Abstract
Background
Geobacter species in a phylogenetic cluster known as subsurface clade 1 are often the predominant microorganisms in subsurface environments in which Fe(III) reduction is the primary electron-accepting process. Geobacter bemidjiensis, a member of this clade, was isolated from hydrocarbon-contaminated subsurface sediments in Bemidji, Minnesota, and is closely related to Geobacter species found to be abundant at other subsurface sites. This study examines whether there are significant differences in the metabolism and physiology of G. bemidjiensis compared to non-subsurface Geobacter species.
Results
Annotation of the genome sequence of G. bemidjiensis indicates several differences in metabolism compared to previously sequenced non-subsurface Geobacteraceae, which will be useful for in silico metabolic modeling of subsurface bioremediation processes involving Geobacter species. Pathways can now be predicted for the use of various carbon sources such as propionate by G. bemidjiensis. Additional metabolic capabilities such as carbon dioxide fixation and growth on glucose were predicted from the genome annotation. The presence of different dicarboxylic acid transporters and two oxaloacetate decarboxylases in G. bemidjiensis may explain its ability to grow by disproportionation of fumarate. Although benzoate is the only aromatic compound that G. bemidjiensis is known or predicted to utilize as an electron donor and carbon source, the genome suggests that this species may be able to detoxify other aromatic pollutants without degrading them. Furthermore, G. bemidjiensis is auxotrophic for 4-aminobenzoate, which makes it the first Geobacter species identified as having a vitamin requirement. Several features of the genome indicated that G. bemidjiensis has enhanced abilities to respire, detoxify and avoid oxygen.
Conclusion
Overall, the genome sequence of G. bemidjiensis offers surprising insights into the metabolism and physiology of Geobacteraceae in subsurface environments, compared to non-subsurface Geobacter species, such as the ability to disproportionate fumarate, more efficient oxidation of propionate, enhanced responses to oxygen stress, and dependence on the environment for a vitamin requirement. Therefore, an understanding of the activity of Geobacter species in the subsurface is more likely to benefit from studies of subsurface isolates such as G. bemidjiensis than from the non-subsurface model species studied so far.
Background
Geobacter bemidjiensis is a member of the Geobacteraceae, a family of Fe(III)-respiring Deltaproteobacteria that are of interest for their role in cycling of carbon and metals in aquatic sediments and subsurface environments as well as the bioremediation of organic- and metal-contaminated groundwater and the harvesting of electricity from complex organic matter [1,2]. It was isolated from subsurface sediments in Bemidji, Minnesota, near a site where aromatic hydrocarbons were being degraded naturally [3]. G. bemidjiensis is a member of the phylogenetic cluster designated subsurface clade 1, which predominates in a diversity of subsurface environments in which dissimilatory Fe(III) reduction is an important process [4]. Environmental proteomic studies have demonstrated that Geobacter species closely related to G. bemidjiensis were metabolically active during the in situ bioremediation of uranium-contaminated groundwater [5].
Preliminary studies have suggested that genome-scale metabolic modeling of Geobacter species [6,7] may aid in predicting the response of subsurface Geobacter species to subsurface bioremediation strategies [8,9]. However, it is not known whether the metabolic potential of subsurface Geobacter species is essentially the same as that of non-subsurface Geobacter species, or significantly different. Therefore, comparative analysis of the genome of a representative of the subsurface clade 1 Geobacter species with the curated genomes of two non-subsurface Geobacter species, Geobacter sulfurreducens and Geobacter metallireducens [10,11], was carried out to improve predictive modeling of the responses of Geobacteraceae to efforts to stimulate bioremediation of organic and metal contaminants in the subsurface.
Results and Discussion
Contents of the G. bemidjiensis genome
The automated annotation process identified 4040 protein-coding genes and 76 ribonucleic acid (RNA) genes in the genome of 4615150 bp. During manual curation, 56 genes were discarded, 40 genes were reannotated as pseudogenes, and another 79 protein-coding genes, 28 pseudogenes, and 778 non-protein-coding features were identified. Protein sequence alignments demonstrated that 27 pseudogenes were frameshifted within runs of five or more identical bases, where DNA replication is most error-prone, and seven of these are polymorphisms where the minor alleles contain no frameshift, indicating a subpopulation of cells that can produce functional proteins from these genes (Additional file 1: Table S1). Of the 4023 intact protein-coding genes in the G. bemidjiensis genome, 148 hypothetical proteins (3.6%) have no match in any other genome, including that of Geobacter sp. M21; a further 87 conserved hypothetical proteins were found only in these two closely related genomes.
Metabolism of pyruvate
An appropriate place to begin to compare metabolism between the subsurface exemplar G. bemidjiensis and the representative non-subsurface species G. sulfurreducens and G. metallireducens is with the central metabolic reactions that interconvert pyruvate and acetyl-CoA. Like other Geobacteraceae, G. bemidjiensis possesses two sets of genes encoding pyruvate dehydrogenase complexes (Gbem_2257 and Gbem_2251-Gbem_2250; Gbem_0459-Gbem_0461), which irreversibly convert pyruvate to acetyl-CoA. The ability of G. sulfurreducens to reverse this reaction and derive biomass from acetyl-CoA has been attributed to a homodimeric pyruvate:ferredoxin/flavodoxin oxidoreductase [12], for which homologs exist in all Geobacteraceae including G. bemidjiensis (Gbem_0209). G. bemidjiensis has an additional pyruvate:ferredoxin oxidoreductase (Gbem_4034), more closely related to those of sulfate-reducing bacteria. In addition to gluconeogenesis and oxidative decarboxylation to acetyl-CoA, a third fate of pyruvate in G. bemidjiensis may be oxidative decarboxylation to acetate by a putative quinone-reducing pyruvate decarboxylase (Gbem_0287) that is 32% identical to the E. coli enzyme [13]. Thus, metabolism of pyruvate may be more complex in G. bemidjiensis than in non-subsurface Geobacter species.
Metabolism of propionate
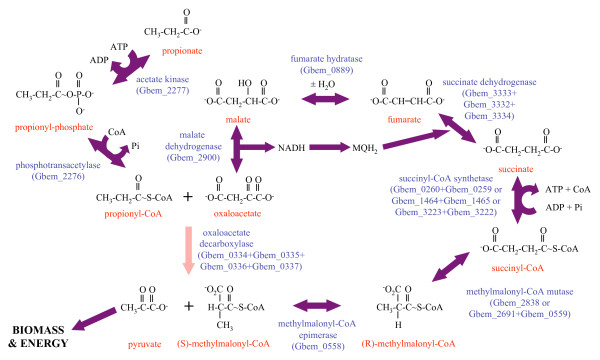
G. bemidjiensis and G. metallireducens utilize propionate as an electron donor, whereas G. sulfurreducens cannot. Analysis of the genome suggests that G. bemidjiensis utilizes propionate by converting it to pyruvate in ten steps (Figure 1): (1-2) activation to propionyl-CoA by the same enzymes that activate acetate; (3) carboxylation to (S)-methylmalonyl-CoA by a biotin-dependent propionyl-CoA carboxyltransferase (Gbem_0335) that is 51% identical to the 12 S subunit of the Propionibacterium freudenreichii methylmalonyl-CoA:pyruvate carboxyltransferase enzyme complex [14]; (4-5) epimerization and rearrangement to succinyl-CoA; (6) hydrolysis to succinate; (7-9) oxidation to oxaloacetate; and (10) decarboxylation to pyruvate by an oxaloacetate decarboxylase (Gbem_0334) that is 60% identical to the 5 S subunit of the P. freudenreichii methylmalonyl-CoA:pyruvate carboxyltransferase complex [14], with concomitant transfer of the carboxyl group to propionyl-CoA (see step 3 above). The carboxyltransferase reaction, possibly involving a conserved hypothetical protein (Gbem_0336) and a biotin attachment domain protein (Gbem_0337) encoded within the same predicted operon as the two enzymes, avoids the energetic cost of 1 ATP associated with other biotin-dependent carboxylations of acyl-CoA substrates. The energy recovered from hydrolysis of succinyl-CoA to succinate may be used for the initial activation of propionate, either in the form of ATP or possibly through direct transfer of CoA by one of several uncharacterized acyl-CoA:carboxylate CoA transferases (Gbem_1430, Gbem_1439, Gbem_3573). This energy-efficient pathway contrasts with the proposed pathway in the non-subsurface species G. metallireducens [10,15], in which no energy is recovered from thioester hydrolysis of propionyl-CoA by 2-methylcitrate synthase. Accordingly, reliable predictions of the metabolic activity of Geobacter species in subsurface environments amended with propionate will depend on models based on G. bemidjiensis rather than G. metallireducens.
Figure 1.
Predicted pathway for conversion of propionate to pyruvate by G. bemidjiensis. Eight steps are catalyzed by enzymes that have orthologs in both G. sulfurreducens and G. metallireducens (purple arrows). The only enzyme unique to G. bemidjiensis (pink arrow) is oxaloacetate decarboxylase, which simultaneously functions as propionyl-CoA carboxylase to carry out an earlier step of the cyclical pathway.
Fatty acid metabolism
The G. bemidjiensis genome encodes many enzymes of acyl-CoA metabolism (Additional file 2: Table S2). This multiplicity of genes suggests that in addition to its known short-chain organic acid electron donors (butyrate, isobutyrate and valerate), G. bemidjiensis may also be able to utilize longer fatty acids as sources of carbon and electrons. Indeed, G. bemidjiensis and the other genomes of subsurface Geobacter species possess a very-long-chain fatty acyl-CoA dehydrogenase (fadE Gbem_2128) 50% identical to that of Bacillus subtilis [16], which is absent from G. sulfurreducens and G. metallireducens. Thus, the metabolism of fatty acids by subsurface Geobacter species may be better understood by examining G. bemidjiensis rather than non-subsurface relatives.
