes
CD8αα+ homodimers are expressed on IELs within the small intestine (Traver et al., 2000). To determine if the hCD2 gene is co-expressed on cells bearing CD8αα+ homodimers, IELs were isolated from both L2aWT and L2aD transgenic mice and tested for reporter expression (Fig. 2C, Table I, Suppl. Fig. 4 and 8). Reporter expression on both CD8αα+ and CD8αβ+ IELs isolated from the L2aWT strains showed similar silencing as CD8+ T cells isolated from inguinal LN. Furthermore, variegated reporter expression was also observed in IELs from L2aWT mice. In IELs isolated from L2aD mice, which lack the L2a element in the transgene, hCD2 reporter expression was observed on both CD8αα+ and CD8αβ+ subsets at levels comparable to those in lymph node CD8+ T cells (Fig. 2B and Table I). Thus, L2a acts as a silencer element that controls CD8α transgene reporter expression in CD8αα+ IELs as well as in CD8αβ thymocytes and peripheral T cells.
3.5 Variegated expression of CD8α reporter activity in CD8 T cells
Although our six transgenic L2aWT strains demonstrated suppressed reporter expression in thymic CD8SP and CD8+ peripheral T cells, four independent strains exhibited significant hCD2 expression levels within a small population (1–5%) of CD8SP thymocytes (Figs. 2A and 2B, Table 1, and Suppl. Fig. 2). This small subset had similar reporter levels (MFI: 42 – 91) as CD8SP thymocytes in L2aD mice (see below) and they formed a population clearly distinct from the majority of reporter-negative cells. A small population of CD8+, hCD2-positive T cells (2–9%) with significant MFI (61–107) was observed in LN as well (Fig. 1C, Table 1, Suppl. Fig. 3).
These populations are reminiscent of E8I–mediated variegated expression observed previously (Ellmeier et al., 1997; Ellmeier et al., 1998; Ellmeier et al., 2002; Garefalaki et al., 2002; Hostert et al., 1998; Hostert et al., 1997b; Kioussis and Ellmeier, 2002). Sorted CD8SP transgenic thymocytes were treated with pronase to remove surface hCD2 and placed in culture at 37°C for 18 hours. Reporter expression on both L2aWT5 and L2aD5 sorted cells was restored within 18 hours of treatment (Suppl. Fig. 5). This indicated that the transgenes have retained their ability to re-express the reporter molecules and that the L2aWT hCD2 reporter activity in the small fraction of CD8+ T cells is the product of stable, variegated expression. Further consistent with this conclusion, four independent lines of transgenic mice that were bred for several generations expressed a reporter positive variegated reporter population of CD8+ T cells in thymus and the periphery.
These results indicated that rare cells have bypassed L2a-mediated silencing and exhibit normal CD8 cell surface protein expression, resulting in variegated expression of the hCD2 reporter within the majority of the repressed CD8+ T cell subset.
3.6 Differential recognition and binding modalities of SATB1 and CDP for L2a
The L2a element was first identified as a MAR which bound SATB1 and CDP at the A/T-rich L site (Fig. 1A) (Banan et al., 1997). We reasoned that determining the mode by which SATB1 and CDP bind to L2a might help to explain how the L2a region represses CD8 reporter transcription.
Banan et al (1997) had suggested that CDP binding required both the L and S motifs (Fig. 1A), whereas SATB1 binding required only L, indicating that the spacing and/or composition of the intervening region (INTER-LS spacer) was critical. We created L2a mutants in which the INTER-LS spacer was expanded by variable GC-rich, 22 bp insertions (Fig. 3A). These were evaluated in EMSAs using nuclear extracts from BW5147, a DN thymoma that expresses CDP, but not SATB1, and in VL3, a CD8+ T cell lymphoma that expresses both CDP and SATB1 (Banan, et al, 1997). EMSA performed in BW5147 revealed that expanding the INTER-LS spacer markedly decreased CDP binding (Fig. 3B). Consistent with this observation, unlabeled spacer mutant fragments competed poorly compared to wild type for CDP binding (Fig. 3C). Spacer insertions significantly reduced CDP binding in VL3 (Fig. 3D), although the inclusion of non-specific (Fig. 3E pre-immune lanes) or anti-SATB1 (Fig. 3E anti-SATB1 Ab lanes) antibodies appeared to ameliorate inhibition, particularly with the shorter (GC-2 and GC-4) insertions.
