Abstract
Background
Serine proteases (SPs) and serine proteases homologs (SPHs) are a large group of proteolytic enzymes, with important roles in a variety of physiological processes, such as cell signalling, defense and development. Genome-wide identification and expression analysis of serine proteases and their homologs in the silkworm might provide valuable information about their biological functions.
Results
In this study, 51 SP genes and 92 SPH genes were systematically identified in the genome of the silkworm Bombyx mori. Phylogenetic analysis indicated that six gene families have been amplified species-specifically in the silkworm, and the members of them showed chromosomal distribution of tandem repeats. Microarray analysis suggests that many silkworm-specific genes, such as members of SP_fam12, 13, 14 and 15, show expression patterns that are specific to tissues or developmental stages. The roles of SPs and SPHs in resisting pathogens were investigated in silkworms when they were infected by Escherichia coli, Bacillus bombysepticus, Batrytis bassiana and B. mori nucleopolyhedrovirus, respectively. Microarray experiment and real-time quantitative RT-PCR showed that 18 SP or SPH genes were significantly up-regulated after pathogen induction, suggesting that SP and SPH genes might participate in pathogenic microorganism resistance in B. mori.
Conclusion
Silkworm SP and SPH genes were identified. Comparative genomics showed that SP and SPH genes belong to a large family, whose members are generated mainly by tandem repeat evolution. We found that silkworm has species-specific SP and SPH genes. Phylogenetic and microarray analyses provide an overview of the silkworm SP and SPHs, and facilitate future functional studies on these enzymes.
Background
Serine proteases (SPs) in the S1 family are involved in physiological processes including digestion, development and defense [1-3]. X-ray crystallography studies show that the active center of bovine chymotrypsin is Ser195, His57, and Asp102 [4,5]. A substrate binding cleft near the active site is the predominant factor in determining SP substrate specificity. SPs are divided into several types, including the trypsin, chymotrypsin and elastase, based on the target scissile bond. SPs are usually produced as zymogens. SP zymogens are sequentially activated in a cascade pathway, with a classic example being the blood clotting cascade and complement system in mammalian plasma. Serine protease homologs (SPHs) are similar to SPs in amino acid sequence, but they have no protease activity because of losing one or more of the catalytic residue [3,4]. In insects, SPHs participate in the innate immune response [6,7].
Genome-wide analysis has been performed for SPs and SPHs in Drosophila melanogaster [5]. Immunity-related SPs and SPHs have been investigated in Anopheles gambiae and Apis mellifera [3,8]. However, little is known about these proteins in the silkworm, even though its SPs and SPHs are involved in biological processes that include digestion, development and immune response [9-12]. Tanaka et al. identified potential immunity-related silkworm SPs and SPHs [13], but their roles require further investigation. Changes in mRNA level during immune response are considered evidence that a gene is involved in an immune process. In this paper, we report the genome-wide classification, distribution and expression profiling of B.mori SPs and SPHs based on the silkworm genome and microarray data [14,15]. The role of SP and SPH genes in immunity has aslo been investigated. The results are expected to stimulate in-depth analysis of silkworm SPs and SPHs.
Results and discussion
Identification and chromosome distribution of silkworm SP and SPH genes
SP and SPH genes from Manduca sexta and other insects were used to search the silkworm genomic sequence, which revealed 143 serine protease genes. Analysis of active sites showed that 51 serine protease genes have the three active site residues intact (SP). The rest 92 serine protease genes had mutations in the catalytic residues (SPH), so they may have lost catalytic function (Additional file 1 and 2). Compared to 147 SP genes and 57 SPH genes in Drosophila, silkworm has 96 fewer SP genes and 35 more SPH genes. The genes were named BmSP(H)1-BmSP(H)143 (Additional file 1), 20 of which had previously been submitted to GenBank, including ovarian serine protease (GenBank Accession No. AAL62027), cocoonase (GenBank Accession No. ABR14241), vitellin-degrading protease precursor (GenBank Accession No. BAA03758), and alkaliphilic serine protease P-Iic (GenBank Accession No. AAB26023) (Additional file 3), which function in digestion, ovary and embryo development, and formation of fibroin. The remaining 123 genes were not reported previously. The genes reported here represent the first catalog of silkworm SP and SPH genes. Approximately 80 (55.9%) were found to be expressed using expressed sequence tag (EST) data according to stringent criteria (see Materials and Methods). The molecular weights and isoelectric points of silkworm SPs and SPHs were widely distributed. Predicted molecular weights were 6.3 kDa-203.6 kDa, and predicted isoelectric points were 3.5-11.3.
In arthropods, clip-domain serine proteases and homologs (clip-SPs and clip-SPHs) mediate innate immunity and embryonic development [13,14], and are involved in signal-amplifying reactions [2]. Previously, these proteases had been found only in invertebrates. We identified 7 clip-SPs and 11 clip-SPHs in the silkworm [13] (Additional file 4), which are consisted of a chymotrypsin-like SP domain, and one or more clip domain(s) at the N-terminus. The SP domain amplies signals by cleaving and activating a downstream SP [16]. Activation of the Toll pathway and melanization mediated by SP cascades are the major defense mechanisms in insects [17,18].