Growth of G. bemidjiensis by disproportionation of fumarate
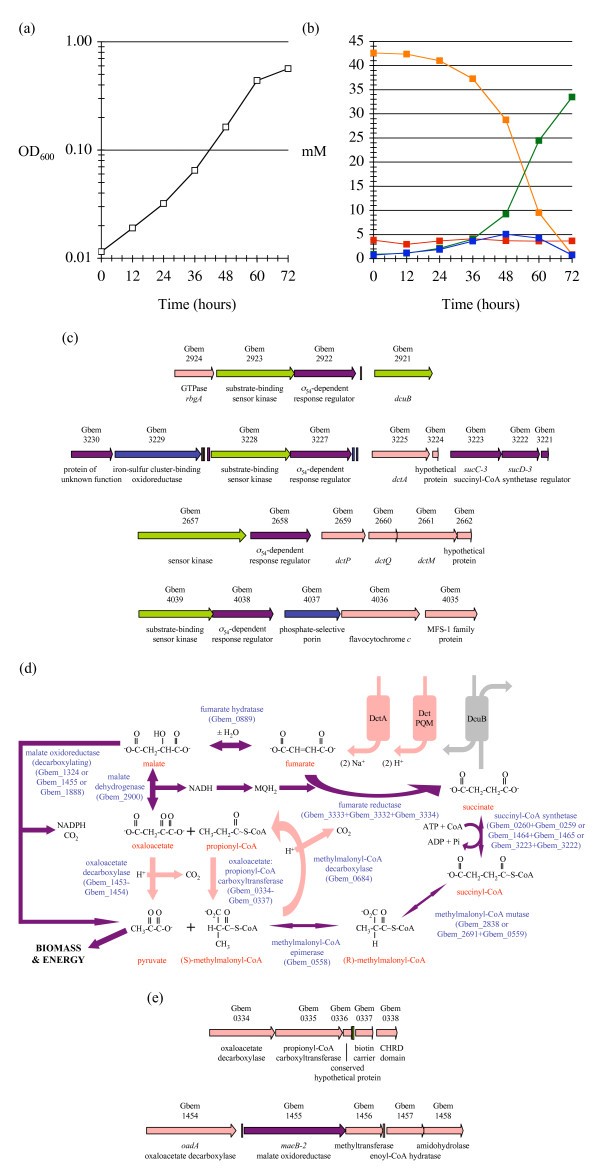
G. bemidjiensis, like G. sulfurreducens, can utilize fumarate as an electron acceptor in combination with acetate and other electron donors [3]. Consistent with its previously described ability to utilize malate as an electron donor and carbon source, G. bemidjiensis also grows when provided with only fumarate (Figure 2a), as was previously reported for Geobacter bremensis [17]. Existing metabolic models of (non-subsurface) Geobacter species do not include this capability; therefore, we examined it more closely. Chromatographic analysis of culture filtrates indicates that 79% of the supply of fumarate serves as electron acceptor and is excreted as succinate, implying that another 13% of the fumarate is completely oxidized to carbon dioxide (Figure 2b). Biomass is inferred to derive from the remaining 8% of the fumarate supply. A small amount of malate is transiently excreted when consumption of fumarate is steepest (Figure 2b). Unlike Desulfovibrio vulgaris [18] and Rhodoferax ferrireducens [19], G. bemidjiensis does not excrete acetate, even at early stages (Figure 2b), indicating that complete oxidation of fumarate is energetically more favourable than partial oxidation to acetyl-CoA followed by substrate-level phosphorylation.
Figure 2.
Growth of G. bemidjiensis by disproportionation of fumarate. (a) Growth of G. bemidjiensis with fumarate as sole electron donor and electron acceptor (white squares). The mean of quadruplicate measurements is shown; variation was negligible. (b) High-pressure liquid chromatography measurements showing consumption of fumarate (orange squares), excretion of succinate (green squares), transient excretion of malate (blue squares) and unaffected trace amounts of extracellular acetate (red squares) during growth of G. bemidjiensis with fumarate alone. (c) Organization of fumarate transporter genes. Genes unique to G. bemidjiensis (pink arrows) are found in proximity to genes that have orthologs in either G. sulfurreducens (green arrows) or G. metallireducens (blue arrows), or both (purple arrows), along with multicopy nucleotide sequence features (rectangles; same colour code). The fumarate transporters are encoded by dcuB (dicarboxylic acid exchanger), dctA (sodium-dependent), and dctPQM (proton-dependent). (d) Predicted pathways of fumarate disproportionation by conversion to pyruvate. Enzymes that have orthologs in both G. sulfurreducens and G. metallireducens (purple arrows) constitute the decarboxylating malate oxidoreductase pathway, which is inactive when the only means of fumarate transport is the fumarate/succinate exchanger DcuB, as in G. sulfurreducens (grey arrows). The unique fumarate transporters and enzymes of G. bemidjiensis (pink arrows) make possible two other pathways involving different oxaloacetate decarboxylases. (e) Organization of oxaloacetate decarboxylase genes; the colour code is the same as in (c).
Either the sodium/fumarate symporter DctA (Gbem_3225) that is 61% identical to the Salmonella typhimurium transporter [20] or the proton/fumarate symporter complex DctPQM (Gbem_2659-Gbem_2661) with 40% to 63% sequence identity to characterized homologs in Rhodobacter capsulatus [21] may be a prerequisite for disproportionation of fumarate. Both transporters, which are absent from G. sulfurreducens, allow import of fumarate without the concomitant export of succinate required by the dicarboxylate exchange transporter DcuB (Gbem_2921, orthologous to GSU2751). Notably, the dcuB and dctA genes are each located 3' of a pair of genes encoding a periplasmic substrate-binding sensor histidine kinase and response regulator that are highly similar (Figure 2c); a third pair is located 5' of a phosphate-selective porin (Gbem_4037, orthologous to Gmet_1042). The three sensor kinases (Gbem_2923; Gbem_3228; Gbem_4039) are 47% to 56% identical and the three response regulators (Gbem_2922; Gbem_3227; Gbem_4038) are 65% to 69% identical, suggesting that G. bemidjiensis has developed parallel signalling pathways possibly linked to dicarboxylate transport.
In D. vulgaris, oxidation of fumarate proceeds through the decarboxylating malate oxidoreductase reaction (B. Giles and J. Wall, personal communication), but it would be surprising if this were the predominant pathway in G. bemidjiensis (Figure 2d). When G. sulfurreducens respires fumarate, the activity of its two malate oxidoreductases (NADP-dependent maeB GSU1700 and NAD-dependent mleA GSU2308) must be kept much lower than that of malate dehydrogenase, which converts malate to oxaloacetate, because the equal exchange of succinate for fumarate by DcuB requires that any malate that is decarboxylated to pyruvate must be replaced by carboxylation of pyruvate to oxaloacetate at the expense of one ATP, which is prohibitive [12]. G. bemidjiensis possesses three maeB genes, all closely related to GSU1700, and no mleA gene. It is possible that one or more of these isozymes are upregulated during disproportionation of fumarate. However, complete oxidation of fumarate through acetyl-CoA using the malate oxidoreductases rather than malate dehydrogenase would result in NADH and NADPH being produced in a ratio of 3:2 rather than 4:1, requiring rerouting of reducing equivalents to meet energy demand. A more reasonable hypothesis is that conversion of fumarate to pyruvate is accomplished through decarboxylation of oxaloacetate by two parallel pathways (Figure 2d; gene diagrams in Figure 2e).
One oxaloacetate decarboxylase complex transfers the carboxyl group to propionyl-CoA as detailed above, forming (S)-methylmalonyl-CoA. In contrast with this same enzyme complex's predicted cyclic involvement in oxidation of propionate to pyruvate (see above, Figure 1), its catalysis of this step in the fumarate oxidation pathway requires that (S)-methylmalonyl-CoA be recycled to propionyl-CoA by methylmalonyl-CoA decarboxylase (Gbem_0684) (Figure 2d). The other oxaloacetate decarboxylase (oadA Gbem_1454) is 60% identical to the catalytic subunit of the sodium-translocating oxaloacetate decarboxylase of Klebsiella pneumoniae [22]. No homologs of the other two subunits were found in G. bemidjiensis, indicating that decarboxylation of oxaloacetate is not coupled to a sodium pump.
The malate oxidoreductase and oxaloacetate decarboxylase pathways also account for the ability of G. bemidjiensis to grow with malate and succinate as sources of carbon and electrons. Thus, the genome annotation of G. bemidjiensis offers insight into its unique capability to metabolize dicarboxylic acids without excreting acetate, which could not be predicted correctly from studies of either the non-subsurface Geobacter genomes or non-Geobacter species.
Possible carbon dioxide fixation via citrate lyase
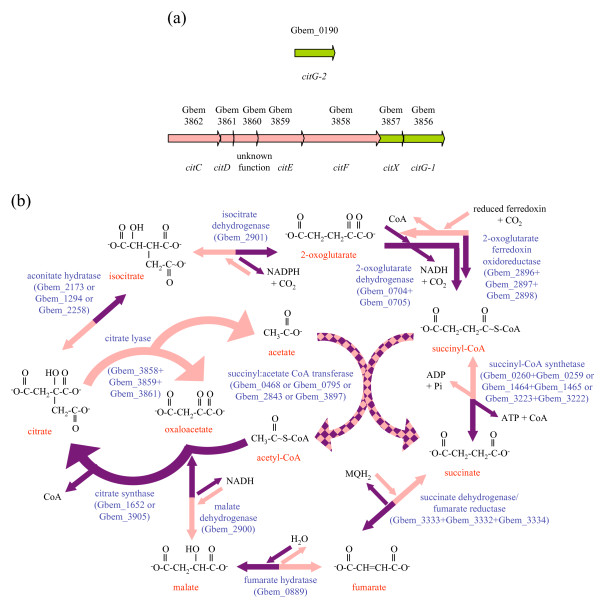
Addition of acetate to the subsurface selectively stimulates the growth of Geobacteraceae, which derive electrons by conversion of acetate to carbon dioxide through the TCA cycle [23]. The presence of a citCDEFXG gene cluster (Figure 3a) encoding the citrate lyase enzyme in G. bemidjiensis (Gbem_3862-Gbem_3856) suggests that this species might also operate the TCA cycle in reverse, fixing two molecules of carbon dioxide and generating acetate (Figure 3b). This gene cluster is not found in the genomes of the non-subsurface species G. sulfurreducens and G. metallireducens. Preliminary data indicate that G. bemidjiensis can grow on Fe(III) oxides plus hydrogen, without any carbon source other than carbon dioxide (D. Holmes and C. Risso, unpublished), although the involvement of citrate lyase remains to be established.