In contrast, SATB1 bound to all of the mutant probes (Fig. 3D), often forming stronger complexes with the insertions (Fig. 3E). Notably, SATB1 bound most strongly to the most extended (GC-4) spacer to which CDP bound most poorly (Fig. 3D). Furthermore, the mobilities of the SATB-1 complexes were retarded as spacer length was increased (Figs. 3D, E and re-addressed below). Anti-CDP and anti-SATB1 antibody ablations eliminated their respective complexes, but had no reciprocal effects, suggesting that binding of SATB1 and CDP is not cooperative (Fig. 3E). Two experiments support this interpretation: First, when the VL3 extract was immunodepleted (Materials and Methods) of the CDP-DNA complex and EMSA carried out with the spacer mutated (GC-1 and GC-2) oligos as probes, the pattern of SATB1 binding (Fig. 3E) was similar to that observed in Fig 3D (data not shown). Second, DNAse footprinting of these same probes indicated that CDP binds to both L and S wild-type L2a, but did not induce any conformational alterations, and binding is lost as the spacer is extended; SATB1 binds primarily to L in all probes with strong hypersensitivity induced within the INTER-LS (Suppl. Fig. 9 and 10)
These results are consistent with the predictions of Banan et al (1997) and indicate that CDP binding to L2a is influenced by spatial constraints imposed by the distance or conformational relationships between the L and S motifs. SATB1 appears to bind to the L-region independently of CDP regardless of context alteration (ie, DNase I hypersensitivity) achieved by INTER-LS insertions.
3.7 SATB1 outcompetes CDP in binding to the L2a element in vitro
A potential consequence of the differential L2a binding modes of CDP and SATB1 would be that one of the two factors may be able to efficiently displace the other from occupied L motifs. To test this possibility in vitro, protein competition-EMSA assays were performed using nuclear extracts from the VL3 cell line that had been immunodepleted of one or the other factor (Materials and Methods). This allowed us to compare relative binding of the two factors and their T cell-associated complexes semi-quantitatively using equivalent amounts of L2a binding activity (Fig. 4A, lanes 8–9; Fig 4B, lanes 1–2). When extracts containing binding equivalents of CDP and SATB1 were mixed 1:1, SATB1, but not CDP bound limiting amounts of radiolabeled L+S (WT200) probe; Fig. 4A, lanes 1–4). CDP-DNA complexes were observed only where the probe was no longer limiting (Fig. 4A, lanes 5–7). The same results were observed when two-fold excesses of either CDP (Fig 4B, lanes 3–8) or SATB1 (data not shown) binding activity were analyzed in similar mixing experiments.
3.8 SATB1 is recruited to L2a L-site chromatin in T cells
The above observations indicate that SATB1, a transcriptional activator in thymocytes, binds to L2a with significantly higher affinity than the general transcriptional repressor, CDP. We performed ChIP assays using primers designed to specifically amplify the entire L2a region or the L-site-bound chromatin. As shown in Fig. 4C, L2a-site-containing chromatin prepared from unfractionated, DP and CD8SP thymocytes was specifically immunoprecipitated by anti-SATB1 (Fig. 4C). Specific SATB1 recruitment was also observed for the L-site. However, recruitment of SATB1 to the S-site or recruitment of CDP to L2a of either subsite was undetectable (data not shown).
We conclude that L2a, and specifically the L region of L2a, is an in vivo target of SATB1 in thymocyte populations where functional repression via L2a was observed.
3.9 L2a-dependent CD8 variegated expression requires SATB1 in vivo
We reasoned that if SATB1 promotes L2a-mediated CD8 silencing, reducing SATB1 levels should increase the variegated CD8 expression observed in L2a WT transgenics (Fig. 2, Table 1, Suppl. Figs. 2–4). Alternatively, if SATB1 overcomes L2a-depending silencing mediated by CDP (or some unknown repressor), variegated expression levels should decrease. SATB1 homozygous knockout (KO) mice survive to only 3 weeks after birth with T cell development blocked mainly at the DP stage; the few SP T cells remaining undergo rapid apoptosis (Alvarez et al., 2000). To circumvent this problem, we bred our L2aWT and L2aD transgenes onto strains (Nie et al, 2005) in which T-cell specific expression of SATB1 is reduced to ~62% of WT levels by homozygous expression of an LCK-driven SATB1 antisense (As+/+) transgene. We achieved stronger SATB1 reduction (~32% of WT) by crossing L2aWT/ As+/+ or L2aD/As+/+ mice onto heterozygous SATB1 KO (SATB1+/−) mice, yielding L2aWT+As+/+SATB1+/− and L2aWT+As+/+SATB1+/− test mice.