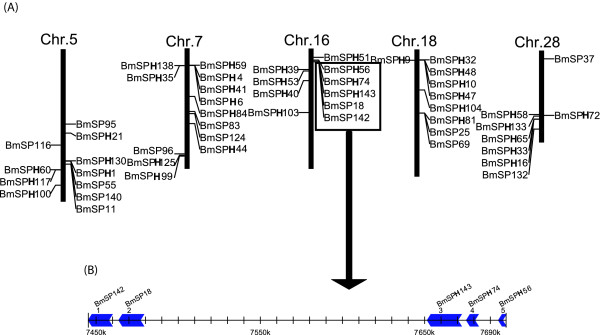
The updated silkworm genome has 128 SP and SPH genes (89.5%) on 28 chromosomes, with an uneven distribution. For example, chromosome 1 and 19 have no SP and SPH genes, while both chromosomes 17 and 22 have one. However, chromosome 5 has 11 genes, chromosome 7 has 13, and chromosome 16 has 10 (Figure 1). Additional analysis showed that SP and SPH genes exhibit a tandem repeat distribution on the chromosomes. For example, there are 10 genes in nscaf3058 of chromosome 16, and BmSP18, BmSPH56, BmSPH74, BmSP142 and BmSPH143 showed tandem repeat distribution on this chromosome (Figure 1). Of these, BmSPH74 and BmSP143 were closely linked. The distance between the two genes is only 3 kb, and their sequence identities were up to 96%. Therefore, these genes probably arose by gene duplication.
Figure 1.
Distribution of SP and SPH genes on silkworm chromosomes . (A) The five Chromosomes that have the most number of SP and SPH genes. Many SP and SPH genes are in tandem clusters. (B) An example of SP and SPH genes clustered on chromosome 16. These five genes are distributed in a cluster within 250 kb.The arrowhead indicates the transcriptional orientation of genes.
Species-specific SP and SPH genes in the silkworm
The TRIBE-MCL clustering algorithm was used to group the SP and SPH genes into families. Caenorhabditis elegans, Homo sapiens, Drosophila melanogaster, Apis mellifera and Anopheles gambiae were included to find members of evolutionarily conserved families, which were defined as containing silkworm genes, and at least one gene in another species. The results uncovered 99 SP and SPH silkworm genes, belonging to 17 gene families, 11 of which were evolutionally conserved (Table 1). SP_fam1 contains the most number of SP and SPH genes. These exist in C. elegans, insects and humans, indicating that SP_fam1 existed before the divergence of these species. SP_fam1 has two members in C. elegans, and shows great expansion in insects and mammals. B. mori and A. mellifera contain 24 and 28 members of SP_fam1, respectively. However, the Diptera insects D. melanogaster and A. gambiae, have 65 and 49 SP_fam1 members, respectively. Interestingly, the other 10 evolutionarily conserved SP and SPH gene families are insect-specific.
Table 1.
The gene number of SP and SPH gene families in different species
| Family | C. elegan | B. mori | D. melanogaster | A. gambiae | A. mellifera | Human |
|---|---|---|---|---|---|---|
| SP_fam1 | 2 | 24 | 65 | 49 | 28 | 61 |
| SP_fam2 | - | 9 | 12 | 37 | 4 | - |
| SP_fam3 | - | 6 | 18 | 22 | 2 | - |
| SP_fam4 | - | 5 | 17 | 19 | 4 | - |
| SP_fam5 | - | 4 | 2 | 29 | 0 | - |
| SP_fam6 | - | 2 | 9 | 9 | 1 | - |
| SP_fam7 | - | 1 | 3 | 4 | 2 | - |
| SP_fam8 | - | 5 | - | 1 | 0 | - |
| SP_fam9 | - | 2 | 1 | 1 | 1 | - |
| SP_fam10 | - | 1 | 1 | 1 | 1 | - |
| SP_fam11 | - | 1 | 1 | 1 | 1 | - |
| SP_fam12 | - | 15 | - | - | - | - |
| SP_fam13 | - | 7 | - | - | - | - |
| SP_fam14 | - | 6 | - | - | - | - |
| SP_fam15 | - | 5 | - | - | - | - |
| SP_fam16 | - | 3 | - | - | - | - |
| SP_fam17 | - | 3 | - | - | - | - |
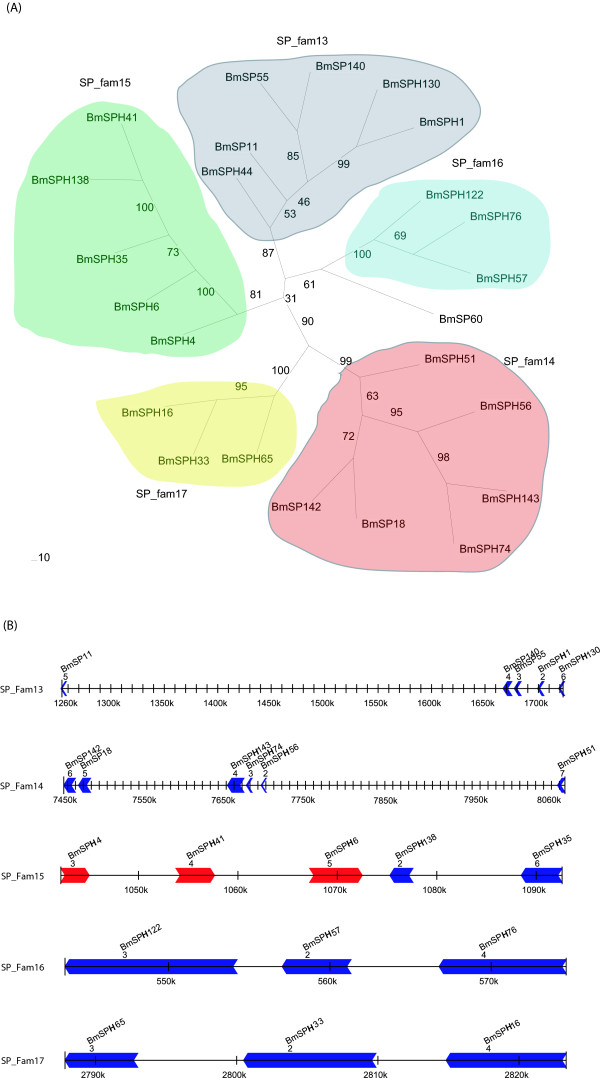