Figure 3.
Carbon dioxide fixation in G. bemidjiensis. (a) The citrate lyase gene cluster and separately located gene Gbem_0190; some components are unique to G. bemidjiensis (pink arrows) and others have orthologs in G. sulfurreducens (green arrows). The catalytic subunits are encoded by citEF, the acyl carrier protein by citD, the cofactor synthesis enzyme by citG-1 and citG-2, the cofactor-transferring enzyme by citX, and the priming enzyme by citC. (b) The oxidative and reductive TCA cycles. Most enzymes that catalyze reductive reactions unique to G. bemidjiensis (pink arrows) also catalyze the oxidative reactions common to G. sulfurreducens and G. metallireducens (purple arrows). The exceptions are citrate lyase (reductive direction only) and citrate synthase and 2-oxoglutarate dehydrogenase (oxidative direction only).
A reverse TCA cycle requires enzymes capable of carrying out two carboxylations: conversion of succinyl-CoA to 2-oxoglutarate and conversion of 2-oxoglutarate to isocitrate (Figure 3b). The first of these conversions has been inferred from a carbon flux analysis study of G. metallireducens [24] and may be attributed to 2-oxoglutarate:ferredoxin oxidoreductase (Gbem_2896-Gbem_2899). The second conversion may be catalyzed by isocitrate dehydrogenase (Gbem_2901), which is 71% identical to the Chlorobium limicola enzyme that is known to be reversible [25]. These genes form a cluster that includes malate dehydrogenase (Gbem_2900), suggesting that flux of oxaloacetate through the reverse TCA cycle may be coordinated.
The presence of homologous gene clusters in Pelobacter propionicus and Desulfuromonas acetoxidans suggests that citrate lyase was present in the common ancestor of the Geobacteraceae, and was lost by most species of the genus Geobacter. Interestingly, the 2'-(5''-triphosphoribosyl)-3'-dephospho-CoA synthase gene of the cluster, which is duplicated in G. bemidjiensis (citG-1 Gbem_3856, citG-2 Gbem_0190), is present in G. sulfurreducens (GSU0806), along with the gene for the enzyme that transfers 2'-(5''-triphosphoribosyl)-3'-dephospho-CoA to the acyl carrier protein of citrate lyase (citX Gbem_3857 = GSU0807), but the genes encoding structural components of citrate lyase are absent, suggesting that 2'-(5''-triphosphoribosyl)-3'-dephospho-CoA may have a second function unrelated to citrate lyase.
Both succinyl:acetate CoA transferase isozymes of G. sulfurreducens are doubly present in G. bemidjiensis (ato-3 Gbem_0795 is a duplicate of ato-1 Gbem_0468, and ato-4 Gbem_3897 is a duplicate of ato-2 Gbem_2843). One possible explanation for this is that the duplicates have distinct functions in the oxidative and reductive TCA cycles (Figure 3b). Another possibility is that each of the duplicate citrate synthases (Gbem_1652, Gbem_3905) utilizes the acetyl-CoA produced by a different pair of isozymes. Although these details remain to be worked out, the TCA cycle on the whole appears to be more complex in the subsurface isolate G. bemidjiensis than in the non-subsurface Geobacter species examined to date.
Carbon monoxide dehydrogenases and associated hydrogenase
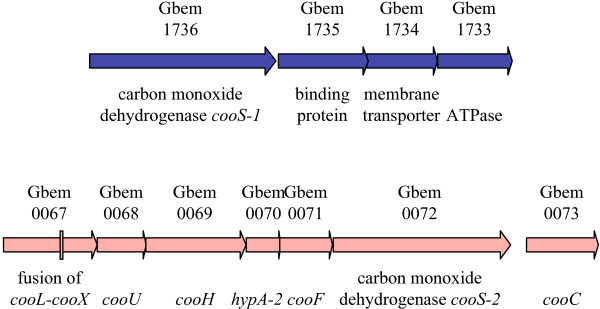
Like the G. metallireducens genome, that of G. bemidjiensis encodes a carbon monoxide dehydrogenase (cooS-1 Gbem_1736) alongside an ABC transporter complex (Gbem_1735-Gbem_1733) of unknown substrate specificity (Figure 4). The presence of this gene suggests that G. bemidjiensis and G. metallireducens may be capable of carbon monoxide cycling (fermentative production of carbon monoxide followed by re-oxidation) under some conditions, as has been reported for D. vulgaris [26]. In addition, G. bemidjiensis possesses a cluster of genes (cooLUH-hypA-cooFSC Gbem_0067-Gbem_0074) with closely related homologs in Carboxydothermus hydrogenoformans [27], encoding a carbon monoxide dehydrogenase and associated hydrogenase (Figure 4). Thus, G. bemidjiensis may couple oxidation of exogenous carbon monoxide to formation of hydrogen from intracellular protons to establish a proton gradient. The hydrogen and some of the carbon dioxide released by this reaction may be fixed subsequently by the reverse TCA cycle. Genome-based metabolic models of the non-subsurface Geobacter species lack this aspect of carbon metabolism of G. bemidjiensis.
Figure 4.
The carbon monoxide dehydrogenase gene clusters. One cluster has orthologs in G. metallireducens (blue arrows), and the other is unique to G. bemidjiensis (pink arrows) and contains an intragenic multicopy nucleotide sequence (pink rectangle). The carbon monoxide dehydrogenase-associated hydrogenase subunits are encoded by cooLXUH, the nickel insertion protein by hypA-2, an accessory protein by cooC, and an iron-sulfur cluster-binding oxidoreductase that transfers electrons from carbon monoxide dehydrogenase to the hydrogenase is encoded by cooF.
Glucose as electron donor
Several unique genes discovered in the G. bemidjiensis genome suggested that G. bemidjiensis should be able to utilize glucose and galactose as carbon sources. These genes encode a glucose/galactose transporter (gluP Gbem_3671) 55% identical to that of Brucella abortus [28], a putative glucose 6-kinase (Gbem_2002) 33% identical to that of E. coli [29], a galactose 1-kinase (Gbem_4019) 35% identical to that of E. coli [30], and a uridine 5'-diphosphate (UDP)-glucose:galactose-1-phosphate uridylyltransferase (Gbem_4017) 32% identical to that of Thermotoga maritima [31]. Most Geobacteraceae, which do not utilize glucose and galactose, possess only a putative glucose 6-kinase (Gbem_1326) similar to those of Streptomyces lividans and Streptomyces coelicolor [32,33], a UDP-glucose/galactose 4-epimerase (Gbem_3215) and a different putative galactose-1-phosphate uridylyltransferase. G. bemidjiensis was able to grow with glucose, but not galactose, as electron donor and carbon source, using Fe(III) oxides as the terminal electron acceptor (D. Holmes, unpublished). This discovery illustrates the need for subsurface metabolic models to be based on subsurface genomes such as that of G. bemidjiensis, rather than approximations based on genomes of non-subsurface species.
Carbohydrate osmoprotectants and cell wall components
Like G. sulfurreducens and G. metallireducens, G. bemidjiensis is predicted to make trehalose from glucose storage polymers by the sequential action of maltooligosyltrehalose synthase (Gbem_0134) and maltooligosyltrehalose trehalohydrolase (Gbem_0132) [10]. G. bemidjiensis lacks homologs of the enzymes predicted to make trehalose from glucose-6-phosphate in G. sulfurreducens [10], but may be able to isomerize maltose to trehalose by means of a maltose-active trehalose synthase (Gbem_0136) that is 33% identical to that of T. thermophilus [34]. The presence of a fructose/mannose 6-kinase (mak Gbem_0370), 39% identical to that of E. coli [35] and a mannitol dehydrogenase (Gbem_0401) with 47% identity to that of Apium graveolens [36] suggests that G. bemidjiensis may synthesize and break down D-mannitol as an additional osmoprotectant.
The lipopolysaccharide of G. sulfurreducens contains no O-antigen [37]. In contrast, the genome of G. bemidjiensis reveals many pathways for the production of various sugars that may be components of the cell wall (Additional file 3: Table S3). Thus, not only central metabolism of carbon but many specialized branch pathways appear to differ between subsurface and non-subsurface Geobacter species.
