Abstract
Background
Carbonic anhydrases (CAs) are a family of enzymes that regulate pH homeostasis in various tissues. CA IX is an exceptional member of this family because in addition to the basic CA function, it has been implicated in several other physiological and pathological processes. Functions suggested for CA IX include roles in cell adhesion and malignant cell invasion. In addition, CA IX likely regulates cell proliferation and differentiation, which was demonstrated in Car9-/- mice. These mice had gastric pit cell hyperplasia and depletion of chief cells; however, the specific molecular mechanisms behind the observed phenotypes remain unknown. Therefore, we wanted to study the effect of CA IX deficiency on whole-genome gene expression in gastric mucosa. This was done using Illumina Sentrix®Mouse-6 Expression BeadChip arrays. The expression of several genes with notable fold change values was confirmed by QRT-PCR.
Results
CA IX deficiency caused the induction of 86 genes and repression of 46 genes in the gastric mucosa. There was 92.9% concordance between the results obtained by microarray analysis and QRT-PCR. The differentially expressed genes included those involved in developmental processes and cell differentiation. In addition, CA IX deficiency altered the expression of genes responsible for immune responses and downregulated the expression of several digestive enzymes.
Conclusions
Microarray analysis identified several potential genes whose altered expression could explain the disturbed cell lineage phenotype in the Car9-/- gastric mucosa. The results also indicated a novel role for CA IX in the regulation of immunologic processes and digestion. These findings reinforce the concept that the main role of CA IX is not the regulation of pH in the stomach mucosa. Instead, it is needed for proper function of several physiological processes.
Background
The carbonic anhydrases (CAs) are a family of zinc-containing metalloenzymes that catalyze the reversible hydration of carbon dioxide in the reaction: CO2 + H2O ↔ H+ + HCO3-. They participate in several physiological processes, such as acid-base balance, CO2 and HCO3- transport, respiration, bone resorption, ureagenesis, gluconeogenesis, lipogenesis, production of body fluids, and fertilization [1,2]. The CA family consists of 13 active isozymes in mammals, 12 of which are expressed and function in humans [3]. The CA isozymes have diverse tissue expression patterns, characteristic subcellular localizations, as well as unique kinetic and inhibitory properties.
CA IX is a dimeric protein associated with the cell membrane [4,5]. In its mature form, CA IX is composed of an N-terminal proteoglycan (PG) domain, a CA catalytic domain, a transmembrane region, and a short intracytoplasmic tail at the C-terminus [6]. CA IX is the only member of the CA family containing a PG domain in addition to the CA domain. Consequently, CA IX has been suggested to participate in cell adhesion processes. In fact, using MDCK (Madin-Darby canine kidney) epithelial cells, it was shown that CA IX reduces E-cadherin-mediated cell-cell adhesion by interacting with β-catenin [7].
CA IX is expressed in only few normal tissues with the expression being strongest in human, rat, and mouse gastric mucosa, where it is present from the gastric pits to the deep gastric glands [8,9]. CA IX is confined to the basolateral surface of epithelial cells and is produced by all major cell types of the gastric epithelium [8]. CA IX is an exceptional member of the CA family because it is expressed in several cancers that arise from CA IX negative tissues including renal, lung, cervical, ovarian, esophageal, and breast carcinomas [6]. However, gastric cancer and premalignant lesions have shown decreased expression of CA IX [10]. In tumor tissues, CA IX is linked with the hypoxic phenotype mediated by the hypoxia-inducible transcription factor 1 (HIF-1), which binds to the hypoxia responsive element, HRE, of the CA9 promoter [11]. In hypoxic conditions, cancer cells are dependent mainly on anaerobic metabolism in their energy production. This anaerobic tumor metabolism generates excesses of acidic products, such as lactic acid and H+ that have to be extruded from the cell interior. It has been shown that CA IX can contribute to the acidification of the hypoxic extracellular milieu, thus helping tumor cells to neutralize the intracellular pH [12]. Accordingly, CA IX overexpression often indicates poor prognosis and resistance to classical radio- and chemotherapies [13]. However, CA IX is not only confined to the hypoxic regions of the tumors, indicating that there might be some other pathways that regulate its expression. In fact, the expression of CA IX can be induced under normoxic conditions by high cell density, and this regulation is mediated by phosphatidylinositol 3-kinase (PI3K) signaling [14]. Moreover, the mitogen-activated protein kinase (MAPK) pathway is involved in the regulation of CA IX expression via control of the CA9 promoter by both HIF-1-dependent and -independent signals [15]. This pathway is also interrelated with the PI3K pathway and the SP1 (specificity protein 1) transcription factor.
The generation of Car9-/- mice has revealed that in addition to pH regulation and cell adhesion, CA IX possesses other functional roles [16]. These Car9 knockout mice showed no abnormalities in growth, reproductive potential, and life span. However, CA IX deficiency resulted in hyperplasia of the gastric mucosa. The hyperplastic mucosa contained an increased number and proportion of the mucus-producing pit cells whereas the number and proportion of chief cells was reduced. The total number of parietal cells remained unchanged, and accordingly, CA IX-deficient mice had normal gastric pH, acid secretion, and serum gastrin levels. Thus, these findings demonstrate that CA IX contributes to the control of differentiation and proliferation of epithelial cell lineages in the gastric mucosa. A previous study examined whether the effects of CA IX deficiency could be modified by a high-salt diet, a known co-factor of carcinogenesis [17]. These results showed that the high-salt diet slightly increased the glandular atrophy in the body mucosa in Car9-/- C57BL/6 mice, whereas this effect was not observed in BALB/c mice. However, no metaplastic or dysplastic abnormalities were seen in the gastrointestinal epithelium of CA IX-deficient C57BL/6 mice. Thus, these observations suggest that CA IX deficiency alone may not be a significant carcinogenic factor but could initiate a carcinogenic process by affecting cell proliferation and/or differentiation in the gastric mucosa.
This study was designed to better understand the hyperplastic phenotype of the stomach mucosa of Car9-/- mice because the molecular mechanisms behind it are currently unknown. In addition, we wanted to more specifically elucidate the functional roles of CA IX, as there is a growing body of evidence showing that they extend far beyond pH regulation. For this purpose, a genome-wide expression analysis was performed. Microarray data analysis revealed several genes whose expression was changed due to Car9 gene disruption in the gastric mucosa.
Results
Transcriptional response to Car9 deficiency in the stomach wall
Stomach RNA from 6 Car9-/- mice and 6 wild-type mice was analyzed by microarray. The analysis revealed 86 upregulated genes and 46 downregulated genes, using a fold change cut-off of ± 1.4 for up- and downregulated expression, respectively (See additional file 1: List of genes differentially expressed in the stomach of Car9-/- mice). This cut-off value has been proposed as an adequate level above which there is a high correlation between microarray and QRT-PCR data, regardless of other factors such as spot intensity and cycle threshold [18]. The fold changes ranged from 10.46 to -12.14. Herein, a list of genes using a cut-off value of ± 2.5-fold is shown (Tables 1 and 2). When using these criteria, all the genes with significantly (p < 0.05) altered expression are displayed, that is, 14 genes with induced expression and 21 genes with repressed expression. The list of all the regulated genes was functionally annotated (see additional file 2: Functional annotation of genes regulated in the stomach of Car9 knockout mice), showing enrichment of hydrolase activity, developmental processes, cell differentiation, proteolysis, peptidase activity, structural molecule activity, and immune system process, among others. The functional annotation categories and gene numbers in each category are shown in Table 3.