The silkworm genome contains 39 genes in 6 silkworm-specific SP and SPH gene families (Table 1 and Additional file 5). If the 44 singleton SP genes are considered, 83 species-specific SP and SPH genes exist, accounting for 56.8% of the silkworm SP and SPH genes. This indicated that the genes that came from the same species-specific SP and SPH gene families were the result of tandem repeats (Figure 2). For example, SP_fam13 has 7 members, and 5 occur as tandem repeats on chromosome 10. Internal members of the same family showed tandem repeat distribution in SP_fam14 (chr.16), SP_fam15 (chr.7), SP_fam16 (chr.26) and SP_fam17 (chr.28). Thus, tandem repeat distribution has an important effect on the expansion of species-specific SP and SPH genes in silkworm. In fact, silkworm genome analysis shows that many silkworm-specific gene families have the typical features of a chromosomal tandem repeat distribution [2,19,20]. In addition, gene sequences in the same cluster show high identity. For example, the chromosomal location of BmSPH74 and BmSPH143 is a typical tandem repeat, with the genes closely linked on the chromosome, only 3 kb apart, and sequence identity of up to 96%. This indicated that one of the genes may have emerged from the duplication of the other, and the duplication event happened recently. Therefore, these specific genes may come from a specific gene duplication family. Genes that originated by duplication might have adapted during silkworm evolution and may cut substrates specific to silkworm. However, this hypothesis need to be valitated by further experiments.
Figure 2.
Phylogenetic analysis of silkworm-specific SP and SPH genes . (A) Neighbor-joining tree of silkworm-specific SP and SPH genes. Genes in SP_fam13, SP_fam14, SP_fam15, SP_fam16, SP_fam17 are in gray, red, green, blue and yellow, respectively. (B) Schematic picture of the distribution on chromosome for genes of SP_fam13, SP_fam14, SP_fam15, SP_fam16 and SP_fam17. The arrowhead indicates the transcriptional orientation of genes.
Expression profile of silkworm SP and SPH genes
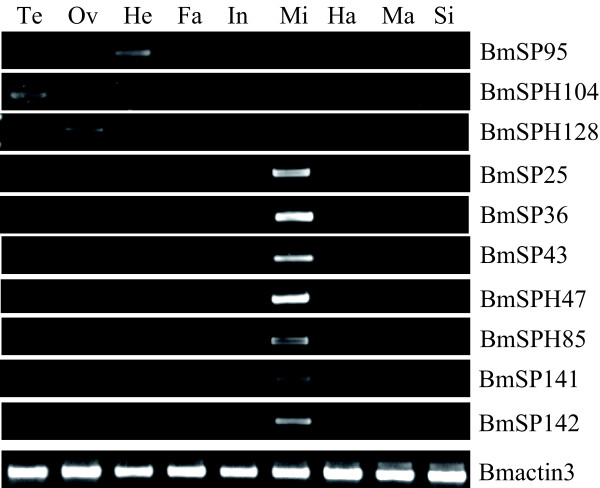
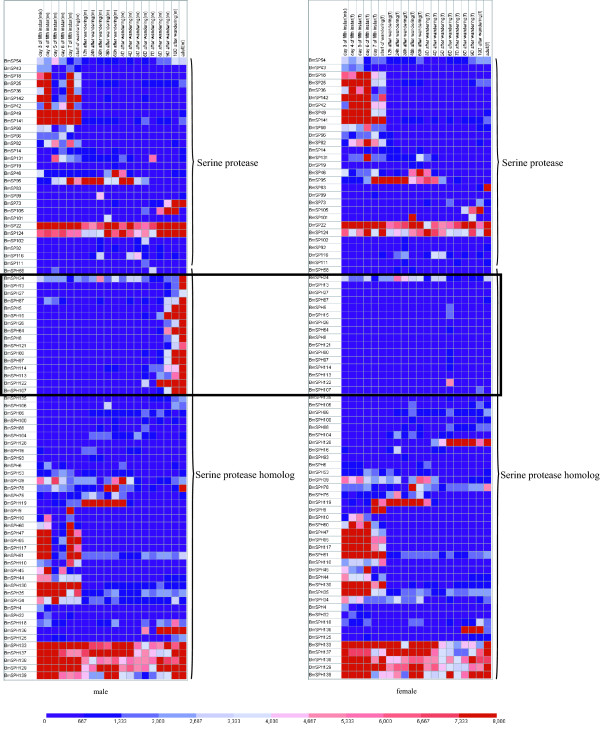
Microarray analysis showed that 64 SP and SPH genes have transcriptional activity in at least one tissue or organ according to a stringent criterion for definition of expressed genes [15]. Midgut had the highest number (38) of expressed genes among the 10 tissues or organs, while the number of expressed genes in testis, ovary, head, integument, fat body, hemocytes, Malpighian tubule, A/MSG and PSG was 26, 24, 32, 30, 15, 22, 20, 7 and 6, respectively (Figure 3). Further analysis reveals that most of genes expressed highly and exclusively in the midgut belong to silkworm-specific families, such as SP_fam12, SP_fam13, SP_fam14 and SP_fam15. These gene families showed unique midgut expression feature that are likely to reflect the special food habit of silkworm, and the process of accumulating abundant amino acids for producing silk. It may also be associated with resistance with pathogenic microorganisms. Some genes had unique expression features in other tissues on the third day of the silkworm fifth instar larva according to microarray analysis, for example, exclusive expression was seen for BmSP95 in the head, BmSPH104 in the testis, and BmSPH128 in the ovaries. Semi-quantitative RT-PCR was also performed to confirm tissue expression patterns of some representative SP and SPH genes for day-3 fifth instar larvae. The results were the same as those for the microarray data (Figure 4).