Biosynthesis of chorismate and folate in G. bemidjiensis
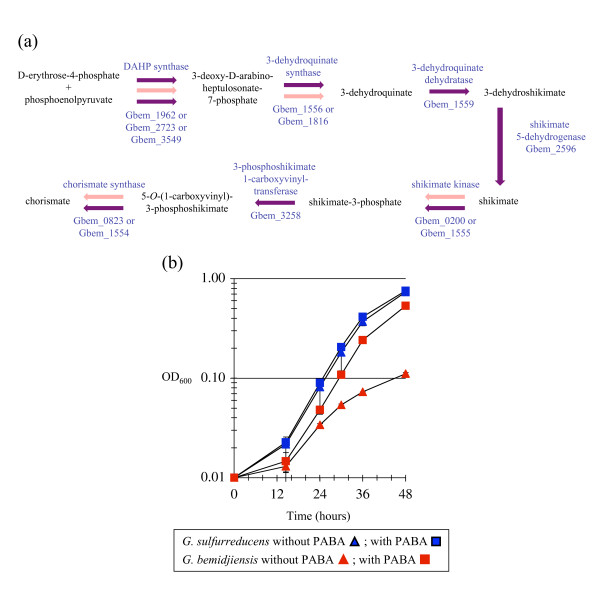
Chorismate is the common precursor of folate, menaquinone, and the aromatic amino acids phenylalanine, tyrosine and tryptophan. Geobacter species have multiple isozymes for the first reaction of the chorismate biosynthesis pathway [10], which may respond to negative feedback from different end products as in E. coli [38]. The chorismate biosynthesis pathway in G. bemidjiensis and Geobacter sp. M21 is notable for the presence of isozymes to catalyze several subsequent reactions as well (Figure 5a), differing in this aspect from G. sulfurreducens and G. metallireducens. Scattered genes encode a 3-dehydroquinate synthase with homologs in G. daltonii and cyanobacteria, a shikimate kinase with homologs in Marinobacter species, methanogens and Clostridia, and a chorismate synthase with homologs in sulfate-reducers and fungi. There is also a cluster of three genes encoding the same functions, as in G. sulfurreducens and G. metallireducens. The presence of these isozymes at multiple steps suggests that regulation of chorismate biosynthesis may be more complex in subsurface Geobacter species.
Figure 5.
Chorismate biosynthesis pathway and 4-aminobenzoate auxotrophy of G. bemidjiensis. (a) Multiplicity of enzymes of chorismate biosynthesis in G. bemidjiensis. Every step may be catalyzed by enzymes shared with G. sulfurreducens and G. metallireducens (purple arrows), but isozymes for some steps are unique to G. bemidjiensis (pink arrows). (b) Growth of G. bemidjiensis on NBAF medium on the first transfer without 4-aminobenzoate (PABA) supplementation (red triangles) compared to growth with PABA (red squares); the controls were G. sulfurreducens without PABA (blue triangles) and with PABA (blue squares).
The G. bemidjiensis genome encodes no homolog of the putative 4-aminodeoxychorismate synthase/lyase of G. metallireducens and G. sulfurreducens (Gmet_3010 = GSU0523), and an attempt to grow G. bemidjiensis without vitamin supplementation confirmed that it is auxotrophic solely for the 4-aminobenzoate (PABA) moiety of folate (Figure 5b). This is the first report of a Geobacter species with any vitamin requirement, and suggests that the metabolic activity of subsurface Geobacter species may be stimulated by adding PABA.
Degradation of benzoate and other aromatic pollutants
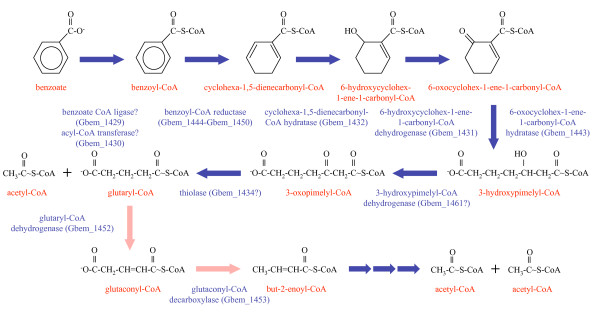
The ability to degrade benzoate is found in both subsurface and non-subsurface Geobacter species. G. bemidjiensis possesses orthologs of the G. metallireducens genes implicated in degradation of benzoate to 3-hydroxypimelyl-CoA (Figure 6) [39-42]. The benzoate--CoA ligase (bamY Gbem_1429) gene sequence contains an internal stop codon (TAG) at position 448, where those of other Geobacter species specify glutamate (GAG). However, the gene next to bamY (Gbem_1430) encodes an acyl-CoA hydrolase/transferase of unknown specificity. The possibility that benzoate can be activated to benzoyl-CoA by this enzyme, perhaps by transferring CoA from succinyl-CoA or acetyl-CoA, should be explored. Like G. metallireducens, G. bemidjiensis possesses bamB-2 and bamC-2 genes (Gbem_2620-Gbem_2619) paralogous to bamB and bamC of the putative benzoyl-CoA reductase, and a ferredoxin gene (Gbem_2621) closely related to the N-terminal domain of bamB.
Figure 6.
Benzoate degradation pathway in G. bemidjiensis. Early enzymes have orthologs in G. metallireducens (blue arrows), but glutaryl-CoA dehydrogenase and glutaconyl-CoA decarboxylase are unique to G. bemidjiensis (pink arrows).
Degradation of 3-hydroxypimelyl-CoA in G. bemidjiensis (Figure 6) is predicted to involve a non-decarboxylating glutaryl-CoA dehydrogenase (Gbem_1452) 44% identical to that of Desulfococcus multivorans, rather than a homolog of the decarboxylating glutaryl-CoA dehydrogenase of G. metallireducens [43]. Subsequent decarboxylation of glutaconyl-CoA may take place through the product of the adjacent gene (gcdA Gbem_1453), which is 52% identical to the catalytic subunit of sodium-translocating glutaconyl-CoA decarboxylase of Acidaminococcus fermentans [44]. No homologs of the other three subunits were found in the G. bemidjiensis genome, indicating that decarboxylation of glutaconyl-CoA is not coupled to a sodium pump. Although the GcdA protein of A. fermentans on its own is capable of decarboxylating glutaconyl-CoA with free biotin as a cofactor [45], it is notable that the oxaloacetate decarboxylase encoded by the gene adjacent to gcdA in G. bemidjiensis (oadA Gbem_1454) contains two biotin attachment domains, whereas its sodium pump-associated homologs contain only one. The possibility that the two decarboxylases cooperate as a complex deserves investigation.
Although the genome of G. bemidjiensis corroborates the observation that it cannot degrade as many aromatic compounds as G. metallireducens [39,42,46], it also suggests that G. bemidjiensis can detoxify some aromatic pollutants without degrading them. A homolog of the broad-specificity aldo-keto reductase YvgN of B. subtilis [47] is present (Gbem_3980, 45% sequence identity), suggesting that G. bemidjiensis may convert chloro- and nitro- derivatives of benzaldehyde to the corresponding benzol derivatives. Although YvgN was previously described as a methylglyoxal reductase [48], detoxification of methylglyoxal, a byproduct of carbohydrate and lipid metabolism, may be of minor importance in G. bemidjiensis, as a methylglyoxal synthase was not found in G. bemidjiensis, but only in G. lovleyi (Glov_0611).
The fact that G. bemidjiensis is auxotrophic for PABA (Figure 5b) indicates that it has grown accustomed to an environment in which PABA is readily available, possibly in the form of 4-azobenzoate. G. bemidjiensis may convert azoaromatic compounds to arylamines by means of an azoreductase (azoR Gbem_2529) 30% identical to that of E. coli [49], which is not present in G. metallireducens. Furthermore, an arylamine N-acetyltransferase (Gbem_0306) not found in other Geobacteraceae may act in detoxification of aromatic compounds by G. bemidjiensis, and a putative amidohydrolase in the benzoate degradation gene cluster (Gbem_1458 = Gmet_2056) may also be involved in metabolism of aromatic compounds. Further studies of subsurface Geobacter species such as G. bemidjiensis are necessary to characterize their abilities to transform aromatic compounds.
Alkylmercury lyase
The presence of an alkylmercury lyase (Gbem_0319) 36% identical to that of S. lividans [50]) suggests that G. bemidjiensis possesses broad-spectrum resistance to mercury in various organic forms. A homolog of this enzyme was not found in any other Geobacter species, but genes that may encode mercuric reductases (e.g. Gbem_0457, Gbem_0640) exhibit vertical inheritance in the family, indicating that Geobacteraceae may generally have the ability to detoxify inorganic Hg(II) ions, whereas subsurface Geobacter species such as G. bemidjiensis may have acquired additional resistance to organomercuric compounds.
Expansion of transport systems for phosphate and molybdate
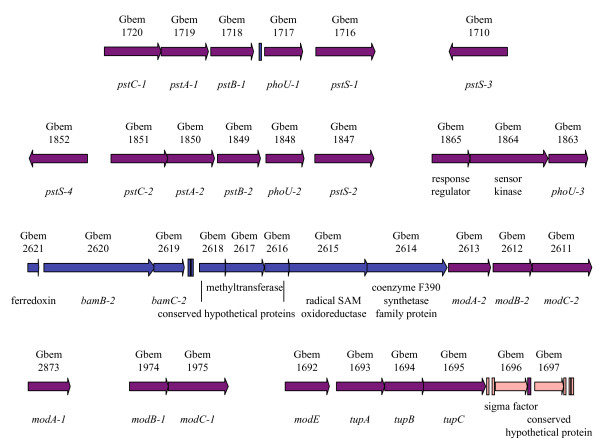
The ATP-binding cassette (ABC) transport system for phosphate consists of a periplasmic phosphate-binding protein (PstS), two membrane proteins (PstC and PstA), an ATP-binding protein (PstB) and a regulatory protein (PhoU). Whereas these are encoded by single-copy genes in G. sulfurreducens and G. metallireducens, the G. bemidjiensis genome contains duplicate transporters, triplicate regulators, and quadruplicate phosphate-binding proteins (Figure 7). This expansion may confer an advantage in the subsurface environment, where Geobacter species experience phosphate limitation [51].
Figure 7.