Table 1.
Genes with upregulated expression using a cut-off value of 2.5-fold.
| Gene symbol | Description | GenBank number | FC | QRT-PCR |
|---|---|---|---|---|
| Cym | chymosin | NM_001111143 | 10.46 | 19.66 |
| Slc9a3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | NM_001081060 | 8.07 | 10.72 |
| U46068 | cDNA sequence U46068, transcript variant 2 | NM_153418 | 5.95 | |
| Dmbt1 | deleted in malignant brain tumors 1 | NM_007769 | 5.68 | 5.29 |
| Il1rl1 | interleukin 1 receptor-like 1, transcript variant 2 | NM_010743 | 5.38 | 8.95 |
| Tm4sf5 | transmembrane 4 superfamily member 5 | NM_029360 | 4.26 | |
| 9130204L05Rik | RIKEN cDNA 9130204L05 gene | NM_001101461 | 4.19 | |
| Sftpd | surfactant associated protein D | NM_009160 | 4.11 | 3.69 |
| Nccrp1 | non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) | NM_001081115 | 3.81 | |
| Pkp4 | plakophilin 4, transcript variant 1 | NM_026361 | 3.54 | 1.09 |
| Sprr2d | small proline-rich protein 2D | NM_011470 | 3.46 | |
| Gm14446 | predicted gene 14446, transcript variant 2 | NM_001101605 | 3.45 | |
| Sprr3 | small proline-rich protein 3 | NM_011478 | 3.39 | |
| Sprr2i | small proline-rich protein 2I | NM_011475 | 3.31 | |
| Sprr1a | small proline-rich protein 1A | NM_009264 | 3.30 | |
| Ivl | involucrin | NM_008412 | 3.08 | |
| Serpinb12 | serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 | NM_027971 | 3.02 | |
| Krt10 | keratin 10 | NM_010660 | 3.01 | |
| Gm94 | predicted gene 94 | NM_001033280 | 2.94 | |
| Krt13 | keratin 13 | NM_010662 | 2.94 | |
| Gsdmc2 | gasdermin C2, transcript variant 2 | NM_177912 | 2.88 | |
| Lor | loricrin | NM_008508 | 2.84 | |
| Dmkn | dermokine, transcript variant 2 | NM_172899 | 2.78 | |
| Ly6d | lymphocyte antigen 6 complex, locus D | NM_010742 | 2.69 | |
| Krt1 | keratin 1 | NM_008473 | 2.52 | |
Table 2.
Genes with downregulated expression using a cut-off value of -2.5-fold.
| Gene symbol | Description | GenBank number | FC | QRT-PCR |
|---|---|---|---|---|
| Try4 | trypsin 4 | NM_011646 | -12.14 | |
| Prss1 | protease, serine, 1 (trypsin 1) | NM_053243 | -10.57 | |
| Amy2a5 | amylase 2a5, pancreatic | NM_001042711 | -9.85 | |
| Cela2a | chymotrypsin-like elastase family, member 2A | NM_007919 | -7.59 | -16.23 |
| Gm12888 | predicted gene 12888 | NM_001033791 | -7.12 | |
| Gdf9 | growth differentiation factor 9 | NM_008110 | -7.04 | |
| Blm | bloom syndrome homolog (human), transcript variant 1 | NM_007550 | -6.27 | |
| Pnlip | pancreatic lipase | NM_026925 | -6.23 | -23.88 |
| Cfd | complement factor D (adipsin) | NM_013459 | -5.35 | -27.45 |
| Ctrb1 | chymotrypsinogen B1 | NM_025583 | -5.00 | |
| Mug1 | murinoglobulin 1 | NM_008645 | -4.30 | |
| Sostdc1 | sclerostin domain containing 1 | NM_025312 | -4.17 | |
| Tmed6 | transmembrane emp24 protein transport domain containing 6 | NM_025458 | -4.07 | |
| Cpa1 | carboxypeptidase A1 | NM_025350 | -3.95 | |
| Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 | NM_011978 | -3.80 | |
| Adipoq | adiponectin, C1Q and collagen domain containing | NM_009605 | -3.74 | -20.51 |
| Car3 | carbonic anhydrase 3 | NM_007606 | -3.72 | -15.39 |
| Egf | epidermal growth factor | NM_010113 | -3.68 | -3.71 |
| Abpg | androgen binding protein gamma | NM_178308 | -3.67 | |
| Slc38a5 | solute carrier family 38, member 5 | NM_172479 | -3.64 | |
| Chia | chitinase, acidic | NM_023186 | -3.52 | |
| LOC638418 | PREDICTED: similar to Ela3 protein | XM_914439 | -3.48 | |
| LOC100043836 | PREDICTED: similar to lacrimal androgen-binding protein delta, transcript variant 1 | XM_001481113 | -3.41 | |
| EG640530 | PREDICTED: predicted gene, EG640530 | XM_917532 | -3.37 | |
| Nrn1 | neuritin 1 | NM_153529 | -3.36 | |
| Cela3b | chymotrypsin-like elastase family, member 3B | NM_026419 | -3.32 | -75.74 |
| Spink3 | serine peptidase inhibitor, Kazal type 3 | NM_009258 | -3.32 | |
| Scd1 | stearoyl-Coenzyme A desaturase 1 | NM_009127 | -3.27 | |
| Ctrl | chymotrypsin-like | NM_023182 | -3.19 | |
| Gper | G protein-coupled estrogen receptor 1 | NM_029771 | -2.96 | |
| Sycn | syncollin | NM_026716 | -2.91 | |
| Bhlha15 | basic helix-loop-helix family, member a15 | NM_010800 | -2.77 | |
| Cpb1 | carboxypeptidase B1 (tissue) | NM_029706 | -2.73 | -18.22 |
| Slc5a8 | solute carrier family 5 (iodide transporter), member 8 | NM_145423 | -2.68 | |
| Cyp2e1 | cytochrome P450, family 2, subfamily e, polypeptide 1 | NM_021282 | -2.65 | |
| Zg16 | zymogen granule protein 16 | NM_026918 | -2.57 | |
Table 3.
The functional annotation categories for the genes regulated by CA IX deficiency.
| Functional category | Total regulated genes | Upregulated genes | Downregulated genes |
|---|---|---|---|
| Developmental processes* | 20 | 13 | 7 |
| Keratinization and keratinocyte differentiation | 8 | 8 | 0 |
| Structural molecule activity | 13 | 13 | 0 |
| Embryo implantation, female pregnancy, and menstrual cycle | 3 | 3 | 0 |
| Rhythmic process | 4 | 3 | 1 |
| Multi-organism process | 5 | 5 | 0 |
| Cell differentiation | 16 | 12 | 4 |
| Serine-type endopeptidase activity, serine hydrolase activity, and serine-type peptidase activity | 9 | 3 | 6 |
| Proteolysis | 15 | 6 | 9 |
| Peptidase activity | 14 | 5 | 9 |
| Endopeptidase activity | 10 | 4 | 6 |
| Trypsin and chymotrypsin activity | 4 | 1 | 3 |
| Hydrolase activity | 22 | 8 | 14 |
| Structural constituent of cytoskeleton | 5 | 5 | 0 |
| Cell communication | 4 | 4 | 0 |
| Metallocarboxypeptidase activity, carboxypeptidase activity, and metalloexopeptidase activity | 3 | 0 | 3 |
| Serine-type endopeptidase inhibitor activity, endopeptidase inhibitor activity, and protease inhibitor activity | 4 | 2 | 2 |
| Taxis and chemotaxis | 4 | 3 | 1 |
| Response to external stimulus | 7 | 4 | 3 |
| Immune system process | 10 | 5 | 5 |
| Defense response | 7 | 5 | 2 |
| Positive regulation of cell differentiation | 3 | 2 | 1 |
| PPAR signaling pathway | 3 | 0 | 3 |
| Leukocyte migration | 3 | 2 | 1 |
*A representative category for the following overlapping categories: epidermis morphogenesis, tissue morphogenesis, epidermis development, ectoderm development, tissue development, organ development, anatomical structure development, anatomical structure morphogenesis, cellular developmental process, multicellular organismal development, and epidermal cell differentiation.