Under highly reduced (L2a+As+/+SATB1+/−) SATB1 conditions, variegated expression of L2aWT was reduced significantly in both thymic (~3-fold) and lymph node (~2-fold) derived CD8+ T cells (Fig. 5, upper panels). No effect of SATB1 reduction was observed in L2aD mutant T cell subsets (Fig. 5, lower panels), confirming the effect required L2a. These results suggest that SATB1 is involved in overcoming silenced CD8 expression mediated by L2a.
Figure 5. L2a-dependent variegated CD8 reporter expression requires SATB1 in vivo.
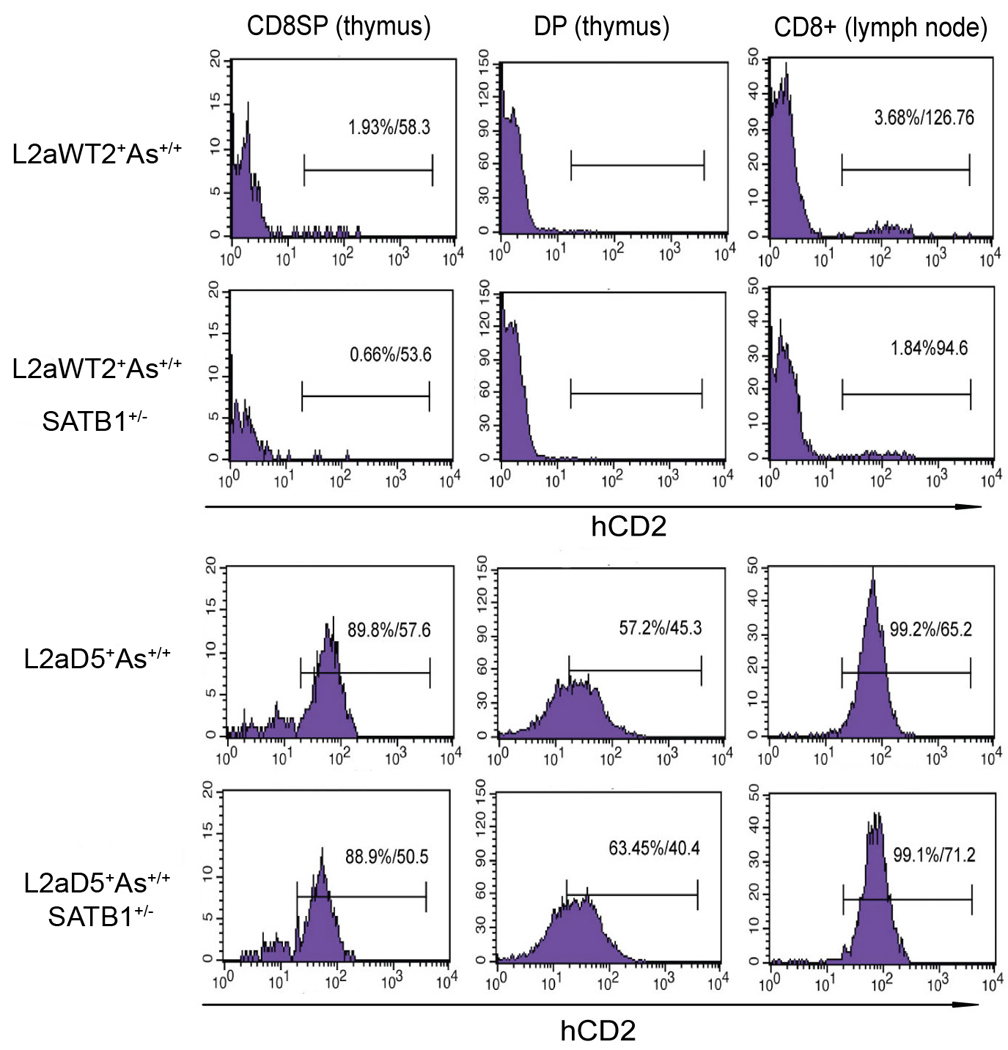
L2aWT2 (top panels) and L2aD5 (lower panels) transgenic mice were crossed to SATB1 “reduced” mice that exhibit ~62% (As+/+) or ~32% (As+/+SATB1+/−) of WT SATB1 levels in thymic and peripheral T cells (Nie et. al, 2005; Materials and Methods). Expression of the hCD2 reporter on thymic CD8SP and DP or from CD8+ lymph node T cells was analyzed and percentages and MFIs of hCD2 positive (>1.0%) subsets are shown as representative of 3 independent experiments. Under highly reduced (L2a+As+/+SATB1+/−) SATB1 conditions, variegated expression of L2aWT was reduced significantly whereas no effect of SATB1 reduction was observed on L2aD mutant mice.