Multiplicity of phosphate and molybdate transport system genes of G. bemidjiensis. Some genes are unique to G. bemidjiensis (pink arrows), some have orthologs in G. metallireducens (blue arrows), and some are shared with both G. sulfurreducens and G. metallireducens (purple arrows). Multicopy nucleotide sequences (rectangles) are found in some intergenic regions. The periplasmic phosphate-binding proteins are encoded by pstS genes, the transporter membrane proteins by pstA and pstC genes, ATP-binding proteins by pstB genes, and regulators by phoU genes. The periplasmic molybdate-binding proteins are encoded by modA genes, the transporter membrane proteins by modB genes, the ATP-binding proteins by modC genes, and a regulator by modE. The periplasmic tungstate-binding protein is encoded by tupA, the transporter membrane protein by tupB, and the ATP-binding protein by tupC. The bamB-2 gene encodes a paralog of benzoyl-CoA reductase, and the bamC-2 gene encodes a paralog of a putative electron transfer protein of benzoyl-CoA reductase.
The ABC transport system for molybdate, consisting of a molybdate-binding protein (ModA), membrane protein (ModB) and ATP-binding protein (ModC), has also expanded in G. bemidjiensis (Figure 7): the modB1C1 genes are located apart from the modA1 gene, while the modA2B2C2 genes remain an intact operon. The regulatory gene modE is located on the 5' side of the tungstate transporter genes tupABC (which are phylogenetically distinct from those of G. sulfurreducens and G. metallireducens; data not shown) in contrast with its location on the 5' side of modABC in G. sulfurreducens. The possibility that these expansions and rearrangements are a response by subsurface Geobacter species to molybdate limitation deserves to be investigated.
Oxygen respiration, oxygen detoxification, and possible anaerotaxis in G. bemidjiensis
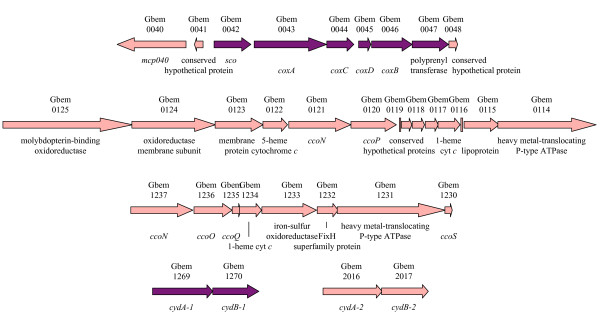
Genome sequencing led to the surprising discovery that G. sulfurreducens and G. metallireducens are capable of oxygen respiration using a cytochrome caa3 oxidase complex, which is also found in G. bemidjiensis (Gbem_0042-Gbem_0047). Near this operon in G. bemidjiensis only (Figure 8) is a gene encoding a methyl-accepting chemotaxis protein (Gbem_0040) with a hemerythrin-like domain, possibly to sense oxygen, by analogy with the much larger DcrH protein of D. vulgaris [52]. In addition, G. bemidjiensis has two other operons encoding components of cytochrome cbb3 oxidases: (Gbem_1237-Gbem_1230; Gbem_0121-Gbem_0120). The G. bemidjiensis genome contains two pairs of genes encoding cytochrome bd quinol oxidases: one (Gbem_1269-Gbem_1270) is closely related to its counterparts in G. sulfurreducens and G. metallireducens, while the other (Gbem_2016-Gbem_2017) is not. Thus, G. bemidjiensis may have enhanced capabilities for oxygen respiration compared to G. sulfurreducens and G. metallireducens, due to oxygen stress in its subsurface niche, and may have the ability to respond to oxygen anaerotactically through chemotaxis-type signalling.
Figure 8.
Oxygen respiration genes of G. bemidjiensis. The cytochrome caa3 oxidase complex (coxACDB genes) and one cytochrome bd quinol oxidase complex (cydAB-1 genes) are encoded by genes with orthologs in G. sulfurreducens and G. metallireducens (purple arrows), while the hemerythrin domain-containing methyl-accepting chemotaxis sensory transducer (mcp040), the other pair of cytochrome bd quinol oxidase complex genes (cydAB-2), and the two operons encoding cytochrome bb3 oxidase components (cco genes) are unique to G. bemidjiensis (pink arrows).
Eight hemerythrin family proteins (Gbem_1252, Gbem_2241, Gbem_2255, Gbem_2262, Gbem_2701, Gbem_2773, Gbem_3870, Gbem_4009) were predicted from the genome of G. bemidjiensis, suggesting that it may have expanded its ability to sequester molecular oxygen and deliver it to respiratory or detoxifying enzymes, in contrast to G. metallireducens with two hemerythrin homologs; G. sulfurreducens has six hemerythrin homologs. To detoxify reactive oxygen species, G. bemidjiensis possesses a desulfoferrodoxin (Gbem_3292) 60% identical to that of Desulfoarculus baarsii [53] and a rubredoxin:oxygen/nitric oxide oxidoreductase (Gbem_0186) 31% identical to that of D. gigas [54], in addition to the superoxide dismutase (Gbem_2204), peroxiredoxins (Gbem_0154, Gbem_0221, Gbem_1338, Gbem_2956, Gbem_4010) and two rubrerythrins (Gbem_2313, Gbem_3325) also present in G. sulfurreducens and G. metallireducens. Phylogenetic analysis (not shown) indicates that although the characterized cytochrome c peroxidase of G. sulfurreducens [55] has an excellent homolog in G. bemidjiensis (Gbem_0020), this is actually an ortholog of MacA, implicated in Fe(III) reduction [56]. As in G. metallireducens, there is no catalase in G. bemidjiensis, meaning that no oxygen is produced from detoxification of hydrogen peroxide; detoxification by rubrerythrins produces only water. All Geobacteraceae encode at least one iron-sulfur-oxygen hybrid cluster protein, thought to detoxify an unidentified reactive compound in response to nitric oxide stress [57], as well as hydrogen peroxide stress [58]; G. bemidjiensis and Geobacter sp. M21 alone have three hybrid cluster protein genes (Gbem_1033, Gbem_1168, Gbem_1239), evidently derived by expansion of a single ancestral gene. G. bemidjiensis also has a quinol-oxidizing nitric oxide reductase (norZ Gbem_3901) 40% identical to that of Cupriavidus necator [59], with a distant homolog in G. metallireducens. Overall, the genome annotation indicates that G. bemidjiensis has evolved to cope with many kinds of reactive oxygen species, a finding that should improve models of Geobacter metabolism in the subsurface.
The outer surface: c-type cytochromes, pili, and flagella
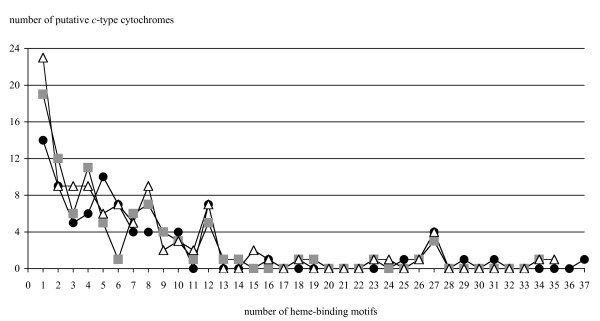
An understanding of subsurface Geobacter metabolism and physiology must include the c-type cytochromes that mediate electron transfer to extracellular electron acceptors such as insoluble Fe(III) oxides. The number of genes encoding c-type cytochromes in the G. bemidjiensis genome is 84, fewer than G. metallireducens (90 genes) and G. sulfurreducens (104 genes), despite the G. bemidjiensis genome being the largest of the three. The distribution of genes with a given number of heme-binding motifs (Figure 9) shows that all three species encode multiple c-type cytochromes with 12 or 27 predicted hemes (due to fusion of four or nine modules of the cytochrome c7 family, respectively), and the most complex c-type cytochrome in each genome contains 34-37 hemes. However, G. sulfurreducens and G. metallireducens encode more monoheme c-type cytochromes than G. bemidjiensis, and while the mode number of hemes in multiheme c-type cytochromes of G. bemidjiensis is 5 hemes, the other two genomes have two modes with 4 and 8 hemes. Of the c-type cytochrome genes implicated in Fe(III) and U(VI) reduction in G. sulfurreducens, G. bemidjiensis possesses macA (Gbem_0020) [56,60,61], three members of the c7 family (ppcB Gbem_4049, ppcD Gbem_4043, and ppcG Gbem_3455) [62], five members of the omcS family (Gbem_1116, Gbem_1117, Gbem_1131, Gbem_2679, Gbem_2680) [63], one member of the omcB family (Gbem_3379) [64], and two omcF-related genes (Gbem_1585, Gbem_2183) [65]. There is no homolog of omcE [63]. Although G. bemidjiensis was reported not to grow with a graphite electrode as electron acceptor [3], it possesses a homolog of omcZ (Gbem_3056), which is required for G. sulfurreducens to grow by transferring electrons to an electrode [66]. Overall, 41 c-type cytochromes of G. bemidjiensis (49%) have full-length homologs in G. sulfurreducens and/or G. metallireducens (Additional file 4: Table S4). Larger c-type cytochromes (13 or more heme-binding motifs) of G. bemidjiensis are absent from G. sulfurreducens and G. metallireducens with one exception (Gbem_1124 = GSU2495; 26 heme-binding motifs). Of the c-type cytochromes of G. bemidjiensis that have one, two or five heme-binding motifs, approximately two-thirds in each category have no match in the other two genomes, whereas of those with three, four or six to twelve heme-binding motifs, at least half in each category are shared. Thus, extensive similarities as well as notable differences in the complement of c-type cytochromes exist between the subsurface species G. bemidjiensis and its non-subsurface relatives.
Figure 9.
Distribution of c-type cytochromes with different numbers of heme-binding motifs in three Geobacter genomes. The pattern for G. bemidjiensis (black circles) shows some differences from G. sulfurreducens (white triangles) and G. metallireducens (grey squares).