Figure 3.
Tissue expression profile of silkworm SP and SPH genes . Gene expression levels are represented by red (higher expression) and blue (lower expression) boxes. The columns represents ten different tissue or organ samples: testis, ovary, head, integument, fat body, midgut, hemocyte, malpighian tubule, A/MSG(anterior/median silk gland) and PSG(posterior silk gland). The column name is composed as "sample name-labeling dye- experiment order".
Figure 4.
Tissue expression patterns of some silkworm SP and SPH genes showing tissue specificity on day 3 of the fifth instar. Semi-quantitative RT-PCR amplification of total RNA with BmSP(H)-specific oligonucleotides. Te, testis; Ov, ovary; He, head; Fa, fat body; In, integument; Mi, midgut; Ha, haemocyte; Ma, Malpighian tubules; Si, silk gland. The silkworm cytoplasmic actin A3 gene (Bmactin3; GenBank accession no. U49854) was used as internal control, and denoted by Bmactin3.
More than 20 genes are expressed exclusively at the fifth instar larval stage, and about 7 genes are expressed highly over the entire metamorphosis process (Figure 5). The majority of SP and SPH genes express in both male and female silkworms. However, a group of SPH genes show differences in expression between males and females during the metamorphosis in which a pupa turns into an adult. These genes express highly and exclusively in males, but are minimally expressed in females during metamorphosis (Figure 5).
Figure 5.
Expression profile of SP and SPH genes during different developmental stages. Gene expression levels are represented by red (higher) and blue (lower) boxes. Genes that express only in male silkworm during metamorphosis are marked by a rectangle. The columns represents twenty different sample time points: day 3, 4, 5, 6, 7 of the fifth instar, start of wandering, 13 different times after wandering: 12 hour, 24 hours, 36 hours, 48 hours, 60 hours, 3 days, 4 days, 5 days, 6 days, 7 days, 8 days, 9 days, 10 days and adult.Word "mix" in the column name represents that the sample came from both gender; word "m" in the column name represents that the sample came from male silkworm; word "f" in the column name represents that the sample came from female silkworm.
We analyzed the expression pattern of repeated genes using information on gene location and expression data from tissue microarray experiments, and found that repeated SP and SPH gene groups in silkworm do not have identical expression patterns. For example, the repeated gene group on chr.16 includes several genes, but only BmSP18 and BmSP142 have a similar expression pattern. Of 23 SP and SPH genes expressed exclusively in the midgut, only BmSP18 and BmSP142 are tandem repeat genes, with too much distance between the other genes (Figure 2), although some were located on the same chromosome, such as BmSPH35, BmSPH4, BmSPH44 and BmSPH6 on chr.7; BmSPH10, BmSP25, BmSPH47 and BmSPH81 on chr.18; BmS142, BmS18 and BmSPH51 on chr.16; and BmSP141 and BmSP42 on chr.12. These results were confirmed by developmental expression patterns (data not shown)
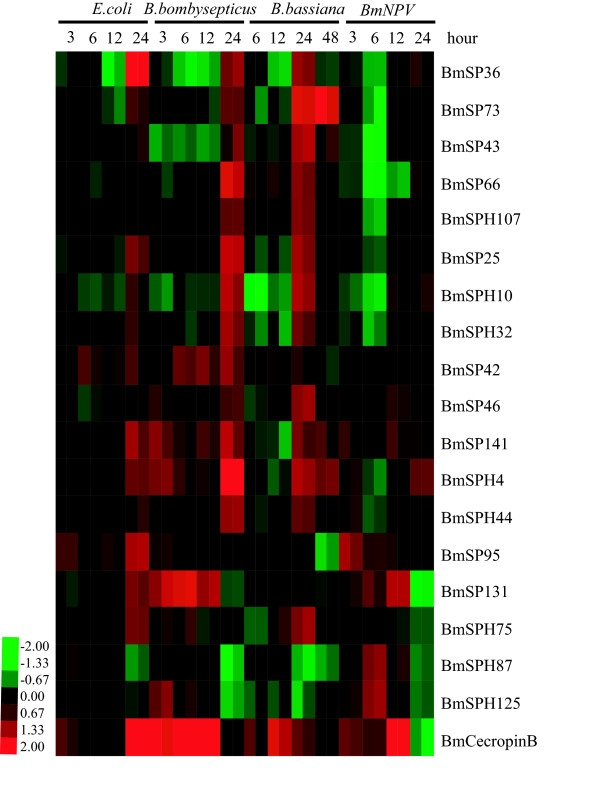
To investigate whether silkworm SP genes are involved in the immune response, SP and SPH expression changes were analyzed after insects infected by four different microorganisms. We found that expression of 65 SP and SPH genes could be detected. Among these, 18 were up-regulated more than 2.0 times in the infected group compared to the control group (Figure 6 and Additional file 6). Most were up-regulated most strongly during the late stage, 24 hours after infection. For example, BmSP36 expression was up to 4 times higher than the control at 24 hours after E. coli infection, and twice as high as the 24-h point for Bacillus bombysepticus or Batrytis bassiana infection. Expression of BmSPH4 exceeded 1.5 times the control after infection with any of the four microorganisms. A few genes showed expression regulation that was different, depending on the infecting microorganism. For example, BmSPH125 was up-regulated after infection with E.coli, B.bombyspticus or B. mori nucleopolyhedrovirus (BmNPV), but did not change significantly after infection with B.bassiana. However, when we compared the magnitude in induction of gene expression between antibacterial proteins BmcecropinB and silkworm SP or SPH genes, we found that BmcecropinB