4. Discussion
The transgenic studies reported here indicate that the L2a element is a cis regulatory silencer of CD8α. In mice containing the L2a wild type transgene (L2aWT), CD8 reporter (hCD2) expression is repressed in both DP and CD8SP thymocytes (summarized in Table 1), suggesting that the L2a silencing occurs at an early stage in CD8 T cell development. CD8 reporter expression within a small population (~5%) of CD8SP thymocytes and peripheral CD8+ T cells bypassed silencing, resulting in variegated reporter expression in these two populations. When the L2a element was deleted (L2aD mice), robust expression of the CD8 reporter was observed in all thymic and lymph node CD8 expressing cells, including DP thymocytes (Table 1). Parallel results were observed for L2a-mediated CD8 repression with variegated expression in CD8αα+ IELs (Table 1).
Our results are consistent with those of Garefalaki et al. (2002) in which a DH cluster II, L2a-lacking transgene allowed E8I to activate a CD8 reporter in DP thymocytes. Our data suggest that thymocytes and IELs share similar CD8 regulatory mechanisms and are consistent with the view that the thymus is the critical source of IEL CD8αα T cells (Cheroutre and Lambolez, 2008).
Our results appear to conflict with observations showing that the L2a-containing DH cluster II region facilitates E8I enhancer-driven reporter expression in DP thymocytes (Hostert et al., 1998) and IELs (Ellmeier et al., 1998). An explanation may be revealed by a closer comparison of the transgenes employed. Hostert et al. (1998) placed the DH cluster II downstream of the hCD2 reporter, in the opposite direction with respect to CD8 transcription and with respect to the germline configuration employed in our constructs (Fig. 1A, Suppl. Fig. 11A). Although the orientation of E8I enhancer does not affect reporter expression (Ellmeier et al., 1997; Ellmeier et al., 1998; Hostert et al., 1998; Hostert et al., 1997b), this is not the case for for L2a. In the single line (L2aR) that we created in which the orientation of the HindIII fragment (containing DH Cluster II and L2a) was reversed (Suppl. Fig. 11A), L2a silencing of hCD2 reporter expression was de-repressed in both DP and CD8SP thymocytes as well as in CD8+ splenocytes (Suppl. Fig. 11B). These results indicate that the silencing function and SATB1-mediated transactivation of the L2a element is sensitive to positional orientation
The modest yet stable levels of variegated CD8 reporter expression observed in L2aWT constructs was CD8-restricted, as predicted by previous studies employing E8I–driven transgenes (Ellmeier et al., 1997; Ellmeier et al., 1998; Hostert et al., 1998; Hostert et al., 1997b). However, we found that loss of the L2a silencing element seemed to compromise subset specificity of the E8I enhancer, allowing a modest but consistently detectable level of CD8 reporter expression in a small fraction of CD4SP thymocytes but not in CD4+ peripheral T cells (Fig. 2, Suppl. Figure 6–8). Breech of E8I specificity has been previously attributed to transgene integration artifacts (Ellmeier et al., 1997; Garefalaki et al., 2002), but this seems an unlikely explanation for our results given the multiple L2aD transgenic lines tested. The small CD4+ population we observed may represent CD4+CD8αα+ thymus-derived IELs with Treg-like properties that are generally absent in the periphery (Das et al., 2003). Alternatively, they may represent CD4hiCD8low DP T cells that have been blocked through L2a-mediated silencing from undergoing co-receptor reversal transition to CD8hiSP (Singer et al., 2008).