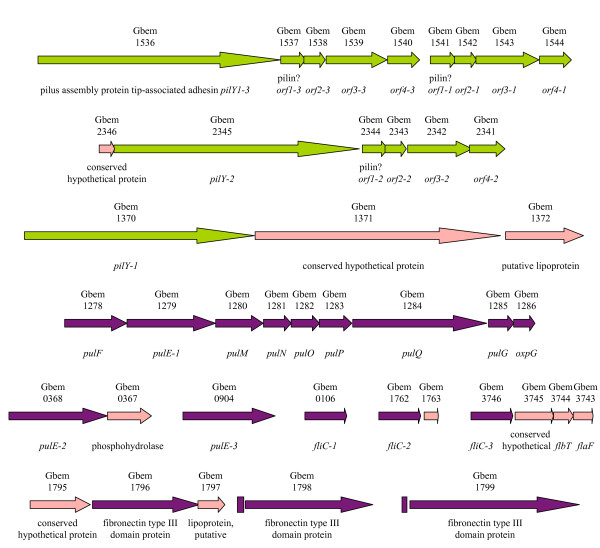
Several genes of pilus biogenesis are present in triplicate in the G. bemidjiensis genome (Figure 10), all apparently expanded from ancestral single-copy genes found in G. sulfurreducens and other Geobacteraceae (but not G. metallireducens). The ancestral flagellin gene has also expanded into triplicates (Gbem_0106, Gbem_1762, Gbem_3746), as has an ancestral GEMM riboswitch-regulated gene encoding a fibronectin type III domain protein (Gbem_1796, Gbem_1798, Gbem_1799) that may be localized to the outer surface of the cell. Interestingly, phylogenetic analysis (not shown) indicates that G. bemidjiensis has also recently triplicated (as Gbem_1116, Gbem_2659 and Gbem_2680) an ancestral gene related to omcS of G. sulfurreducens, which encodes an outer surface c-type cytochrome that is important for reduction of insoluble Fe(III) [63]. Thus, many proteins of the outer surface, with possible roles in electron transfer to insoluble extracellular electron acceptors, are triply present in G. bemidjiensis, although the significance of this expansion to life in subsurface environments is unknown.
Figure 10.
Triplicated genes for biogenesis of pili and flagella in G. bemidjiensis. The pilus assembly protein tip-associated adhesin genes (pilY1) and associated genes (orf1, orf2, orf3, orf4) have single orthologs in G. sulfurreducens (green arrows); those of G. metallireducens are of different origin. The flagellin genes (fliC) and pilus biogenesis ATPase genes (pulE) have single orthologs in both G. sulfurreducens and G. metallireducens (purple arrows), while genes predicted to be in the same operons are often unique to G. bemidjiensis (pink arrows). GEMM riboswitches (purple rectangles) are found near the recently triplicated fibronectin type III domain protein genes.
Sigma factors and signalling proteins
The G. bemidjiensis genome was examined for features of gene regulation conserved between it and its non-subsurface relatives. Of the six sigma factors of RNA polymerase in G. sulfurreducens, G. bemidjiensis has orthologs of five: RpoD (Gbem_3694), RpoS (Gbem_2683), RpoN (Gbem_0869), RpoH (Gbem_0573), and FliA (Gbem_3764). No homolog of the putative stress response sigma factor RpoE was found. There are also two additional sigma factors (Gbem_1696 and Gbem_3169) unrelated to the unique sigma factor of G. metallireducens.
The G. bemidjiensis genome encodes 127 putative sensor histidine kinases containing HATPase_c domains (Additional file 5: Table S5), including 8 chemotaxis-type kinases (cheA genes), of which 47 genes (37%) have full-length homologs in G. sulfurreducens and/or G. metallireducens. There are 163 proteins with response receiver (REC) domains (Additional file 5: Table S5), including 19 that may belong to chemotaxis-type signalling pathways; of these, 82 genes (50%) have full-length homologs in G. sulfurreducens and/or G. metallireducens. Thus, G. bemidjiensis has a different and much larger repertoire of phosphorylation-dependent signalling proteins than either G. sulfurreducens or G. metallireducens. The G. bemidjiensis genome encodes 21 GGDEF domain proteins that may synthesize the intracellular messenger cyclic diguanylate (Additional file 5: Table S5), a similar number to G. sulfurreducens and G. metallireducens, but only 10 of these are conserved. These differences in the repertoire of predicted signalling proteins among subsurface and non-subsurface Geobacter species are remarkable, especially considering that some ancestral genes encoding signalling proteins appear to have undergone duplication or triplication in G. bemidjiensis (Additional file 5: Table S5).
Non-protein-coding features of the G. bemidjiensis genome
Riboswitches that have been identified in the non-subsurface species G. sulfurreducens and G. metallireducens [10,67] were found in the subsurface G. bemidjiensis genome also. In addition, several families of multicopy nucleotide sequences were noted in G. bemidjiensis (Additional file 6: Table S6; Additional files 7, 8, 9, 10, 11, 12, 13, 14 and 15: Figures S1-S9), most of which have no counterparts in the G. sulfurreducens or G. metallireducens genomes. Some of these families are based on palindromic sequences, and others consist of direct repeats of 6 to 42 nucleotides that occupy intergenic regions throughout the genome. Multicopy sequences (other than rRNA and tRNA genes) are found in 12% of regions between protein-coding genes in G. bemidjiensis, and multiple sequences are present in 31% of such intergenic regions, indicating that insertion is not random. One nucleotide sequence family was found inserted into protein-coding genes (on both strands and in all reading frames, without causing frameshifts) as well as between genes, as previously observed for a different family in G. metallireducens [10]. The implications of so many more multicopy sequences being present in the genome of a subsurface Geobacter species than in those of its non-subsurface relatives remain to be elucidated.
Conclusions
The complete genome sequence of G. bemidjiensis reveals many differences from the previously published genomes of non-subsurface Geobacter species. Enzymes that account for the metabolic versatility of G. bemidjiensis were identified, and further metabolic, physiological and genomic peculiarities were discovered, including a more efficient pathway for oxidation of propionate, a pathway of fumarate disproportionation without excretion of acetate, a reductive TCA cycle, utilization of glucose, a defective folate biosynthesis pathway, and enhanced abilities to respond to oxygen stress. This information is of utmost value for an understanding of the activity of Geobacteraceae in subsurface environments undergoing bioremediation accompanied by reduction of Fe(III).
Methods
Sequence analysis and annotation
The genome of G. bemidjiensis Bem(T) [3] was sequenced at the Joint Genome Institute (JGI) using a combination of 3 kb, 6 kb and 35 kb DNA libraries. Inserts were sequenced from both ends using the standard Sanger method. All three libraries provided 11-fold coverage of the genome. The Phred/Phrap/Consed software package http://www.phrap.com was used for sequence assembly and quality assessment [68-70]. After the shotgun stage, 65888 reads were assembled with parallel Phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher (Han, 2006) or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, by custom primer walks, or by PCR amplification (Roche Applied Science, Indianapolis, IN). A total of 2059 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The completed genome sequence of G. bemidjiensis Bem(T) contains 67990 reads, achieving an average of 11-fold sequence coverage per base with an error rate less than 1 in 100,000.
The protein-coding genes were predicted using Prodigal V1.0 [71]. A BLASTP search of the translations vs. Genbank's non-redundant database (NR of Nov. 2007) at 1e-05 evalue was conducted. Matches to the Geobacter genus were excluded and the alignment of the N-terminus of each gene model vs. the best NR match was used to pick a preferred gene model. The gene/protein set was searched using BLASTP, hmmer, RPS-BLAST and Interpro. BLASTP searches were done vs. Swiss-Prot/TrEMBL, NR, and KEGG databases with a cutoff evalue of 1e-05. Hmmer searches were done vs. Pfam and TIGRfam databases using the trusted cutoff. RPS-BLAST searches against PRIAM used its 1e-30 high-confidence cutoff and searches against COGS used a 1e-10 cutoff. Interpro was run using its default cutoffs. Automated product assignment was made using the following hierarchy of data sources: PRIAM, TIGRFam, Pfam, Interpro profiles, Swiss-Prot/TrEMBL, KEGG, and COG group. tRNAs were annotated using tRNAscan-SE (v1.23). rRNAs were annotated using RNAmmer v 1.1 [72]. The srpRNA was located using the SRPscan website. The rnpB and ssrA genes were located using the Rfam database and Infernal.
Manual curation
The automated genome annotation of G. bemidjiensis and the manually curated genome annotations of G. sulfurreducens and G. metallireducens [10] were queried reciprocally with the protein BLAST algorithm [73] as implemented by OrthoMCL [74] using the default inflation parameter value (1.5), to identify mutual best hits as potential orthologs. The functional annotations of G. bemidjiensis genes were emended for consistency with their counterparts in G. sulfurreducens and G. metallireducens. The coordinates of numerous genes were adjusted according to the criteria of full-length alignment, plausible ribosome-binding sites, and minimal overlap between genes on opposite DNA strands. The annotations of G. bemidjiensis genes that were not matched to genes in G. sulfurreducens or G. metallireducens were checked by BLAST searches of NR and the Swiss-Prot database. Functional annotations in all three genomes were updated to match the experimental characterization of highly similar full-length homologs, with extensive reference to the EcoSal online textbook http://www.ecosal.org and the MetaCyc database [75]. Genes that had no protein-level homologs in NR were checked (together with flanking intergenic sequences) by translated nucleotide BLAST in all six reading frames, and by nucleotide BLAST to ensure that conserved protein-coding or non-protein-coding features had not been missed. All intergenic regions of 30 bp or larger were also checked, which led to the annotation of numerous conserved nucleotide sequences.