Confirmation of microarray results by QRT-PCR
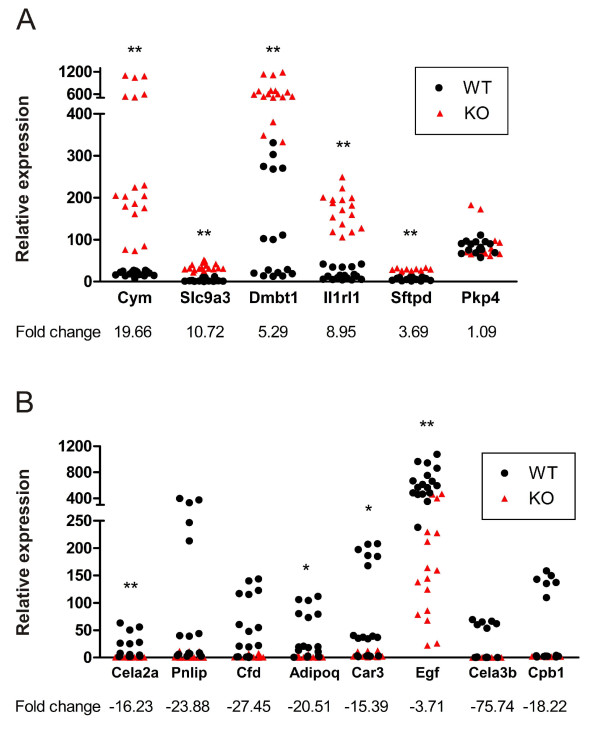
Changes in the expression levels of selected genes were confirmed, and microarray results were validated by QRT-PCR using the same RNA samples as those used for the microarray. Fourteen genes with notable fold change values were selected for validation based on the results of functional annotation. The selected genes involved representatives from different functional categories. Thirteen (92.9%) out of 14 genes showed concordant results between microarray analysis and QRT-PCR (Tables 1 and 2, Figure 1). The sole discrepant result was Pkp4, which was upregulated according to the microarray and did not change according to the QRT-PCR (1.09-fold).
Figure 1.
Verification of microarray data from Car9-/- mice by QRT-PCR. The expression of various transcripts in 6 Car9-/- mice was compared to that in 6 wild-type controls. The normalized values of triplicate experiments are shown. A, QRT-PCR analysis of 6 genes with induced expression. B, QRT-PCR evaluation of 8 genes with repressed expression. Statistically significant differences relative to wild-type mice were determined. *p < 0.05; **p < 0.01.
Discussion
A previous study showed a hyperplastic phenotype of gastric body mucosa in Car9-/- mice [16]. CA IX deficiency led to overproduction of gastric pit cells and reduction of chief cells. Quite surprisingly, the gastric pH of CA IX-deficient mice remained unaltered. Based on these observations, it can be concluded that the main role of CA IX in the stomach mucosa is not related to pH regulation but it is required for normal gastric morphogenesis and homeostasis within the gastric mucosa. In this paper, we report alterations in mRNA expression that might contribute to the phenotypic changes reported in the stomach of Car9-deficient mice. However, one must take into consideration that some of these changes can merely reflect differences in the relative proportions of various cell types within the gastric mucosa.
As expected, the Car9-/- phenotype changed the expression of genes involved in developmental processes and cell differentiation. The depletion of chief cells in Car9-/- gastric mucosa may be explained by the significant downregulation of the basic helix-loop-helix family, member a15/Mist1 (Bhlha15) gene, which is a class B basic helix-loop-helix (bHLH) transcription factor that exhibits acinar cell-specific expression [19]. Mist1 gene expression is observed in a wide array of tissues with serous type secretion including the pancreas, salivary glands, chief cells of the stomach, Paneth cells of the small intestine, seminal vesicles, and lacrimal glands [20]. The deletion of Mist1 blocks normal mucous neck cell redifferentiation into zymogenic chief cells as all basal zymogen-secreting cells in Mist1-/- mice show multiple structural defects [21].
Another interesting candidate with regard to cell lineage disturbance in the gastric epithelium of CA IX-deficient mice is epidermal growth factor (Egf), which has an expression that was significantly decreased when compared to wild-type mice. Among growth factors, the EGF family includes important agents for gastric mucosa. EGF is a single-chain polypeptide of 53 amino acid residues, which is found mainly in the submandibular glands and Brunner's glands of the gastrointestinal tract [22]. EGF binds and activates the epidermal growth factor receptor resulting in cellular proliferation, differentiation, and survival [23]. Interestingly, several investigators have reported that EGF receptors are enriched in rodent chief cells [24,25], suggesting the importance of EGF function in these cells. Therefore alterations in the expression of EGF receptor ligands or, in this case, EGF could contribute to the differentiation and function of chief cells.
One of the most strongly upregulated genes of this study was deleted in malignant brain tumors 1 (Dmbt1), which belongs to the scavenger receptor cysteine-rich (SRCR) superfamily of proteins. This is a superfamily of secreted and membrane-bound proteins with SRCR domains that are highly conserved down to sponges [26,27]. The expression of DMBT1 is the highest in epithelia and is usually observed on the apical cell surface or in luminal exocrine secretions. In mice, DMBT1 is most strongly expressed in the gastrointestinal system [28]. DMBT1 is proposed to be a tumor suppressor [26,29,30] and/or a regulator of epithelial differentiation [31], as well as having roles in innate immune defense and inflammation [32,33]. DMBT1 is located in the crypt cells of the human, mouse, and rabbit small intestine [31,34,35] and the neck region of normal human gastric mucosa [36], which are predominantly composed of stem/progenitor cells. Thus, this gene is potentially involved in the physiological renewing process of gastrointestinal epithelia. A role for DMBT1 in innate immune defense has been demonstrated in various studies. The human DMBT1 glycoprotein, expressed in salivary glands, airways, and genital tract, binds various bacterial pathogens and viruses [37-40]. Additionally, expression of mouse DMBT1 in the gastrointestinal tract is regulated by bacteria [41,42], and its expression is increased during infection [43]. It has also been shown that there is an association of a Dmbt1 variant allele with Crohn's disease and there is a correlation of expression levels of Dmbt1 with inflammatory bowel disease severity [44].