was remarkably upregulated on infection by these four pathogens at all time points except for BmNPV infection at 24 h (Figure 6). The induced expression pattern was consistent with SP or SPH genes at most of the time points. The data suggest that SP or SPH genes might play important roles in the innate immune system of B. mori. The gene expression results from microarray experiments were validated by real-time quantitative RT-PCR (Additional file 7). The results are consistent with the microarray experiments. Only 3 of the 18 up-regulated genes were in GenBank, specifically BmSPH125 (NP_001036891), BmSP131 (NP_001040537) and BmSP141 (NP_001040178) (Additional file 3).Of 18 up-regulated SPs and SPHs, BmSP95 has two clip domains, while BmSPH125 has one clip domain (Additional file 4). Although the function of clip domains is not yet clear, enzymes that contain this structure are of interest, because they may regulate serine protease activity and mediate innate immunity [5].
Figure 6.
Hierarchical cluster analysis of SP and SPH genes after microorganism infection. Red represents that gene expression is up-regulated; black represents that gene expression is not changed; green represents that gene expression is download-regulated; gray represents data missing. Hierarchical clustering was performed using Cluster version 3.0 and visualized by TreeView.
SPs involved in cascade regulation of the insect immune response were found to be highly conserved. Broehan found that chymotrypsin-like protease cascade systems that contain CTLP1 control chitin synthase activity in the tobacco hornworm, Manduca sexta [21]. The chitin portion of the peritrophic matrix in the midgut plays an important role in resistance to pathogenic bacteria and toxins. BmSPH10 has 70% identity to CLIP 1 (Additional file 6), and like CLIP 1, BmSPH10 was also expressed in the midgut, so it may have a similar function. The melanism reaction in insects is mediated by an SP signalling cascade that is highly conserved. In the M. sexta SP signalling cascade, proHP14 is a pattern recognition protein that binds to bacteria and autoactivates and triggers the prophenoloxidase activation system in the hemolymph, which contains a series of proteinases such as HP1, 6 8, 17 and 21. Finally prophenoloxidase-activating proteinase (PAPs) are activated, leading to the melanism reaction [22-24]. Cytotoxic molecules are produced in this process, including quinones and reactive oxygen intermediates, that may kill the invading microorganisms that are trapped by the melanin [25]. BmSP14, BmSPH125 and BmSPH78 in the silkworm are highly similar to HP14, HP17 and PAP, respectively, in the tobacco hornworm (Additional file 6). BmSP14, BmSPH125 and BmSPH78 were markedly up-regulated after induction, suggesting that they may have similar functions to their tobacco hornworm homologs (BmSP14 and BmSPH78, which were up-regulated between 1.5 and 2 times, were not included in the table).
Conclusions
From the updated B. mori silkworm genome, we identified SP and SPH genes and analyzed their expression patterns. Comparative genomics showed that SP and SPH genes belong to a large family whose members were generated mainly by duplication and show a lineage of tandem chromosomal repeats. In addition, we found that silkworm has species-specific SP and SPH genes, and combining the results of express profiles, we hypothesize that members of the silkworm SP and SPH families may participate in biological processes that include digestion, development and the immune response. We hope the data presented in this study facilitates functional studies on SPs and SPHs in B. mori.
Methods
Identification of silkworm SP and SPH genes in the silkworm
SP and SPH genes from M. sexta and other insects were downloaded from GenBank http://www.ncbi.nlm.nih.gov/Genbank/. The sequence homology BLAST alignment tool was downloaded from the ftp site of National Center for Biotechnology Information ftp://ftp.ncbi.nih.gov/blast/. SP and SPH gene sequences from other insects were used as queries in BLAST searches against the silkworm database (E-value: 10-6) [26]. Identified genes were validated by aligning to a non-redundant gene dataset. Identified genes were also validated by BLAST searching against an EST database with threshold E-value <10-30, identities >90%, and match lengths >100 bp.