Nuclear matrix-associated regions (MARs) are short AT-rich DNA sequences that are widespread throughout the eukaryotic genome and have great affinity for the nuclear matrix in vitro (Blasquez et al., 1989; Sander and Hsieh, 1985). MAR-binding proteins SATB1 and CDP were shown previously to specifically interact with the L2a element (Banan et al., 1997). Our in vitro analyses revealed that CDP and SATB1 recognize the L and S motifs within the L2a element in fundamentally different ways when their spacing and, potentially, their conformation are perturbed by elongating the intervening spacer with GC insertions (Fig. 3). We found that CDP binding requires the presence of both L and S sites and is highly sensitive to proper spacing, as strongly suggested by probe mobility and DNase1 hypersensitivity changes (Fig. 3 and Suppl. Fig. 9). The CDP protein contains a homeodomain and three cut repeats, and each of them may have specific and unique DNA binding activities (Maeda et al., 2002; Nepveu, 2001).
In contrast, SATB1 binding was unperturbed or even more avid in direct proportion to spacer elongation (Fig. 3 and Suppl. Fig. 10). SATB1 binding sites normally show a propensity to become stably base-unpaired (Dickinson et al., 1992). In addition, SATB1 can act by itself or as a platform for remodeling factors to induce novel chromatin architecture in the thymocyte nucleus (Cai et al., 2003; Cai et al., 2006; Pavan Kumar et al., 2006; Yasui et al., 2002). These features, along with the significant increase in DNase I hypersensitivity induced within the Inter-LS spacer by SATB1 binding, may underlie both the observed retardation of SATB1-DNA complexes as well as its strong L-site affinity (Figs. 3, 4, and Suppl. Fig. 10). When coupled with previous observations that the CDP repressor associates with DNA in a rapid and unstable fashion (Sansregret and Nepveu, 2008), it was not surprising that SATB1 successfully outcompeted CDP for L-site binding (Fig. 4).
It has been proposed by others that ratios of SATB1 and CDP may determine the outcome of silencing or activation (Liu et al., 1999). Given the strong dominance of SATB1 L-site binding, we tested whether its reduction in vivo promoted or de-repressed L2a-mediated CD8 reporter silencing. When the L2WT mice were crossed onto a background reduced ~5-fold in SATB1 expression, reporter activity within the small fraction of variegated-expressing CD8SP thymocytes and peripheral CD8+ T cells was consistently decreased 2–3-fold (Fig. 5). SATB1 expression required both the presence (Fig. 5) and appropriate orientation (Suppl. Fig. 11B) of L2a. These results suggest that SATB1 is required to re-establish CD8 expression exerted by L2a at the E8I enhancer, or the CD8α promoter, or both. Our findings are also consistent with the previous finding (Nie et al., 2008) that SATB1 is indispensable for re-initiation of CD8 transcription during coreceptor reversal (transition from CD4+CD8low DP to CD8hi SP). We could not address this issue further because our transgenes lack the E8III DP enhancer, which also may account for our inability to detect DP variegated expression (Fig. 2, Table 1). We also point out that studies by Kiefer et al (2002) and our own unpublished EMSA experiments identified a number of additional SATB1 sites scattered across the CD8β-CD8α intergenic region, including two within Cluster II. While these sites played no role in SATB1-mediated variegated expression of our transgenic reporter (Fig. 5), they must be considered in terms of endogenous locus regulation. These issues must await results from L2a and L sub-site knockout mice currently under investigation. Our efforts to test the complimentary prediction--that CDP promotes L2a-mediated silencing-- were uninformative (data not shown), owing we suspect to the maximum reduction of less than 2-fold achievable in the heterozygous CDP knockout background (homozygotes are early embryonic lethal; Sinclair et al., 2001).
We conclude that L2a represents the first identified cis-acting silencer within the CD8 locus. This ~220 bp MAR may act alone or in concert with other, as yet to be identified regulatory control sequences within DH Clusters II and III, to contribute to silencing or activation decisions crucial to CD8SP thymic development and peripheral CD8+ expression. On the basis of our results and earlier studies on SATB1 and CDP and their interaction with the L2a element (Banan et al., 1997; Lee et al., 1994), we propose that SATB1 and CDP operate with L2a as a switch for activating or silencing CD8α transcription.
Supplementary Material
(A) Schematic of the transgenic constructs. Components of the constructs are highlighted in different colors and detailed in the text and in Figure 1. The L2a element (red) resides in the first DH site of DH cluster II. L2aD has a deleted 200 bp (L+S) sequence in the HindIII/HindIII fragment. (B) Southern hybridization was used to identify positive transgenic mice, and the copy numbers were determined by comparing signal intensities of wild type and transgene bands.