Furthermore, DMBT1 binds surfactant proteins SP-D and SP-A. These are collagen- containing, (C-type) calcium-dependent lectins called collectins which interact with glycoconjugates and lipids on the surface of microorganisms mostly through their carbohydrate recognition domains (CRDs) [45]. SP-D and SP-A are involved in a range of immune functions, including viral neutralization, aggregation and killing of bacteria and fungi, and clearance of apoptotic and necrotic cells. In immunologically naïve lungs, they downregulate inflammatory reactions, but when challenged with LPS or apoptotic cells they induce phagocytosis by macrophages, pro-inflammatory cytokine production, and enhancement of adaptive immune responses [46]. It is interesting that in the present study, in addition to Dmbt1, the expression of surfactant associated protein D (Sftpd or SP-D) was highly induced in Car9-/- mice. Thus, the upregulation of both Dmbt1 and Sftpd suggest that CA IX deficiency induced an immune process in the gastric mucosa. It should also be noted that CA IX has been suggested to bind to dendritic cells in a receptor-mediated manner and scavenger receptors appear to play an important role in this process [47]. In addition, CA IX seems to activate an immune response by a mechanism characteristic to heat shock proteins. Thus, CA IX could directly interact with DMBT1 via a scavenger receptor.
In fact, the disruption of Car9 gene caused disregulation of several genes that are involved in immune processes. This corroborates the results from a previous study where gastric submucosal inflammation was detected in the body region in a majority of C57BL/6 Car9-/- mice [17]. In the present study, Il1rl1/ST2 transcript variant 2 mRNA was highly induced due to CA IX deficiency. The Il1rl1/ST2 gene is a member of the interleukin-1 (IL-1) receptor family and produces a soluble secreted form (sST2) and a transmembrane form (ST2L), coded by transcript variants 2 and 1, respectively. The membrane-bound form is expressed primarily by hematopoietic cells and the soluble form is predominantly expressed by fibroblasts [48]. In particular, ST2L is preferentially expressed in murine and human Th2 cells and can be used as a specific marker of Th2 cells in in vitro experiments [49-52]. Therefore, the function of ST2L has been suggested to correlate with Th2 cell-mediated immunological responses and IL-33, a newly discovered member of the IL-1 cytokine family, has been reported as a specific ligand for ST2L [53]. Interestingly, several studies have shown that the level of soluble ST2 in sera is elevated in asthmatic disease [54,55]. Therefore, it has been suggested that soluble ST2 may also play a critical role in Th2 cell-mediated diseases. In fact, it has been demonstrated that soluble ST2 acts as an antagonistic decoy receptor for IL-33 using a murine thymoma cell line, EL-4, which stably expresses ST2L, and a murine model of asthma [56]. This suggests that soluble ST2 negatively modulates the production of Th2 cytokines through IL-33 signaling in allergic airway inflammation. It is interesting to note that in the present study, the mRNA level of IL-33 was also elevated ~ 2-fold, although this change was not statistically significant. Thus, it seems plausible that CA IX is somehow involved in regulation of the Th2 response.
CA IX could also contribute to the development of the immune system as CA IX deficiency significantly downregulated the bloom syndrome homolog (human) gene (Blm), which encodes an ATP-dependent DNA helicase that belongs to an evolutionarily conserved family of RecQ helicases [57]. Mutation of Blm causes a rare human autosomal recessive disorder called Bloom's syndrome (BS), which is characterized by marked genomic instability. BS is associated with growth retardation, profound susceptibility to most cancer types, and immunodeficiency [58], with the latter two features accounting for early death [59]. The functions of BLM have been only partially characterized. However, it has been shown that BLM has a role in the proliferation and survival of both developing and mature T lymphocytes, and its deletion leads to defective T-cell immunity [60]. Additionally, the conditional knockout of Blm contributed to compromised B cell development and maintenance, strongly impaired T cell-dependent and -independent antibody responses after immunization, and a propensity for developing B cell lymphomas [61]. Therefore, it is conceivable that Car9-/- mice have compromised acquired immunity responses.
An unexpected finding was that several small proline-rich proteins (Sprr) were notably upregulated including Sprr1a, Sprr2d, Sprr2e, Sprr2i, and Sprr3. However, only the induction of Sprr1a was statistically significant. SPRR proteins were originally identified as markers for terminal squamous cell differentiation where they are precursors of the cornified envelope [62]. Additionally, SPRR proteins are expressed in various nonsquamous tissues, and their biological function is not restricted to structural proteins of the cornified envelope [63]. It has been shown that SPRR proteins participate in the response to various stresses in many tissues without a stratified epithelium. In the biliary tract, SPRR2 members appear to contribute to modification of the biliary barrier under conditions of stress [64]. Likewise, SPRR1A has been identified as a novel stress-inducible downstream mediator of gp130 cytokines in cardiomyocytes with a cardioprotective effect against ischemic stress [65]. Furthermore, in mice, SPRR2A protein was reported to be highly induced in gastric mucosa infected by Helicobacter heilmannii [66]. In addition, it has been suggested that SPRR1A is a regeneration-associated protein because its induction in neuronal injury correlates with regenerative capacity, with virtually all injured dorsal root ganglion neurons expressing SPRR1A one week after sciatic nerve injury [67]. Therefore, SPRR genes are presumably induced in response to different stress conditions and contribute to cell protection, tissue remodeling, and/or tissue regeneration. In the light of these findings it seems plausible that CA IX deficiency creates a stress condition in the gastric mucosa, which leads to upregulation of some protective factors.
Our analysis identified mRNAs from four members of solute carrier family proteins to be misregulated in the Car9-/- mice stomach mucosa. These solute carrier proteins are involved in membrane transport of various molecules. The expression of Slc9a3 was strongly induced, whereas the expression of Slc27a2, Slc38a5, and Slc5a8 was repressed. The basic function of SLC9A3 or NHE3 is the exchange of extracellular Na+ for intracellular H+, thus causing either a rise in intracellular pH or, if coupled to the action of other transporters, an increase in cell volume [68]. In the gastrointestinal tract, SLC9A3 is expressed in the apical membrane and recycling endosomes of the ileum, jejunum, colon, and stomach [69]. Further studies are needed to elucidate the possible interplay between CA IX and SLC9A3 in the gastric mucosa.
Surprisingly, among the most downregulated genes we found several genes involved in digestion. These included trypsin 1, amylase 2a5, chymotrypsin-like elastase family member 2A, and pancreatic lipase with statistically significant p-values and others, such as chymotrypsinogen B1, carboxypeptidase A1, and carboxypeptidase B1 with nonsignificant p-values. The implication of this finding remains unclear. A possible explanation for this might be that downregulation of these enzymes is a secondary event. If it is assumed that these enzymes are produced by chief cells, the reduced number of chief cells in the Car9-/- mice gastric mucosa can also cause a decrease in the amount of the enzymes they produce. However, the exact mechanism behind this phenomenon remains to be elucidated.
Conclusions
In conclusion, CA IX deficiency was shown to alter the expression of various genes in the gastric mucosa. The number of affected genes and the magnitude of changes were relatively high, indicating that CA IX has a remarkable role in gastric functions. Microarray analysis revealed several genes involved in developmental processes and cell differentiation, which could explain the cell lineage disturbance in the gastric epithelium of Car9-/- mice. Interestingly, some of the regulated genes identified in this study are involved in digestion as well as functions of the immune and defense responses. This finding suggests that CA IX deficiency compromises the immune system in the gastric epithelium implying yet another role for this multifunctional enzyme.
Methods
Animal model and tissue preparation
Generation of the targeted disruption of the Car9 gene has been previously described by Ortova Gut et al. [16]. These Car9-deficient C57BL/6 mice were produced and maintained in the animal facility of the University of Oulu and then delivered to the animal facility of the University of Tampere. Permissions for use of the experimental mice were obtained from the Animal Care Committees of the Universities of Oulu and Tampere. Six Car9-/- mice (3 males and 3 females) and six wild-type control mice (3 males and 3 females) were kept under tightly controlled specific pathogen-free conditions and fed identical diets. The mice were sacrificed at approximately 11 (SD = ± 1.35) months of age. Tissue specimens from the body of the stomach were immediately collected, immersed in RNAlater solution (Ambion, Austin, TX, USA), and frozen at -80°C.