Phylogenetic analysis
Phylogenetic analysis of selected proteins was performed. In each case, the protein sequence of interest was included, along with its relatives, as identified by BLAST [76], and the set of sequences was aligned by TCoffee [77]. ProtTest [78] was used to select a model of molecular evolution and MrBayes [79] was used to create a Bayesian estimation of the phylogeny.
Growth experiments
To monitor the disproportionation of fumarate, G. bemidjiensis was cultured under strictly anaerobic conditions at 30°C in an atmosphere of N2 and CO2 (80%:20%), as previously described for G. sulfurreducens [80], in rubber-stoppered 156 ml bottles containing NBAF medium [81] from which sodium acetate was omitted. Samples of 1 ml were removed aseptically using anoxic syringes to monitor growth, then diluted 50-fold, passed through a 0.22 μm filter to remove cells, and stored at 4°C until high-pressure liquid chromatography analysis was performed as described previously [19]. The 4-aminobenzoate requirement of G. bemidjiensis was tested in rubber-stoppered 26 ml glass tubes containing 10 ml of NBAF medium [81] from which the vitamin solution and resazurin were omitted, with 4-aminobenzoate (Sigma Aldrich) added to individual tubes to a final concentration of 100 μg/L.
List of Abbreviations used
ABC: ATP-binding cassette; ATP: adenosine 5'-triphosphate; CoA: coenzyme A; DNA: deoxyribonucleic acid; FAD: flavin adenine dinucleotide; GTP: guanosine 5'-triphosphate; NAD(H): nicotinamide adenine dinucleotide (reduced); NADP(H): nicotinamide adenine dinucleotide 2'-phosphate (reduced); PABA: 4-aminobenzoate; rRNA: ribosomal RNA; RNA: ribonucleic acid; TCA: tricarboxylic acid; tRNA: transfer RNA; UDP: uridine 5'-diphosphate.
Authors' contributions
CH supervised the genome sequencing, HK performed genome sequence finishing, and ML oversaw the automated annotation process. MA performed manual curation of the genome annotation (assisted by MC and NY) and wrote the manuscript. NY did the phylogenetic analyses. DH, CR and MA conducted physiological experiments. DL conceived of the study and offered guidance with the writing. All authors read, assisted with editing, and approved the final manuscript.
Supplementary Material
Table S1. Genes of G. bemidjiensis in which frameshifts occur in homopolymeric regions.
Table S2. Enzymes of acyl-CoA metabolism in G. bemidjiensis.
Table S3. Selected sugar interconversion genes of G. bemidjiensis.
Table S4. Predicted c-type cytochromes of G. bemidjiensis.
Table S5. Signalling proteins of G. bemidjiensis.
Table S6. Multicopy nucleotide sequences in G. bemidjiensis.
Figure S1. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S2. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S3. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S4. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S5. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S6. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S7. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S8. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S9. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Contributor Information
Muktak Aklujkar, Email: muktak@microbio.umass.edu.
Nelson D Young, Email: nyoung@microbio.umass.edu.
Dawn Holmes, Email: dholmes@microbio.umass.edu.
Milind Chavan, Email: chavanm@gmail.com.
Carla Risso, Email: crisso@microbio.umass.edu.
Hajnalka E Kiss, Email: hajkis@lanl.gov.
Cliff S Han, Email: han_cliff@lanl.gov.
Miriam L Land, Email: landml@ornl.gov.
Derek R Lovley, Email: dlovley@microbio.umass.edu.
Acknowledgements
We thank Mounir Izallalen for helpful discussions and P. Brown, T. Woodard, K. Nevin, T. Brettin, C. Detter, and C. Kuske for technical assistance. This research was supported by the Office of Science (Biological and Environmental Research), U.S. Department of Energy (Grant No. DE-FC02-02ER63446). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References
- Lovley DR. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev. 1991;55(2):259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- Nevin KP, Holmes DE, Woodard TL, Hinlein ES, Ostendorf DW, Lovley DR. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int J Syst Evol Microbiol. 2005;55(Pt 4):1667–1674. doi: 10.1099/ijs.0.63417-0. [DOI] [PubMed] [Google Scholar]
- Holmes DE, O'Neil RA, Vrionis HA, N'Guessan LA, Ortiz-Bernad I, Larrahondo MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP. et al. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 2007;1(8):663–677. doi: 10.1038/ismej.2007.85. [DOI] [PubMed] [Google Scholar]
- Wilkins MJ, Verberkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H, N'Guessan AL, Thomas BC, Nicora CD, Shaht MB. et al. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol. 2009;75(20):6591–6599. doi: 10.1128/AEM.01064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Bond DR, Butler JE, Esteve-Nunez A, Coppi MV, Palsson BO, Schilling CH, Lovley DR. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol. 2006;72(2):1558–1568. doi: 10.1128/AEM.72.2.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Sayyar B, Butler JE, Pharkya P, Fahland TR, Famili I, Schilling CH, Lovley DR, Mahadevan R. Genome-scale constraint-based modeling of Geobacter metallireducens. BMC Syst Biol. 2009;3:15. doi: 10.1186/1752-0509-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe TD, Mahadevan R, Fang Y, Garg S, Long PE, Lovley DR. Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microbial Biotechnology. 2009;2(2):274–286. doi: 10.1111/j.1751-7915.2009.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Fang Y, Scheibe TD, Lovley DR, Mahadevan R. Modeling and sensitivity analysis of electron capacitance for Geobacter in sedimentary environments. J Contam Hydrol. 2010;112(1-4):30–44. doi: 10.1016/j.jconhyd.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Aklujkar M, Krushkal J, DiBartolo G, Lapidus A, Land ML, Lovley DR. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 2009;9:109. doi: 10.1186/1471-2180-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ. et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302(5652):1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Segura D, Mahadevan R, Juarez K, Lovley DR. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput Biol. 2008;4(2):e36. doi: 10.1371/journal.pcbi.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabau C, Cronan JE Jr. Nucleotide sequence and deduced amino acid sequence of Escherichia coli pyruvate oxidase, a lipid-activated flavoprotein. Nucleic Acids Res. 1986;14(13):5449–5460. doi: 10.1093/nar/14.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton CG, Kumar GK, Shenoy BC, Haase FC, Phillips NFB, Park VM, Magner WJ, Hejlik DP, Wood HG, Samols D. Primary structure of the 5 S subunit of transcarboxylase as deduced from the genomic DNA sequence. FEBS Lett. 1993;330:191–196. doi: 10.1016/0014-5793(93)80271-U. [DOI] [PubMed] [Google Scholar]
- Bond DR, Mester T, Nesbo CL, Izquierdo-Lopez AV, Collart FL, Lovley DR. Characterization of citrate synthase from Geobacter sulfurreducens and evidence for a family of citrate synthases similar to those of eukaryotes throughout the Geobacteraceae. Appl Environ Microbiol. 2005;71(7):3858–3865. doi: 10.1128/AEM.71.7.3858-3865.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka H, Hirooka K, Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem. 2007;282(8):5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- Straub KL, Hanzlik M, Buchholz-Cleven BE. The use of biologically produced ferrihydrite for the isolation of novel iron-reducing bacteria. Syst Appl Microbiol. 1998;21(3):442–449. doi: 10.1016/S0723-2020(98)80054-4. [DOI] [PubMed] [Google Scholar]
- Zaunmuller T, Kelly DJ, Glockner FO, Unden G. Succinate dehydrogenase functioning by a reverse redox loop mechanism and fumarate reductase in sulphate-reducing bacteria. Microbiology. 2006;152(Pt 8):2443–2453. doi: 10.1099/mic.0.28849-0. [DOI] [PubMed] [Google Scholar]
- Risso C, Sun J, Zhuang K, Mahadevan R, Deboy R, Ismail W, Shrivastava S, Hout H, Kothari S, Daugherty S. et al. Genome-scale comparison and constraint-based metabolic reconstruction of the facultative anaerobic Fe(III)-reducer Rhodoferax ferrireducens. BMC Genomics. 2009;10(1):447. doi: 10.1186/1471-2164-10-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Ditullio KP, Neuhard J, Kelln RA. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli requires the dicarboxylate transport protein encoded by dctA. J Bacteriol. 1996;178(24):7099–7105. doi: 10.1128/jb.178.24.7099-7105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse Gram-negative bacteria. J Bacteriol. 1997;179(17):5482–5493. doi: 10.1128/jb.179.17.5482-5493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Oesterhelt D, Reinke H, Beyreuther K, Dimroth P. The sodium ion translocating oxalacetate decarboxylase of Klebsiella pneumoniae. Sequence of the biotin-containing alpha-subunit and relationship to other biotin-containing enzymes. J Biol Chem. 1988;263(20):9640–9645. [PubMed] [Google Scholar]
- Holmes DE, Nevin KP, O'Neil RA, Ward JE, Adams LA, Woodard TL, Vrionis HA, Lovley DR. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71(11):6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Chakraborty R, Martin HG, Chu J, Hazen TC, Keasling JD. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe(III)-nitrilotriacetic acid. Appl Environ Microbiol. 2007;73(12):3859–3864. doi: 10.1128/AEM.02986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao T, Kawamura M, Fukui T, Atomi H, Imanaka T. Characterization of isocitrate dehydrogenase from the green sulfur bacterium Chlorobium limicola. A carbon dioxide-fixing enzyme in the reductive tricarboxylic acid cycle. Eur J Biochem. 2002;269(7):1926–1931. doi: 10.1046/j.1432-1033.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- Voordouw G. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J Bacteriol. 2002;184(21):5903–5911. doi: 10.1128/JB.184.21.5903-5911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboh B, Linder D, Hedderich R. Purification and catalytic properties of a CO-oxidizing:H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur J Biochem. 2002;269(22):5712–5721. doi: 10.1046/j.1432-1033.2002.03282.x. [DOI] [PubMed] [Google Scholar]