DNase I footprinting was performed with VL3 nuclear extract, 200(L+S), GC-1 and GC-4 mutated probes. Retarded bands of probe-protein complexes and corresponding free probes were excised from EMSA (DNase I treated) gels for footprinting analysis. Protected regions are labeled as L and S.
(A) Comparison of the L2aWT and L2aR transgenes. The 4.3 kb HindIII fragment containing the DH CII region and L2a site was cloned in reverse in L2aR relative to its germline orientation in L2aWT. (B) Effect of reversed L2a orientation on transgenic hCD2 expression under normal conditions (L2aR+SATB1+/+, black boxes) and under conditions in which SATB1 has been reduced to ~32% wild type levels (L2aR+AS+/+SATB1+/−, gray boxes) in thymocytes (left) and splenocytes (right). Derivation of the normal and SATB1-reduced L2aR transgenic lines, FACS gating and derivation of mean fluorescence intensities (MFI) were described in Materials and Methods. MFIs, quantified as the average of 3 independent experiments, are provided below corresponding columns.
FACS analysis of thymocytes isolated from seven independent L2aWT transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color FACS analysis. Expression of the hCD2 reporter gene on gated DP, CD4SP, and CD8SP thymocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
Fluorescent activated cytometric analysis of lymph node T cells isolated from seven independent L2aWT transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color cytometric analysis. Expression of the hCD2 reporter gene on gated CD4+ and CD8+ lymphocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
IELs were isolated from four L2aWT transgenic mice, and were stained with the combination of anti-CD8α, anti-CD8β and anti-hCD2 antibodies. Gated IEL subsets were analyzed for hCD2 reporter expression. Percentages and MFIs of hCD2 positive (>1.0%) subsets are shown.
FAC sorted CD8+ T cell populations were treated with pronase and placed in culture at 37°C for 18 hr or 4°C as controls. hCD2 and CD8 expression on L2aWT5 sorted cells were upregulated efficiently 18 hour after the pronase treatment. Percentages and MFIs of hCD2 positive subsets are shown.
Fluorescent activated cell analysis of thymocytes isolated from five independent L2aD transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color fluorescent staining analyses. Expression of the hCD2 reporter gene on gated DP, CD4SP and CD8SP thymocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
Fluorescent activated cell analysis of lymph node T cells isolated from five independent L2aD transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color Fluorescent activated cell analysis. Expression of the hCD2 reporter gene on gated CD4+ and CD8+ lymphocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
IELs were isolated from four L2aD transgenic mice, and were stained with anti-CD8α, anti-CD8β and anti-hCD2 antibodies. Gated IEL subsets were analyzed for hCD2 reporter expression. Percentages and MFIs of hCD2 positive (>1.0%) subsets are shown. Lymph node L2aD results of Suppl. Fig. 6 are included for comparative purposes.
DNase I footprinting was performed with BW5147 nuclear extract, 200(L+S), GC-1, and GC-2 mutated probes as described in Materials and Methods. Retarded bands of probe-protein complexes and corresponding free probes were excised from EMSA (DNase I treated) gels for footprinting analysis. Protected regions corresponding to L and S sites (Fig. 1A) are indicated.
Acknowledgments
We thank June Harriss, Chhaya Das and Maya Ghosh for technical assistance. We thank Dr. Wilfried Ellmeier, Institute of Immunology, Medical University of Vienna (Vienna, Austria), for advice and for providing subclones for construction of transgenes. These studies were supported by the NIH (AI47209 and CA31534) and the Marie Betzer Morrow endowment to PWT.
Abbreviations
- DN
double negative
- DP
double positive
- SP
single positive
- MAR
matrix-associated region
- SATB1
Special AT-rich Binding protein 1
- CDP
CCAAT displacement protein
- EMSA
electrophoretic mobility shift assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
All authors concur with the submission and that the material submitted for publication has not been previously reported and is not under consideration for publication elsewhere; the authors have no financial conflict of interest.