RNA isolation
Total RNA was obtained using the RNeasy RNA isolation kit (Qiagen, Valencia, USA) following the manufacturer's instructions. Residual DNA was removed from the samples using RNase-free DNase (Qiagen). The RNA concentration and purity were determined by measurement of the optical density (OD) at 260 and 280 nm. All samples had an OD260/OD280 ratio of 1.95 or higher.
Microarray analysis
All microarray data reported in the present article are described in accordance with MIAME guidelines, have been deposited in NCBI's Gene Expression Omnibus public repository [70], and are accessible through GEO Series accession number GSE20981 [71]. Microarray experiments were performed in the Finnish DNA Microarray Centre at the Turku Centre for Biotechnology. Stomach RNA samples from 6 Car9-/- mice were used. As controls, RNA samples from the stomach of 6 wild-type mice were used. All 12 samples were analyzed individually. 400 ng of total RNA from each sample was amplified using the Illumina® TotalPrep RNA Amplification kit (Ambion) as recommended by the manufacturer. The in vitro transcription reaction, which was conducted for 14.5 h, included labeling of the cRNA by biotinylation.
Hybridization and scanning
Labeled and amplified cRNAs (1.5 μg/array) were hybridized to Illumina's Sentrix® Mouse-6 Expression Bead Chips (Illumina, Inc., San Diego, CA) at 58°C for 18 h according to Illumina® Whole-Genome Gene Expression with IntelliHyb Seal System Manual. The arrays were washed and then stained with 1 μg/ml cyanine3-streptavidin (Amersham Biosciences, Buckinghamshire, UK). The Illumina BeadArray™ Reader was used to scan the arrays according the manufacturer's instructions. The numerical results were extracted with the Illumina's BeadStudio software without any normalization or background subtraction.
Data analysis
Array data were normalized with Chipster (v1.3.0) using the quantile normalization method. The data were filtered according to the SD of the probes. The percentage of data that did not pass through the filter was adjusted to 99.4%, implicating a SD value of almost 3. Statistical analysis was next performed using the empirical Bayes t-test for the comparison of two groups. Finally, the probes were further filtered according to fold change with ± 1.4 as the cut-off for up- and downregulated expression, respectively. The functional annotation tool DAVID (Database for Annotation, Visualization and Integrated Discovery) [72,73] was used to identify enriched biological categories among the regulated genes when compared to all the genes present on Illumina's Sentrix Mouse-6 Expression Bead Chip. The annotation groupings analyzed were Gene Ontology biological process and molecular functions, SwissProt comments, SwissProt Protein Information Resources Keywords, Kyoto Encyclopedia of Genes and Genomes pathway, and Biocarta pathway. Results were filtered to remove categories with EASE (Expression Analysis Systematic Explorer) scores greater than 0.05. Overlapping categories with the same gene members were removed to yield a single representative category.
Quantitative real-time PCR
For quantitative real-time PCR (QRT-PCR), the same samples were used as for the microarray analysis. 5 μg of each RNA sample was converted into first strand cDNA using a First Strand cDNA Synthesis Kit (Fermentas, Burlington, Canada) using random hexamer primers following the manufacturer's instructions. The relative expression levels of the target genes in the stomach wall were assessed by QRT-PCR using the LightCycler detection system (Roche, Rotkreuz, Switzerland). The primer sets for the target genes (Table 4) were designed using Primer3 [74], based on the complete cDNA sequences deposited in GenBank. The specificity of the primers was verified using NCBI Basic Local Alignment Search Tool (BLAST) [20]. The house-keeping genes β-actin (Actb), hypoxanthine guanine phosphoribosyl transferase (Hprt), and succinate dehydrogenase complex, subunit A (Sdha) were used as internal controls to normalize the cDNA samples for possible differences in quality and quantity (Table 4). The Actb and Hprt primers are available in the public Quantitative PCR Primer Database [75] under the identification numbers 634 and 10050, respectively. The Sdha primers were obtained from the public PrimerBank database [76] with the identification number 15030102a2. To avoid amplification of genomic DNA, the primers from each primer pair were located in different exons, when possible.
Table 4.
Sequences of the QRT-PCR primers used in this study.
| Gene symbol | Description | GenBank number | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|---|---|
| Adipoq | adiponectin, C1Q and collagen domain containing | NM_009605 | TCCTGCTTTGGTCCCTCCAC | TCCTTTCTCTCCCTTCTCTCC |
| Car3 | carbonic anhydrase 3 | NM_007606 | GCTCTGCTAAGACCATCC | ATTGGCGAAGTCGGTAGG |
| Cela2a | chymotrypsin-like elastase family, member 2A | NM_007919 | TGATGTGAGCAGGGTAGTTGG | CACTCGGTAGGTCTGATAGTTG |
| Cela3b | chymotrypsin-like elastase family, member 3B | NM_026419 | TGCCTGTGGTGGACTATGAA | CAGCCCAAGGAGGACACAA |
| Cfd | complement factor D (adipsin) | NM_013459 | AACCGGACAACCTGCAATC | CCCACGTAACCACACCTTC |
| Cpb1 | carboxypeptidase B1 (tissue) | NM_029706 | GGTTTCCACGCAAGAGAG | GTTGACCACAGGCAGAACA |
| Cym | chymosin | NM_001111143 | ATGAGCAGGAATGAGCAG | TGACAAGCCACCACTTCACC |
| Dmbt1 | deleted in malignant brain tumors 1 | NM_007769 | GCACAAGTCCTCCATCATTC | AGACAGAGCAGAGCCACAAC |
| Egf | epidermal growth factor | NM_010113 | GCTCGGTGTTTGTGTCGTG | CTGTCCCATCATCGTCTGG |
| Il1rl1 | interleukin 1 receptor-like 1, transcript variant 2 | NM_010743 | ATTCTCTCCAGCCCTTCATC | AAGCCCAAAGTCCCATTCTC |
| Pkp4 | plakophilin 4, transcript variant 1 | NM_026361 | GAACATAACCAAAGGCAGAGG | GGTGGACAGAGAAGGGTGTG |
| Pnlip | pancreatic lipase | NM_026925 | CCCGCTTTCTCCTCTACACC | TCACACTCTCCACTCGGAAC |
| Sftpd | surfactant associated protein D | NM_009160 | CCAACAAGGAAGCAATCTGAC | TCTCCCATCCCGTCCATCAC |
| Slc9a3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | NM_001081060 | TGACTGGCGTGGATTGTGTG | ACCAAGGACAGCAGGAAGG |
| Actb | actin, beta | NM_007393 | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT |
| Hprt | hypoxanthine guanine phosphoribosyl transferase | NM_013556 | AGCTACTGTAATGATCAGTCAACG | AGAGGTCCTTTTCACCAGCA |
| Sdha | succinate dehydrogenase complex, subunit A | NM_023281 | GCTTGCGAGCTGCATTTGG | CATCTCCAGTTGTCCTCTTCCA |
Each PCR reaction was performed in a total volume of 20 μl containing 0.5 μl of first strand cDNA, 1x of QuantiTect SYBR Green PCR Master Mix (Qiagen, Hilden, Germany), and 0.5 μM of each primer. Amplification and detection were carried out as follows: after an initial 15 min activation step at 95°C, amplification was performed in a three-step cycling procedure for 45 cycles: denaturation at 95°C for 15 s, annealing at a temperature determined according to the Tm for each primer pair for 20 s, and elongation at 72°C for 15 s (the ramp rate was 20°C/s for all the steps), followed by a final cooling step. Melting curve analysis was always performed after the amplification to examine the specificity of the PCR. To quantify the levels of the target transcripts, a standard curve was established for each gene using five-fold serial dilutions of known concentrations of purified PCR products generated with the same primer pairs. Every cDNA sample was tested in triplicate. The mean and SD of the 3 crossing point (Cp) values were calculated for each sample and a SD cut-off of 0.2 was set. Accordingly, when the SD of the triplicates of a sample was greater than 0.2, the most outlying replicate was excluded and the analysis was continued with the two remaining replicates. Using a specific standard curve, the Cp values were then transformed by the LightCycler software into copy numbers. The expression value for each sample was the mean of the copy numbers for the sample's replicates. The geometric mean of the three internal control genes was used as an accurate normalization factor for gene expression levels [77]. The final relative mRNA expression was given as the expression value of the sample divided by the corresponding normalization factor and multiplied by 103.