Evolutionary analysis of silkworm SP and SPH genes
SP and SPH sequences of D. melanogaster, C. elegans and A. gambiae were included in an all-against-all BLAST search using BLASTP (E-value 1e-30), with the results converted into a Markov Martrix [27]. The TRIBE-MCL clustering algorithm was used to detect family members [27]. Clustering operations were implemented at inflation values of 1.2. Identified SP and SPH genes were used for phylogenetic analyses. Multiple sequence alignments of protein sequences were made using ClustalX 1.83 [28], and neighbor-joining used Phylip software [29], with a bootstrap value of 1000 to reconstruct the phylogenetic tree.
Expression of silkworm SP and SPH genes from whole-genome microarrays
In 2007, Xia et al. constructed a genome-wide oligonucleotide microarray with more than 22,000 probes [15], including 107 SP- and SPH-specific oligonucleotide probes. We analyzed tissue expression of SP and SPH genes at the third day of the fifth instar larvae of Dazao. In brief, insects were reared at a stable temperature of 25°C until the third day of the fifth instar. Then testis, ovary, head, integument, fat body, midgut, hemocytes, Malpighian tubules, A/MSG (anterior/median silk gland) and PSG (posterior silk gland) from this developmental point were hand-dissected on ice. To determine the developmental expression patterns, individuals at 20 different time points from day 3 of the fifth instar to the moth stage were collected for both genders. Gene expression levels were visualized using GeneCluster 2.0 [30]. The detailed experimental procedures for microarray and data analyses were as previously reported [15].
For insects used for microorganism induction, newly hatched Dazao larvae were reared to the third day of the fifth instar with an artificial diet, in constant-temperature incubators. The gram-negative E. coli, gram-positive B. bombysepticus, B. bassiana and BmNPV were used for feeding induction, and Dazao fed with physiological saline were used as negative control. Whole body of individual samples induced with B. bassiana were collected after 6, 12, 24 and 48 hours, and whole body of individual samples from induction with the other microorganisms were collected after 3, 6, 12 and 24 hours. Microarray procedures and data analysis were as described [15]. Genes were considered to be up-regulated if the change in gene expression was greater than two fold on at least one occasion over the four time points in any of the four microorganism induction experiments [31-35]. Hierarchical clustering of gene expression patterns was performed using the Cluster (version 3.0) and visualized by TreeView programs [36].
Tissue expression analysis of silkworm SP and SPH genes by semi-quantitative RT-PCR
Total RNA was isolated from nine tissues (testis, ovary, head, fat body, integument, midgut, hemocytes, Malpighian tubules and silk gland) from day 3 fifth instar larvae using TRIzol reagent (Invitrogen, USA). Contaminating genomic DNA was digested using RNase-free DNase I (Promega) for 30 min at 37°C. RNA samples were diluted with RNase-free water and stored at -80°C. The concentration of total RNA was estimated by measuring the absorbance at 260 nm. Total RNA (10 μg) was reverse-transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen, USA) at 42°C. All cDNA samples were normalized using B. mori actin A3 as an internal control (forward primer: 5'-AAC ACC CCG TCC TGC TCA CTG-3'; reverse primer: 5'-GGG CGA GAC GTG TGA TTT CCT-3'). All SP and SPH primers for semi-quantitative RT-PCR detection are listed in Table Additional file 8. PCR amplification was performed in a total reaction volume of 25 μL containing normalized cDNA, 10 pmol of each primer, 2 mM MgCl2, 0.25 mM dNTP, 1× buffer and 2.5 U of Taq DNA polymerase. Semi-quantitative RT-PCR reactions were performed in a volume of 25 μL using the following program: initial incubation at 94°C for 4 min, followed by 24-28 cycles (BmSP95, BmSPH104, BmSPH128, BmSP36, BmSPH47, BmSPH85, BmSP142 for 24 cycles; BmSP43 and BmSP141 for 26 cycles; BmSP25 for 28 cycles) of 40 s at 94°C, 40 s of annealing (temperatures listed in Additional file 8), 45 s of extension (72°C), and a final extension at 72°C for 10 min. Aliquots of 4 μL of the PCR products were separated on 2.0% agarose gels and stained with EB.
Induced expression analysis of silkworm SP and SPH genes by real-time quantitative RT-PCR
Quantitative RT-PCR was performed using an ABI PRISM 7000 sequence detection system (Applied Biosystems). The 15 μl mixture included 1.5 μl of cDNA, 0.5 mM of each primer and 1× SYBR Premix Ex Taq (TaKaRa) in each well of a 96-well plate. PCR was carreied out with initial denaturation at 94°C for 10 s, followed by 40 cycles at 95°C for 5 s and 60°C for 31 s. The primer sequences for all genes are listed in Additional file 9. Relative gene expression data were normalized against Ct values for the housekeeping sw22934 gene in silkworm and the fold change (2-ΔΔCt) was determined by comparison with average expression levels for control samples, with the index dened as 1.0. Four larvae were used for each treatment. Each expression assay was repeated three times. Student's t-test was used to evaluate statistical signicance (P < 0.01).
Authors' contributions
PZ conceived and designed the study and wrote the manuscript. GHW performed microarray experiments and generated array data. ZMD and JD identified the SPs and SPHs and analyzed the sequences. PZX and TCC performed microorganism induction experiments. QYX participated in designing the study, analyzed the results and revised the manuscript. ZHX supervised the study. All authors read and approved the final manuscript.
