Abstract
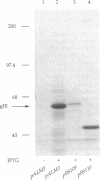
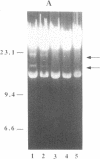
A simple, efficient method for introducing recombinant DNA into a herpesvirus vector and retrieving it at a later time has been developed. By using the Cre-lox site-specific recombination system of coliphage P1, DNA can be readily inserted in vitro into a pseudorabies virus (PRV) vector containing the lox recombination site. The vector PRV42 contains a lox site within the nonessential gIII gene, which encodes a virion envelope glycoprotein. Incubation in vitro of PRV42 DNA with Cre protein and a circular plasmid containing a lox site generates approximately 5% recombinant molecules having the plasmid integrated into the PRV genome at the lox site. Transfection of the reaction mixture into cultured cells allows recovery of the infectious recombinant virus, which is readily identified by a nondestructive "black-plaque assay" using a gIII-specific monoclonal antibody. PRV42 plaques stain black when treated with the gIII monoclonal antibody and a peroxidase-linked second anti-antibody because the lox site placed within the gIII gene of PRV42 does not destroy the gIII epitope. However, Cre-mediated integration of heterologous DNA at the lox site disrupts the gIII epitope so that the resulting recombinant virus produces white plaques. The recombinant virus is infectious, stable, and grows as well as the parental PRV42 vector. The inserted plasmid can be efficiently excised (greater than 50%) from viral DNA by Cre and recovered by transformation of Escherichia coli.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984 Feb 10;259(3):1509–1514. [PubMed] [Google Scholar]
- Abremski K., Hoess R., Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983 Apr;32(4):1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Kaplan A. S. Synthesis of proteins in cells infected with herpesvirus. V. Viral glycoproteins. Virology. 1970 Jun;41(2):265–273. doi: 10.1016/0042-6822(70)90078-4. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Holland T. C., Homa F. L., Marlin S. D., Levine M., Glorioso J. Herpes simplex virus type 1 glycoprotein C-negative mutants exhibit multiple phenotypes, including secretion of truncated glycoproteins. J Virol. 1984 Nov;52(2):566–574. doi: 10.1128/jvi.52.2.566-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland T. C., Sandri-Goldin R. M., Holland L. E., Marlin S. D., Levine M., Glorioso J. C. Physical mapping of the mutation in an antigenic variant of herpes simplex virus type 1 by use of an immunoreactive plaque assay. J Virol. 1983 May;46(2):649–652. doi: 10.1128/jvi.46.2.649-652.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Watson R. J., Whealy M. E., Hays W. W., Enquist L. W. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex virus type 1 and type 2 glycoprotein C. J Virol. 1986 May;58(2):339–347. doi: 10.1128/jvi.58.2.339-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Whealy M. E., Watson R. J., Enquist L. W. Pseudorabies virus gene encoding glycoprotein gIII is not essential for growth in tissue culture. J Virol. 1986 Sep;59(3):635–645. doi: 10.1128/jvi.59.3.635-645.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Carmichael L. E., Deinhardt F., de-The G., Nahmias A. J., Plowright W., Rapp F., Sheldrick P., Takahashi M., Wolf K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology. 1981;16(4):201–217. doi: 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- Roizman B., Jenkins F. J. Genetic engineering of novel genomes of large DNA viruses. Science. 1985 Sep 20;229(4719):1208–1214. doi: 10.1126/science.2994215. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. O., Kennell W. L., Lamm D. L. Visualization of minute centers of viral infection in unfixed cell cultures by an enzyme-linked antibody assay. J Immunol Methods. 1981;40(3):297–305. doi: 10.1016/0022-1759(81)90361-6. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981 Aug 25;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]