- Essenberg RC, Candler C, Nida SK. Brucella abortus strain 2308 putative glucose and galactose transporter gene: cloning and characterization. Microbiology. 1997;143(Pt 5):1549–1555. doi: 10.1099/00221287-143-5-1549. [DOI] [PubMed] [Google Scholar]
- Meyer D, Schneider-Fresenius C, Horlacher R, Peist R, Boos W. Molecular characterization of glucokinase from Escherichia coli K-12. J Bacteriol. 1997;179(4):1298–1306. doi: 10.1128/jb.179.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck C, Riccio A, Schumperli D, McKenney K, Jeffers J, Hughes C, Rosenberg M, Heusterspreute M, Brunel F, Davison J. Structure of the galactokinase gene of Escherichia coli, the last (?) gene of the gal operon. Nucleic Acids Res. 1985;13(6):1841–1853. doi: 10.1093/nar/13.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl W, Wagner B, Schellhase J. Properties of an alpha-galactosidase, and structure of its gene galA, within an alpha-and beta-galactoside utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst Appl Microbiol. 1998;21(1):1–11. doi: 10.1016/s0723-2020(98)80002-7. [DOI] [PubMed] [Google Scholar]
- Angell S, Schwarz E, Bibb MJ. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992;6(19):2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- Mahr K, van Wezel GP, Svensson C, Krengel U, Bibb MJ, Titgemeyer F. Glucose kinase of Streptomyces coelicolor A3(2): large-scale purification and biochemical analysis. Antonie Van Leeuwenhoek. 2000;78(3-4):253–261. doi: 10.1023/A:1010234916745. [DOI] [PubMed] [Google Scholar]
- Tsusaki K, Nishimoto T, Nakada T, Kubota M, Chaen H, Fukuda S, Sugimoto T, Kurimoto M. Cloning and sequencing of trehalose synthase gene from Thermus aquaticus ATCC33923. Biochim Biophys Acta. 1997;1334(1):28–32. doi: 10.1016/s0304-4165(96)00103-1. [DOI] [PubMed] [Google Scholar]
- Sproul AA, Lambourne LT, Jean-Jacques DJ, Kornberg HL. Genetic control of manno(fructo)kinase activity in Escherichia coli. Proc Natl Acad Sci USA. 2001;98(26):15257–15259. doi: 10.1073/pnas.211569798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JD, Stoop JM, Massel MO, Conkling MA, Pharr DM. Sequence analysis of a mannitol dehydrogenase cDNA from plants reveals a function for the pathogenesis-related protein ELI3. Proc Natl Acad Sci USA. 1995;92(16):7148–7152. doi: 10.1073/pnas.92.16.7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E, Korenevsky A, Lovley DR, Beveridge TJ. The structure of the core region of the lipopolysaccharide from Geobacter sulfurreducens. Carbohydr Res. 2004;339(18):2901–2904. doi: 10.1016/j.carres.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Wallace BJ, Pittard J. Genetic and biochemical analysis of the isoenzymes concerned in the first reaction of aromatic biosynthesis in Escherichia coli. J Bacteriol. 1967;93(1):237–244. doi: 10.1128/jb.93.1.237-244.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, Reski R, van Dorsselaer A, Boll M. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol Microbiol. 2005;58(5):1238–1252. doi: 10.1111/j.1365-2958.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- Kung JW, Loffler C, Dorner K, Heintz D, Gallien S, Van Dorsselaer A, Friedrich T, Boll M. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc Natl Acad Sci USA. 2009;106(42):17687–17692. doi: 10.1073/pnas.0905073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters F, Shinoda Y, McInerney MJ, Boll M. Cyclohexa-1,5-diene-1-carbonyl-coenzyme A (CoA) hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: Evidence for a common benzoyl-CoA degradation pathway in facultative and strict anaerobes. J Bacteriol. 2007;189(3):1055–1060. doi: 10.1128/JB.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, He Q, Nevin KP, He Z, Zhou J, Lovley DR. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics. 2007;8:180. doi: 10.1186/1471-2164-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischgoll S, Taubert M, Peters F, Jehmlich N, von Bergen M, Boll M. Decarboxylating and nondecarboxylating glutaryl-coenzyme A dehydrogenases in the aromatic metabolism of obligately anaerobic bacteria. J Bacteriol. 2009;191(13):4401–4409. doi: 10.1128/JB.00205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendrat K, Buckel W. Cloning, sequencing and expression of the gene encoding the carboxytransferase subunit of the biotin-dependent Na+ pump glutaconyl-CoA decarboxylase from Acidaminococcus fermentans in Escherichia coli. Eur J Biochem. 1993;211(3):697–702. doi: 10.1111/j.1432-1033.1993.tb17598.x. [DOI] [PubMed] [Google Scholar]
- Braune A, Bendrat K, Rospert S, Buckel W. The sodium ion translocating glutaconyl-CoA decarboxylase from Acidaminococcus fermentans: cloning and function of the genes forming a second operon. Mol Microbiol. 1999;31(2):473–487. doi: 10.1046/j.1365-2958.1999.01189.x. [DOI] [PubMed] [Google Scholar]
- Peters F, Heintz D, Johannes J, van Dorsselaer A, Boll M. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J Bacteriol. 2007;189(13):4729–4738. doi: 10.1128/JB.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Zhou YF, Li LF, Su XD. Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis. Protein Sci. 2009;18(8):1792–1800. doi: 10.1002/pro.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Katayama K, Katsuragi T, Tani Y. Glycolaldehyde-forming route in Bacillus subtilis in relation to vitamin B6 biosynthesis. J Biosci Bioeng. 2001;91(2):147–152. doi: 10.1263/jbb.91.147. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Yatome C, Ishida N, Kitade Y. Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J Biol Chem. 2001;276(49):46394–46399. doi: 10.1074/jbc.M104483200. [DOI] [PubMed] [Google Scholar]
- Sedlmeier R, Altenbuchner J. Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol Gen Genet. 1992;236(1):76–85. doi: 10.1007/BF00279645. [DOI] [PubMed] [Google Scholar]
- N'Guessan AL, Elifantz H, Nevin KP, Mouser PJ, Methe B, Woodard TL, Manley K, Williams KH, Wilkins MJ, Larsen JT. et al. Molecular analysis of phosphate limitation in Geobacteraceae during the bioremediation of a uranium-contaminated aquifer. ISME J. 2010;4(2):253–266. doi: 10.1038/ismej.2009.115. [DOI] [PubMed] [Google Scholar]
- Xiong J, Kurtz DM Jr, Ai J, Sanders-Loehr J. A hemerythrin-like domain in a bacterial chemotaxis protein. Biochemistry. 2000;39(17):5117–5125. doi: 10.1021/bi992796o. [DOI] [PubMed] [Google Scholar]
- Pianzzola MJ, Soubes M, Touati D. Overproduction of the rbo gene product from Desulfovibrio species suppresses all deleterious effects of lack of superoxide dismutase in Escherichia coli. J Bacteriol. 1996;178(23):6736–6742. doi: 10.1128/jb.178.23.6736-6742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CM, Silva G, Oliveira S, LeGall J, Liu MY, Xavier AV, Rodrigues-Pousada C, Teixeira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J Biol Chem. 1997;272(36):22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Seidel J, Einsle O. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J Mol Biol. 2009;393(4):951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kaufmann F, Coppi MV, Nunez C, Lovley DR. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J Bacteriol. 2004;186(12):4042–4045. doi: 10.1128/JB.186.12.4042-4045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filenko N, Spiro S, Browning DF, Squire D, Overton TW, Cole J, Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol. 2007;189(12):4410–4417. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CC, Romao CV, Lindley PF, Teixeira M, Saraiva LM. The role of the hybrid cluster protein in oxidative stress defense. J Biol Chem. 2006;281(43):32445–32450. doi: 10.1074/jbc.M605888200. [DOI] [PubMed] [Google Scholar]
- Cramm R, Siddiqui RA, Friedrich B. Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J Bacteriol. 1997;179(21):6769–6777. doi: 10.1128/jb.179.21.6769-6777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Lovley DR. Investigation of direct vs. indirect involvement of the c-type cytochrome MacA in Fe(III) reduction by Geobacter sulfurreducens. FEMS Microbiol Lett. 2008;286(1):39–44. doi: 10.1111/j.1574-6968.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, Xu H, Leang C, Butler JE, Kim BC. et al. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 2007;7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Leang C, Hodges Myerson AL, Coppi MV, Ciufo S, Methé B, Sandler SJ, Lovley DR. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem J. 2003;369(Pt 1):153–161. doi: 10.1042/BJ20020597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T, Coppi MV, Childers SE, Lovley DR. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol. 2005;71(12):8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C, Coppi MV, Lovley DR. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol. 2003;185(7):2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Leang C, Ding YH, Glaven RH, Coppi MV, Lovley DR. OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J Bacteriol. 2005;187(13):4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methe BA, DiDonato RJ, Covalla SF, Franks AE, Liu A. et al. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One. 2009;4(5):e5628. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321(5887):411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008. pp. D623–631. [DOI] [PMC free article] [PubMed]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Caccavo F Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1994;60(10):3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67(7):3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes of G. bemidjiensis in which frameshifts occur in homopolymeric regions.
Table S2. Enzymes of acyl-CoA metabolism in G. bemidjiensis.
Table S3. Selected sugar interconversion genes of G. bemidjiensis.
Table S4. Predicted c-type cytochromes of G. bemidjiensis.
Table S5. Signalling proteins of G. bemidjiensis.
Table S6. Multicopy nucleotide sequences in G. bemidjiensis.
Figure S1. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S2. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S3. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S4. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S5. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S6. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S7. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S8. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).
Figure S9. Multicopy nucleotide sequences of the G. bemidjiensis genome: base coordinates and alignments. (See also Table S6.).