Supplementary Material
The SPs and SPHs predicted in the silkworm . a. The scaffold name of silkworm genome that can be found in the SilkDB http://www.silkdb.org; b. The start position of the SPs and SPHs gene on the silkworm genome; c. The end position of the SPs and SPHs gene on the silkworm genome; d. The strand of the SPs and SPHs gene on the silkworm genome; e. The position of the SPs and SPHs gene on the silkworm chromosomes; f. Whether the SPs and SPHs genes are supported by the EST evidence.
The serine protease genes that contain intact active site . The active site of Ser, His, and Asp play important role in the catalytic processes for SP genes. By analyzing the gene sequences, we found that 51 serine protease genes have the three active site residues intact (SP), and the rest 92 serine protease genes had mutations in the catalytic residues (SPH), so they may have lost catalytic function.
The SPs and SPHs in the silkworm that have been reported . The GenBank accession numbers and description of the reported SPs and SPHs in the silkworm are listed.
The clip domain in the silkworm, Bombyx mori. The conserved six cysteine residues for each CLIP are marked by color. We identified 7 clip-SPs and 11 clip-SPHs in the silkworm, which are consisted of a chymotrypsin-like SP domain, and one or more clip domain(s) at the N-terminus.
Phylogenetic analysis of silkworm-specific SP and SPH genes of silkworm and Drosophila. Most of t he SP and SPH genes were included in the analysis. Except for SP_fam1, we choose three representative genes to analyze. Multiple alignments of protein sequences were made by ClustalW. Then neighbor-joining phylogenetic trees were reconstructed by Phylip. The parameters were chosen as follows: the evolutionary distance was poisson-corrected, gaps were completely deleted, and 100 iterations were used for calculating bootstrap values.
The up-regulated SP and SPHs after silkworm infected by microorganisms . Genes whose expression were up-regulated than two fold on at least one occasion over the four time points in any of the four microorganism induction experiments were listed. The best NCBI BLAST Results of the up-regulated SP and SPHs were also shown in the table.
The induced expression analysis of silkworm SP and SPH genes by quantitative real-time RT-PCR . We chose the time points of infecting 6 h and 24 h to do the expression analysis. The expression of SP or SPH in the control sample was set to 1. The abbreviations are used, the E. coli infected sample (Ec), the B. bombyseptieus infected sample (Bs), the B. bassiana infected sample (Bb) and the B. mori nucleopolyhedrovirus infected sample (NPV). Each expressive assay was replicated by three times. The Student's t-test was used to evaluate statistical signicance (P < 0. 01).
Primers used in semi-quantitative RT-PCR study . Primer sequences, melting temperature, and amplicon size were listed.
Primers used in qRT-PCR study . Primer sequences, melting temperature and amplicon size were listed.
Contributor Information
Ping Zhao, Email: zhaop@swu.edu.cn.
Gen-Hong Wang, Email: wanggh@cqu.edu.cn.
Zhao-Ming Dong, Email: dong-zhaoming@163.com.
Jun Duan, Email: duanj@cqu.edu.cn.
Ping-Zhen Xu, Email: xpz198249@163.com.
Ting-Cai Cheng, Email: chengtc@cqu.edu.cn.
Zhong-Huai Xiang, Email: xbxzh@swu.edu.cn.
Qing-You Xia, Email: xiaqy@swu.edu.cn.
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (No. 2005CB121000), the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT0750), and the National Natural Science Foundation (No. 30972147 and No. 30901054).
References
- Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27(2):67–74. doi: 10.1016/S0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- Zou Z, Lopez DL, Kanost MR, Evans JD, Jiang H. Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol Biol. 2006;15(5):603–614. doi: 10.1111/j.1365-2583.2006.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4(3):337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/S0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci USA. 1997;94(21):11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Baek MJ, Lee MH, Park JW, Lee SY, Soderhall K, Lee BL. A new easter-type serine protease cleaves a masquerade-like protein during prophenoloxidase activation in Holotrichia diomphalia larvae. J Biol Chem. 2002;277(42):39999–40004. doi: 10.1074/jbc.M205508200. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G. et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yaginuma T, Kobayashi M, Yamashita O. cDNA cloning, sequencing and temporal expression of the protease responsible for vitellin degradation in the silkworm, Bombyx mori. Comp Biochem Physiol B. 1991;99(2):405–411. doi: 10.1016/0305-0491(91)90062-I. [DOI] [PubMed] [Google Scholar]
- Maki N, Yamashita O. The 30 kP protease A responsible for 30-kDa yolk protein degradation of the silkworm, Bombyx mori: cDNA structure, developmental change and regulation by feeding. Insect Biochem Mol Biol. 2001;31(4-5):407–413. doi: 10.1016/S0965-1748(00)00135-1. [DOI] [PubMed] [Google Scholar]
- Kaji K, Tomino S, Asano T. A serine protease in the midgut of the silkworm, Bombyx mori: protein sequencing, identification of cDNA, demonstration of its synthesis as zymogen form and activation during midgut remodeling. Insect Biochem Mol Biol. 2009;39(3):207–217. doi: 10.1016/j.ibmb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Nakazawa H, Tsuneishi E, Ponnuvel KM, Furukawa S, Asaoka A, Tanaka H, Ishibashi J, Yamakawa M. Antiviral activity of a serine protease from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Virology. 2004;321(1):154–162. doi: 10.1016/j.virol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T. et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C. et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306(5703):1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Xia Q, Cheng D, Duan J, Wang G, Cheng T, Zha X, Liu C, Zhao P, Dai F, Zhang Z. et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8(8):R162. doi: 10.1186/gb-2007-8-8-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Song YL, Kim JH, Park SY, Park JW, Lee BL, Oh BH, Ha NC. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. Embo J. 2005;24(24):4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- Shi L, Paskewitz S. Proteomics and insect immunity. ISJ. 2006;3:4–17. [Google Scholar]
- Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1138–1146. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Duan J, Xia Q, Cheng D, Zha X, Zhao P, Xiang Z. Species-specific expansion of C2H2 zinc-finger genes and their expression profiles in silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1121–1129. doi: 10.1016/j.ibmb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Broehan G, Zimoch L, Wessels A, Ertas B, Merzendorfer H. A chymotrypsin-like serine protease interacts with the chitin synthase from the midgut of the tobacco hornworm. J Exp Biol. 2007;210(Pt 20):3636–3643. doi: 10.1242/jeb.008334. [DOI] [PubMed] [Google Scholar]
- Ji C, Wang Y, Guo X, Hartson S, Jiang H. A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta. J Biol Chem. 2004;279(33):34101–34106. doi: 10.1074/jbc.M404584200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem Mol Biol. 2005;35(8):931–943. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci USA. 1998;95(21):12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nappi AJ, Ottaviani E. Cytotoxicity and cytotoxic molecules in invertebrates. Bioessays. 2000;22(5):469–480. doi: 10.1002/(SICI)1521-1878(200005)22:5<469::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (PHYLogeny Inference Package) version 3.6a2. Distributed by the author. 1993.
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Aguilar R, Jedlicka AE, Mintz M, Mahairaki V, Scott AL, Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem Mol Biol. 2005;35(7):709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Baton LA, Robertson A, Warr E, Strand MR, Dimopoulos G. Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium berghei infection. BMC Genomics. 2009;10:257. doi: 10.1186/1471-2164-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eum JH, Seo YR, Yoe SM, Kang SW, Han SS. Analysis of the immune-inducible genes of Plutella xylostella using expressed sequence tags and cDNA microarray. Dev Comp Immunol. 2007;31(11):1107–1120. doi: 10.1016/j.dci.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Walker LA, Maccallum DM, Bertram G, Gow NA, Odds FC, Brown AJ. Genome-wide analysis of Candida albicans gene expression patterns during infection of the mammalian kidney. Fungal Genet Biol. 2009;46(2):210–219. doi: 10.1016/j.fgb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Gavotte L, Xie Y, Dobson SL. Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics. 2008;9:1. doi: 10.1186/1471-2164-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SPs and SPHs predicted in the silkworm . a. The scaffold name of silkworm genome that can be found in the SilkDB http://www.silkdb.org; b. The start position of the SPs and SPHs gene on the silkworm genome; c. The end position of the SPs and SPHs gene on the silkworm genome; d. The strand of the SPs and SPHs gene on the silkworm genome; e. The position of the SPs and SPHs gene on the silkworm chromosomes; f. Whether the SPs and SPHs genes are supported by the EST evidence.
The serine protease genes that contain intact active site . The active site of Ser, His, and Asp play important role in the catalytic processes for SP genes. By analyzing the gene sequences, we found that 51 serine protease genes have the three active site residues intact (SP), and the rest 92 serine protease genes had mutations in the catalytic residues (SPH), so they may have lost catalytic function.
The SPs and SPHs in the silkworm that have been reported . The GenBank accession numbers and description of the reported SPs and SPHs in the silkworm are listed.
The clip domain in the silkworm, Bombyx mori. The conserved six cysteine residues for each CLIP are marked by color. We identified 7 clip-SPs and 11 clip-SPHs in the silkworm, which are consisted of a chymotrypsin-like SP domain, and one or more clip domain(s) at the N-terminus.
Phylogenetic analysis of silkworm-specific SP and SPH genes of silkworm and Drosophila. Most of t he SP and SPH genes were included in the analysis. Except for SP_fam1, we choose three representative genes to analyze. Multiple alignments of protein sequences were made by ClustalW. Then neighbor-joining phylogenetic trees were reconstructed by Phylip. The parameters were chosen as follows: the evolutionary distance was poisson-corrected, gaps were completely deleted, and 100 iterations were used for calculating bootstrap values.
The up-regulated SP and SPHs after silkworm infected by microorganisms . Genes whose expression were up-regulated than two fold on at least one occasion over the four time points in any of the four microorganism induction experiments were listed. The best NCBI BLAST Results of the up-regulated SP and SPHs were also shown in the table.
The induced expression analysis of silkworm SP and SPH genes by quantitative real-time RT-PCR . We chose the time points of infecting 6 h and 24 h to do the expression analysis. The expression of SP or SPH in the control sample was set to 1. The abbreviations are used, the E. coli infected sample (Ec), the B. bombyseptieus infected sample (Bs), the B. bassiana infected sample (Bb) and the B. mori nucleopolyhedrovirus infected sample (NPV). Each expressive assay was replicated by three times. The Student's t-test was used to evaluate statistical signicance (P < 0. 01).
Primers used in semi-quantitative RT-PCR study . Primer sequences, melting temperature, and amplicon size were listed.
Primers used in qRT-PCR study . Primer sequences, melting temperature and amplicon size were listed.