Statistical analyses
Statistical analyses of the microarray were performed using the empirical Bayes t-test for comparison of the two groups, and the p-values are shown in Additional file 1. For the QRT-PCR results, the Mann-Whitney test was used to evaluate differences in group values for Car9-/- mice vs. wild-type mice.
List of abbreviations used
CA IX: carbonic anhydrase IX; HIF-1: hypoxia-inducible transcription factor 1; HRE: hypoxia responsive element; MAPK: mitogen-activated protein kinase; PI3K: phosphatidylinositol 3-kinase; QRT-PCR: quantitative real-time PCR.
Authors' contributions
HK prepared the samples, participated in the microarray data analysis and QRT-PCR confirmations, and drafted the manuscript. MH participated in the design of the study and microarray data analysis. AR participated in the experimental design of the study and critically reviewed the manuscript. EHL and AML participated in the QRT-PCR confirmations. SP conceived the study, participated in its design and coordination, and critically reviewed the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of genes differentially expressed in the stomach of Car9-/- mice. The Car9-/- and control groups contained samples from 6 mice. Microarray data were normalized with Chipster (v1.3.0) using the quantile normalization method. Statistical analysis was performed using the empirical Bayes t-test for the comparison of two groups. The probes were filtered according to fold change with cut-off of ± 1.4 for up- and down-regulated expression, respectively.
Functional annotation of genes regulated in the stomach of Car9 knockout mice. The functional annotation tool DAVID was used to identify enriched biological categories among the differentially expressed genes as compared to all genes present in Illumina's Sentrix Mouse-6 Expression Bead Chip.
Contributor Information
Heini Kallio, Email: heini.kallio@uta.fi.
Mika Hilvo, Email: mika.hilvo@vtt.fi.
Alejandra Rodriguez, Email: ltalro@uta.fi.
Eeva-Helena Lappalainen, Email: elappala@abo.fi.
Anna-Maria Lappalainen, Email: anna.lappalainen.2500@student.uu.se.
Seppo Parkkila, Email: seppo.parkkila@uta.fi.
Acknowledgements
We thank the Illumina team at the Finnish DNA Microarray Centre at the Turku Centre for Biotechnology, especially Reija Venho for performing the microarray hybridizations and scanning. This study was supported by the Competitive Research Funding of the Tampere University Hospital (9L071), EU FP6 project (DeZnIT), and the Academy of Finland.
References
- Sly WS, Hu PY. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- Parkkila S. An overview of the distribution and function of carbonic anhydrase in mammals. Exs. 2000. pp. 79–93. [DOI] [PubMed]
- Hilvo M, Tolvanen M, Clark A, Shen B, Shah GN, Waheed A, Halmi P, Hanninen M, Hamalainen JM, Vihinen M, Sly WS, Parkkila S. Characterization of CA XV, a new GPI-anchored form of carbonic anhydrase. Biochem J. 2005;392(Pt 1):83–92. doi: 10.1042/BJ20051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilvo M, Baranauskiene L, Salzano AM, Scaloni A, Matulis D, Innocenti A, Scozzafava A, Monti SM, Di Fiore A, De Simone G, Lindfors M, Janis J, Valjakka J, Pastorekova S, Pastorek J, Kulomaa MS, Nordlund HR, Supuran CT, Parkkila S. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J Biol Chem. 2008;283(41):27799–27809. doi: 10.1074/jbc.M800938200. [DOI] [PubMed] [Google Scholar]
- Alterio V, Hilvo M, Di Fiore A, Supuran CT, Pan P, Parkkila S, Scaloni A, Pastorek J, Pastorekova S, Pedone C, Scozzafava A, Monti SM, De Simone G. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc Natl Acad Sci USA. 2009;106(38):16233–16238. doi: 10.1073/pnas.0908301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorekova S. Carbonic anhydrase IX (CA IX) as a potential target for cancer therapy. Cancer Therapy. 2004;2:245–262. [Google Scholar]
- Svastova E, Zilka N, Zat'ovicova M, Gibadulinova A, Ciampor F, Pastorek J, Pastorekova S. Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with beta-catenin. Exp Cell Res. 2003;290(2):332–345. doi: 10.1016/S0014-4827(03)00351-3. [DOI] [PubMed] [Google Scholar]
- Pastorekova S, Parkkila S, Parkkila AK, Opavsky R, Zelnik V, Saarnio J, Pastorek J. Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology. 1997;112(2):398–408. doi: 10.1053/gast.1997.v112.pm9024293. [DOI] [PubMed] [Google Scholar]
- Hilvo M, Rafajova M, Pastorekova S, Pastorek J, Parkkila S. Expression of carbonic anhydrase IX in mouse tissues. J Histochem Cytochem. 2004;52(10):1313–1322. doi: 10.1369/jhc.3A6225.2004. [DOI] [PubMed] [Google Scholar]
- Leppilampi M, Saarnio J, Karttunen TJ, Kivela J, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila S. Carbonic anhydrase isozymes IX and XII in gastric tumors. World J Gastroenterol. 2003;9(7):1398–1403. doi: 10.3748/wjg.v9.i7.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60(24):7075–7083. [PubMed] [Google Scholar]
- Svastova E, Hulikova A, Rafajova M, Zat'ovicova M, Gibadulinova A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastorekova S. Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett. 2004;577(3):439–445. doi: 10.1016/j.febslet.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Kaluz S, Kaluzova M, Chrastina A, Olive PL, Pastorekova S, Pastorek J, Lerman MI, Stanbridge EJ. Lowered oxygen tension induces expression of the hypoxia marker MN/carbonic anhydrase IX in the absence of hypoxia-inducible factor 1 alpha stabilization: a role for phosphatidylinositol 3'-kinase. Cancer Res. 2002;62(15):4469–4477. [PubMed] [Google Scholar]
- Kopacek J, Barathova M, Dequiedt F, Sepelakova J, Kettmann R, Pastorek J, Pastorekova S. MAPK pathway contributes to density- and hypoxia-induced expression of the tumor-associated carbonic anhydrase IX. Biochim Biophys Acta. 2005;1729(1):41–49. doi: 10.1016/j.bbaexp.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Ortova Gut MO, Parkkila S, Vernerova Z, Rohde E, Zavada J, Hocker M, Pastorek J, Karttunen T, Gibadulinova A, Zavadova Z, Knobeloch KP, Wiedenmann B, Svoboda J, Horak I, Pastorekova S. Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology. 2002;123(6):1889–1903. doi: 10.1053/gast.2002.37052. [DOI] [PubMed] [Google Scholar]
- Leppilampi M, Karttunen TJ, Kivela J, Gut MO, Pastorekova S, Pastorek J, Parkkila S. Gastric pit cell hyperplasia and glandular atrophy in carbonic anhydrase IX knockout mice: studies on two strains C57/BL6 and BALB/C. Transgenic Res. 2005;14(5):655–663. doi: 10.1007/s11248-005-7215-z. [DOI] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C, To RQ, Swanson BJ, Lyons GE, Konieczny SF. Mist1: a novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Dev Biol. 1997;182(1):101–113. doi: 10.1006/dbio.1996.8454. [DOI] [PubMed] [Google Scholar]