References
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Banan M, Rojas IC, Lee WH, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- Bilic I, Koesters C, Unger B, Sekimata M, Hertweck A, Maschek R, Wilson CB, Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasquez VC, Sperry AO, Cockerill PN, Garrard WT. Protein:DNA interactions at chromosomal loop attachment sites. Genome. 1989;31:503–509. doi: 10.1139/g89-098. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F. The thymus chapter in the life of gut-specific intra epithelial lymphocytes. Curr Opin Immunol. 2008;20:185–191. doi: 10.1016/j.coi.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Augustine MM, Das J, Bottomly K, Ray P, Ray A. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc Natl Acad Sci U S A. 2003;100:5324–5329. doi: 10.1073/pnas.0831037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Maschek R, Littman DR. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16:623–634. doi: 10.1016/s1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- Feik N, Bilic I, Tinhofer J, Unger B, Littman DR, Ellmeier W. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Garefalaki A, Coles M, Hirschberg S, Mavria G, Norton T, Hostert A, Kioussis D. Variegated expression of CD8 alpha resulting from in situ deletion of regulatory sequences. Immunity. 2002;16:635–647. doi: 10.1016/s1074-7613(02)00308-4. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Hart CM, Laemmli UK. Facilitation of chromatin dynamics by SARs. Curr Opin Genet Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- Hostert A, Garefalaki A, Mavria G, Tolaini M, Roderick K, Norton T, Mee PJ, Tybulewicz VL, Coles M, Kioussis D. Hierarchical interactions of control elements determine CD8alpha gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- Hostert A, Tolaini M, Festenstein R, McNeill L, Malissen B, Williams O, Zamoyska R, Kioussis D. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J Immunol. 1997a;158:4270–4281. [PubMed] [Google Scholar]
- Hostert A, Tolaini M, Roderick K, Harker N, Norton T, Kioussis D. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity. 1997b;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat Rev Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- Landry DB, Engel JD, Sen R. Functional GATA-3 binding sites within murine CD8 alpha upstream regulatory sequences. J Exp Med. 1993;178:941–949. doi: 10.1084/jem.178.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Banan M, Harriss JV, Hwang I, Woodward E, Youn HJ, Gottlieb PD. Cis-acting DNA elements and cell type-specific nuclear proteins which may play a role in regulation of mouse CD8 alpha (Lyt-2) gene transcription. Int Immunol. 1994;6:1307–1321. doi: 10.1093/intimm/6.9.1307. [DOI] [PubMed] [Google Scholar]
- Liu J, Barnett A, Neufeld EJ, Dudley JP. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Mol Cell Biol. 1999;19:4918–4926. doi: 10.1128/mcb.19.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol Cell Biol. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Maeda M, Stewart AF. TEF-1 transcription factors regulate activity of the mouse mammary tumor virus LTR. Biochem Biophys Res Commun. 2002;296:1279–1285. doi: 10.1016/s0006-291x(02)02085-5. [DOI] [PubMed] [Google Scholar]
- Nakauchi H, Tagawa M, Nolan GP, Herzenberg LA. Isolation and characterization of the gene for the murine T cell differentiation antigen and immunoglobulin-related molecule, Lyt-2. Nucleic Acids Res. 1987;15:4337–4347. doi: 10.1093/nar/15.10.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, Scell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Nie H, Maika SD, Tucker PW, Gottlieb PD. A role for SATB1, a nuclear matrix association region-binding protein, in the development of CD8SP thymocytes and peripheral T lymphocytes. J Immunol. 2005;174:4745–4752. doi: 10.4049/jimmunol.174.8.4745. [DOI] [PubMed] [Google Scholar]
- Nie H, Yao X, Maika SD, Tucker PW. SATB1 is required for CD8 coreceptor reversal. Mol Immunol. 2008;46:207–211. doi: 10.1016/j.molimm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Jenkinson EJ, Brown MH, Sewell WA, Krissansen GW, Crumpton MJ, Owen JJ. Murine CD2 gene expression during fetal thymus ontogeny. Eur J Immunol. 1988;18:187–189. doi: 10.1002/eji.1830180129. [DOI] [PubMed] [Google Scholar]
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Sander M, Hsieh TS. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Sayre PH, Chang HC, Hussey RE, Brown NR, Richardson NE, Spagnoli G, Clayton LK, Reinherz EL. Molecular cloning and expression of T11 cDNAs reveal a receptor-like structure on human T lymphocytes. Proc Natl Acad Sci U S A. 1987;84:2941–2945. doi: 10.1073/pnas.84.9.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann RH, Garrard WT. MARs of antigen receptor and co-receptor genes. Crit Rev Eukaryot Gene Expr. 1999;9:295–310. doi: 10.1615/critreveukargeneexpr.v9.i3-4.140. [DOI] [PubMed] [Google Scholar]
- Sheldon LA, Becker M, Smith CL. Steroid hormone receptor-mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J Biol Chem. 2001;276:32423–32426. doi: 10.1074/jbc.C100315200. [DOI] [PubMed] [Google Scholar]
- Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Akashi K, Manz M, Merad M, Miyamoto T, Engleman EG, Weissman IL. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- Wang Z, Goldstein A, Zong RT, Lin D, Neufeld EJ, Scheuermann RH, Tucker PW. Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Maitra U, Johnston D, Lozano M, Dudley JP. The homeodomain protein CDP regulates mammary-specific gene transcription and tumorigenesis. Mol Cell Biol. 2004;24:4810–4823. doi: 10.1128/MCB.24.11.4810-4823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic of the transgenic constructs. Components of the constructs are highlighted in different colors and detailed in the text and in Figure 1. The L2a element (red) resides in the first DH site of DH cluster II. L2aD has a deleted 200 bp (L+S) sequence in the HindIII/HindIII fragment. (B) Southern hybridization was used to identify positive transgenic mice, and the copy numbers were determined by comparing signal intensities of wild type and transgene bands.