- Pin CL, Bonvissuto AC, Konieczny SF. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat Rec. 2000;259(2):157–167. doi: 10.1002/(SICI)1097-0185(20000601)259:2<157::AID-AR6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134(1):211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- Heitz PU, Kasper M, van Noorden S, Polak JM, Gregory H, Pearse AG. Immunohistochemical localisation of urogastrone to human duodenal and submandibular glands. Gut. 1978;19(5):408–413. doi: 10.1136/gut.19.5.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(2 Suppl):21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Kamimura H, Hosaka M, Torii S, Izumi T, Kuwano H, Takeuchi T. Functional localization of proprotein-convertase furin and its substrate TGFbeta in EGF receptor-expressing gastric chief cells. Growth Factors. 2004;22(1):51–59. doi: 10.1080/0897719042000201659. [DOI] [PubMed] [Google Scholar]
- Kamimura H, Konda Y, Yokota H, Takenoshita S, Nagamachi Y, Kuwano H, Takeuchi T. Kex2 family endoprotease furin is expressed specifically in pit-region parietal cells of the rat gastric mucosa. Am J Physiol. 1999;277(1 Pt 1):G183–190. doi: 10.1152/ajpgi.1999.277.1.G183. [DOI] [PubMed] [Google Scholar]
- Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, von Deimling A, Poustka A. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet. 1997;17(1):32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- Holmskov U, Mollenhauer J, Madsen J, Vitved L, Gronlund J, Tornoe I, Kliem A, Reid KB, Poustka A, Skjodt K. Cloning of gp-340, a putative opsonin receptor for lung surfactant protein D. Proc Natl Acad Sci USA. 1999;96(19):10794–10799. doi: 10.1073/pnas.96.19.10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lisle RC, Petitt M, Huff J, Isom KS, Agbas A. MUCLIN expression in the cystic fibrosis transmembrane conductance regulator knockout mouse. Gastroenterology. 1997;113(2):521–532. doi: 10.1053/gast.1997.v113.pm9247472. [DOI] [PubMed] [Google Scholar]
- Somerville RP, Shoshan Y, Eng C, Barnett G, Miller D, Cowell JK. Molecular analysis of two putative tumour suppressor genes, PTEN and DMBT, which have been implicated in glioblastoma multiforme disease progression. Oncogene. 1998;17(13):1755–1757. doi: 10.1038/sj.onc.1202066. [DOI] [PubMed] [Google Scholar]
- Mori M, Shiraishi T, Tanaka S, Yamagata M, Mafune K, Tanaka Y, Ueo H, Barnard GF, Sugimachi K. Lack of DMBT1 expression in oesophageal, gastric and colon cancers. Br J Cancer. 1999;79(2):211–213. doi: 10.1038/sj.bjc.6690035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takito J, Hikita C, Al-Awqati Q. Hensin, a new collecting duct protein involved in the in vitro plasticity of intercalated cell polarity. J Clin Invest. 1996;98(10):2324–2331. doi: 10.1172/JCI119044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Sina C, End C, Renner M, Lyer S, Till A, Hellmig S, Nikolaus S, Folsch UR, Helmke B, Autschbach F, Schirmacher P, Kioschis P, Hafner M, Poustka A, Mollenhauer J, Schreiber S. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178(12):8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- Kojouharova MS, Tsacheva IG, Tchorbadjieva MI, Reid KB, Kishore U. Localization of ligand-binding sites on human C1q globular head region using recombinant globular head fragments and single-chain antibodies. Biochim Biophys Acta. 2003;1652(1):64–74. doi: 10.1016/j.bbapap.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Cheng H, Bjerknes M, Chen H. CRP-ductin: a gene expressed in intestinal crypts and in pancreatic and hepatic ducts. Anat Rec. 1996;244(3):327–343. doi: 10.1002/(SICI)1097-0185(199603)244:3<327::AID-AR5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Mollenhauer J, Herbertz S, Holmskov U, Tolnay M, Krebs I, Merlo A, Schroder HD, Maier D, Breitling F, Wiemann S, Grone HJ, Poustka A. DMBT1 encodes a protein involved in the immune defense and in epithelial differentiation and is highly unstable in cancer. Cancer Res. 2000;60(6):1704–1710. [PubMed] [Google Scholar]
- Kang W, Nielsen O, Fenger C, Madsen J, Hansen S, Tornoe I, Eggleton P, Reid KB, Holmskov U. The scavenger receptor, cysteine-rich domain-containing molecule gp-340 is differentially regulated in epithelial cell lines by phorbol ester. Clin Exp Immunol. 2002;130(3):449–458. doi: 10.1046/j.1365-2249.2002.01992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, Frangsmyr L, Holmskov U, Leffler H, Nilsson C, Boren T, Wright JR, Stromberg N, Fisher SJ. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275(51):39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- Stoddard E, Cannon G, Ni H, Kariko K, Capodici J, Malamud D, Weissman D. gp340 expressed on human genital epithelia binds HIV-1 envelope protein and facilitates viral transmission. J Immunol. 2007;179(5):3126–3132. doi: 10.4049/jimmunol.179.5.3126. [DOI] [PubMed] [Google Scholar]
- White MR, Crouch E, van Eijk M, Hartshorn M, Pemberton L, Tornoe I, Holmskov U, Hartshorn KL. Cooperative anti-influenza activities of respiratory innate immune proteins and neuraminidase inhibitor. Am J Physiol Lung Cell Mol Physiol. 2005;288(5):L831–840. doi: 10.1152/ajplung.00365.2004. [DOI] [PubMed] [Google Scholar]
- Wu Z, Van Ryk D, Davis C, Abrams WR, Chaiken I, Magnani J, Malamud D. Salivary agglutinin inhibits HIV type 1 infectivity through interaction with viral glycoprotein 120. AIDS Res Hum Retroviruses. 2003;19(3):201–209. doi: 10.1089/088922203763315704. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Sun FJ, Kaur S, Ziemer D, Banerjee S, Samuelson LC, De Lisle RC. Decreased gastric bacterial killing and up-regulation of protective genes in small intestine in gastrin-deficient mouse. Dig Dis Sci. 2003;48(5):976–985. doi: 10.1023/A:1023068116934. [DOI] [PubMed] [Google Scholar]
- Norkina O, Kaur S, Ziemer D, De Lisle RC. Inflammation of the cystic fibrosis mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2004;286(6):G1032–1041. doi: 10.1152/ajpgi.00473.2003. [DOI] [PubMed] [Google Scholar]