DNase I footprinting was performed with VL3 nuclear extract, 200(L+S), GC-1 and GC-4 mutated probes. Retarded bands of probe-protein complexes and corresponding free probes were excised from EMSA (DNase I treated) gels for footprinting analysis. Protected regions are labeled as L and S.
(A) Comparison of the L2aWT and L2aR transgenes. The 4.3 kb HindIII fragment containing the DH CII region and L2a site was cloned in reverse in L2aR relative to its germline orientation in L2aWT. (B) Effect of reversed L2a orientation on transgenic hCD2 expression under normal conditions (L2aR+SATB1+/+, black boxes) and under conditions in which SATB1 has been reduced to ~32% wild type levels (L2aR+AS+/+SATB1+/−, gray boxes) in thymocytes (left) and splenocytes (right). Derivation of the normal and SATB1-reduced L2aR transgenic lines, FACS gating and derivation of mean fluorescence intensities (MFI) were described in Materials and Methods. MFIs, quantified as the average of 3 independent experiments, are provided below corresponding columns.
FACS analysis of thymocytes isolated from seven independent L2aWT transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color FACS analysis. Expression of the hCD2 reporter gene on gated DP, CD4SP, and CD8SP thymocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
Fluorescent activated cytometric analysis of lymph node T cells isolated from seven independent L2aWT transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color cytometric analysis. Expression of the hCD2 reporter gene on gated CD4+ and CD8+ lymphocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
IELs were isolated from four L2aWT transgenic mice, and were stained with the combination of anti-CD8α, anti-CD8β and anti-hCD2 antibodies. Gated IEL subsets were analyzed for hCD2 reporter expression. Percentages and MFIs of hCD2 positive (>1.0%) subsets are shown.
FAC sorted CD8+ T cell populations were treated with pronase and placed in culture at 37°C for 18 hr or 4°C as controls. hCD2 and CD8 expression on L2aWT5 sorted cells were upregulated efficiently 18 hour after the pronase treatment. Percentages and MFIs of hCD2 positive subsets are shown.
Fluorescent activated cell analysis of thymocytes isolated from five independent L2aD transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color fluorescent staining analyses. Expression of the hCD2 reporter gene on gated DP, CD4SP and CD8SP thymocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
Fluorescent activated cell analysis of lymph node T cells isolated from five independent L2aD transgenic mice. Cells were stained with anti-CD4, anti-CD8α and anti-hCD2 antibodies, and were applied to three-color Fluorescent activated cell analysis. Expression of the hCD2 reporter gene on gated CD4+ and CD8+ lymphocytes were analyzed in histograms. Percentages and MFIs of hCD2 positive (>1.0%) subsets and numbers (n) of mice tested are shown.
IELs were isolated from four L2aD transgenic mice, and were stained with anti-CD8α, anti-CD8β and anti-hCD2 antibodies. Gated IEL subsets were analyzed for hCD2 reporter expression. Percentages and MFIs of hCD2 positive (>1.0%) subsets are shown. Lymph node L2aD results of Suppl. Fig. 6 are included for comparative purposes.
DNase I footprinting was performed with BW5147 nuclear extract, 200(L+S), GC-1, and GC-2 mutated probes as described in Materials and Methods. Retarded bands of probe-protein complexes and corresponding free probes were excised from EMSA (DNase I treated) gels for footprinting analysis. Protected regions corresponding to L and S sites (Fig. 1A) are indicated.