- Renner M, Bergmann G, Krebs I, End C, Lyer S, Hilberg F, Helmke B, Gassler N, Autschbach F, Bikker F, Strobel-Freidekind O, Gronert-Sum S, Benner A, Blaich S, Wittig R, Hudler M, Ligtenberg AJ, Madsen J, Holmskov U, Annese V, Latiano A, Schirmacher P, Amerongen AV, D'Amato M, Kioschis P, Hafner M, Poustka A, Mollenhauer J. DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn's disease. Gastroenterology. 2007;133(5):1499–1509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43(9):1293–1315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115(1):13–23. doi: 10.1016/S0092-8674(03)00758-X. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang XY, Subjeck JR, Kim HL. Carbonic anhydrase IX has chaperone-like functions and is an immunoadjuvant. Mol Cancer Ther. 2008;7(12):3867–3877. doi: 10.1158/1535-7163.MCT-08-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachter T, Werenskiold AK, Klemenz R. Transcription of the interleukin-1 receptor-related T1 gene is initiated at different promoters in mast cells and fibroblasts. J Biol Chem. 1996;271(1):124–129. doi: 10.1074/jbc.271.1.124. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Naito Y, Kuroiwa K, Arai T, Furukawa Y, Tomizuka H, Miura Y, Kasahara T, Tetsuka T, Tominaga S. The expression of ST2 gene in helper T cells and the binding of ST2 protein to myeloma-derived RPMI8226 cells. J Biochem. 1997;121(1):95–103. doi: 10.1093/oxfordjournals.jbchem.a021577. [DOI] [PubMed] [Google Scholar]
- Xu D, Chan WL, Leung BP, Huang F, Wheeler R, Piedrafita D, Robinson JH, Liew FY. Selective expression of a stable cell surface molecule on type 2 but not type 1 helper T cells. J Exp Med. 1998;187(5):787–794. doi: 10.1084/jem.187.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez-Ramos JC, Levinson D, Radbruch A, Kamradt T. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA. 1998;95(12):6930–6935. doi: 10.1073/pnas.95.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecart S, Lecointe N, Subramaniam A, Alkan S, Ni D, Chen R, Boulay V, Pene J, Kuroiwa K, Tominaga S, Yssel H. Activated, but not resting human Th2 cells, in contrast to Th1 and T regulatory cells, produce soluble ST2 and express low levels of ST2L at the cell surface. Eur J Immunol. 2002;32(10):2979–2987. doi: 10.1002/1521-4141(2002010)32:10<2979::AID-IMMU2979>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Oshikawa K, Kuroiwa K, Tago K, Iwahana H, Yanagisawa K, Ohno S, Tominaga SI, Sugiyama Y. Elevated soluble ST2 protein levels in sera of patients with asthma with an acute exacerbation. Am J Respir Crit Care Med. 2001;164(2):277–281. doi: 10.1164/ajrccm.164.2.2008120. [DOI] [PubMed] [Google Scholar]
- Oshikawa K, Yanagisawa K, Tominaga S, Sugiyama Y. Expression and function of the ST2 gene in a murine model of allergic airway inflammation. Clin Exp Allergy. 2002;32(10):1520–1526. doi: 10.1046/j.1365-2745.2002.01494.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Hayakawa M, Kume A, Tominaga S. Soluble ST2 blocks interleukin-33 signaling in allergic airway inflammation. J Biol Chem. 2007;282(36):26369–26380. doi: 10.1074/jbc.M704916200. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374(Pt 3):577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72(6):393–406. [PubMed] [Google Scholar]
- German J. Bloom's syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet. 1997;93(1):100–106. doi: 10.1016/S0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- Babbe H, Chester N, Leder P, Reizis B. The Bloom's syndrome helicase is critical for development and function of the alphabeta T-cell lineage. Mol Cell Biol. 2007;27(5):1947–1959. doi: 10.1128/MCB.01402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbe H, McMenamin J, Hobeika E, Wang J, Rodig SJ, Reth M, Leder P. Genomic instability resulting from Blm deficiency compromises development, maintenance, and function of the B cell lineage. J Immunol. 2009;182(1):347–360. doi: 10.4049/jimmunol.182.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden HP, Kubilus J, Phillips SB, Kvedar JC, Tahan SR. A new class of soluble basic protein precursors of the cornified envelope of mammalian epidermis. Biochim Biophys Acta. 1987;925(1):63–73. doi: 10.1016/0304-4165(87)90148-6. [DOI] [PubMed] [Google Scholar]
- Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999;30(2):243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- Nozaki I, Lunz JG, Specht S, Stolz DB, Taguchi K, Subbotin VM, Murase N, Demetris AJ. Small proline-rich proteins 2 are noncoordinately upregulated by IL-6/STAT3 signaling after bile duct ligation. Lab Invest. 2005;85(1):109–123. doi: 10.1038/labinvest.3700213. [DOI] [PubMed] [Google Scholar]
- Pradervand S, Yasukawa H, Muller OG, Kjekshus H, Nakamura T, St Amand TR, Yajima T, Matsumura K, Duplain H, Iwatate M, Woodard S, Pedrazzini T, Ross J, Firsov D, Rossier BC, Hoshijima M, Chien KR. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. EMBO J. 2004;23(22):4517–4525. doi: 10.1038/sj.emboj.7600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A, O'Rourke J, Grimm J, Guillemin K, Dixon MF, Lee A, Falkow S. Distinct gene expression profiles characterize the histopathological stages of disease in Helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Proc Natl Acad Sci USA. 2003;100(3):1292–1297. doi: 10.1073/pnas.242741699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey ML, Davies M, Yip PK, Carter LM, Wong DJ, McMahon SB, Bradbury EJ. Expression of the regeneration-associated protein SPRR1A in primary sensory neurons and spinal cord of the adult mouse following peripheral and central injury. J Comp Neurol. 2009;513(1):51–68. doi: 10.1002/cne.21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Yun CC. Mechanisms of the regulation of the intestinal Na+/H+ exchanger NHE3. J Biomed Biotechnol. 2010. p. 238080. [DOI] [PMC free article] [PubMed]
- Hoogerwerf WA, Tsao SC, Devuyst O, Levine SA, Yun CH, Yip JW, Cohen ME, Wilson PD, Lazenby AJ, Tse CM, Donowitz M. NHE2 and NHE3 are human and rabbit intestinal brush-border proteins. Am J Physiol. 1996;270(1 Pt 1):G29–41. doi: 10.1152/ajpgi.1996.270.1.G29. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEO. Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/
- DAVID Bioinformatics Resources. http://david.abcc.ncifcrf.gov/
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3.. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Primer3. http://frodo.wi.mit.edu/primer3/
- Quantitative PCR Primer Database. http://web.ncifcrf.gov/rtp/gel/primerdb/
- PrimerBank database. http://pga.mgh.harvard.edu/primerbank/
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes differentially expressed in the stomach of Car9-/- mice. The Car9-/- and control groups contained samples from 6 mice. Microarray data were normalized with Chipster (v1.3.0) using the quantile normalization method. Statistical analysis was performed using the empirical Bayes t-test for the comparison of two groups. The probes were filtered according to fold change with cut-off of ± 1.4 for up- and down-regulated expression, respectively.
Functional annotation of genes regulated in the stomach of Car9 knockout mice. The functional annotation tool DAVID was used to identify enriched biological categories among the differentially expressed genes as compared to all genes present in Illumina's Sentrix Mouse-6 Expression Bead Chip.