Abstract
Rationale
Aquaporin-5 (AQP5) can cause mucus overproduction and lower lung function. Genetic variants in the AQP5 gene might be associated with rate of lung function decline in chronic obstructive pulmonary disease (COPD).
Methods
Five single nucleotide polymorphisms (SNPs) in AQP5 were genotyped in 429 European American individuals with COPD randomly selected from the NHLBI Lung Health Study. Mean annual decline in FEV1 % predicted, assessed over five years, was calculated as a linear regression slope, adjusting for potential covariates and stratified by smoking status. Constructs containing the wildtype allele and risk allele of the coding SNP N228K were generated using site-directed mutagenesis, and transfected into HBE-16 (human bronchial epithelial cell line). AQP5 abundance and localization were assessed by immunoblots and confocal immunofluoresence under control, shear stress and cigarette smoke extract (CSE 10%) exposed conditions to test for differential expression or localization.
Results
Among continuous smokers, three of the five SNPs tested showed significant associations (0.02>P>0.004) with rate of lung function decline; no associations were observed among the group of intermittent or former smokers. Haplotype tests revealed multiple association signals (0.012>P>0.0008) consistent with the single-SNP results. In HBE16 cells, shear stress and CSE led to a decrease in AQP5 abundance in the wild-type, but not in the N228K AQP5 plasmid.
Conclusions
Polymorphisms in AQP5 were associated with rate of lung function decline in continuous smokers with COPD. A missense mutation modulates AQP-5 expression in response to cigarette smoke extract and shear stress. These results suggest that AQP5 may be an important candidate gene for COPD.
Introduction
COPD is the fourth leading cause of death in the United States and the fifth leading cause of death worldwide and its prevalence is expected to increase in coming decades.[1], [2] The overwhelming majority of COPD is caused by environmental exposures. In the United States, this exposure is primarily cigarette smoke; however only 15% of smokers develop COPD,[3] suggesting an important role for genetic susceptibility.
COPD is characterized by abnormal mucous production which may promote bacterial adhesion and may impair bacterial clearance leading to chronic inflammation and irreversible airflow limitation.[4], [5] Aquaporins are water-specific membrane channel proteins and aquaporin 5 (AQP5) is found in airway epithelial cells, type I alveolar epithelial cells and submucosal gland acinar cells in the lungs where it plays a key role in water transport.[6] Decreased expression of human AQP5 has been associated with mucus overproduction in the airways of subjects with COPD and lower lung function.[7] Furthermore, smoking has been shown to attenuate the expression of AQP5 in submucosal glands of subjects with COPD.[7] These data support a potential role of AQP5 in severity of airflow obstruction in COPD and suggest that the expression of AQP5 may be modified by smoking exposure.
AQP5 is a single copy gene on human chromosome 12q13.[8] A single nucleotide polymorphism (SNP) in intron 3 (rs3736309) has been associated with the presence of COPD in a Chinese population, but not with cross-sectional measures of lung function or COPD severity.[9] Whether polymorphisms in AQP5 correlate with the decline of pulmonary function, a trait associated with the development and progression of COPD, is unknown. In this study, we examined associations between genetic variants in the AQP5 gene and rate of lung function decline in a randomly selected subset of the multicenter NHLBI-supported Lung Health Study (LHS) cohort. Identifying pathways and novel molecular targets that modify the clinical course of disease is fundamental to developing preventive strategies and novel therapies.
Methods
Ethics Statement
This study has been approved by the Johns Hopkins University Institutional Review Board. Written informed consent for research was obtained from all participants of the LHS and consent for this analysis was waived because the research involved no additional risk to subjects, and the data used was de-identified. Findings from this manuscript were previously presented in abstract form.
Study Subjects
We randomly selected 429 European Americans of the LHS for whom DNA was available. The LHS was a multicenter (10 centers) randomized clinical trial aimed to determine whether a program of smoking intervention and use of an inhaled bronchodilator could slow the rate of decline in pulmonary function over a 5-year follow-up period. Details of LHS methods have been described extensively.[10]–[12] LHS inclusion and exclusion criteria included the following: Patients were all active smokers between the ages of 35 and 60 with mild to moderate airflow obstruction defined as an FEV1/FVC ratio less than 0.7, and an FEV1 between 50%–90% predicted. Lung function was measured annually over five years.[11], [13], [14] Lung function data from Annual Visit 1 to Annual Visit 5 was used for the current analyses and has been shown to have a good linear fit in previous LHS analyses.[11], [14] Subjects with <3 annual lung function measurements were excluded from analysis (n = 15), resulting in a final group of 414 subjects with data available.
SNP Selection and Genotyping
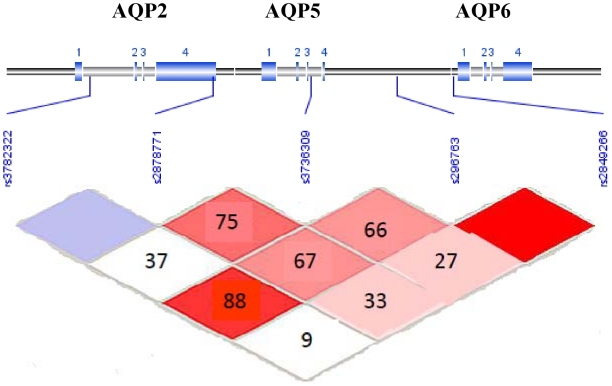
Single nucleotide polymorphisms (SNPs) in the AQP5 gene were selected from Goldenpath (http://genome.ucsc.edu/) and/or NCBI (http://www.ncbi.nlm.nih.gov/). As AQP5 is within the gene cluster of 3 AQP genes (including AQP2 and AQP6; see Figure 1 ), to test whether AQP5 gene itself versus loci in surrounding regions influence disease risk, a total of five SNPs spanning 21,000 bp on human chromosome 12q13 with an average inter-SNP distance of 5.25 kb (ranging from 3.3–7.1 kb) were selected for genotyping using the rationale described below, and genotyped on the Illumina™GoldenGate platform. The study was designed in 2005, prior to conventional LD-tagging approaches for SNP selection. Consequently using standard methodology available at that time, criteria used for SNP selection included 1) SNPs approximating an inter-SNP distance as close to 5 kb as were available at the time of the dbSNP Build 124; 2) representation of SNPs in the promoter, coding and 3′UTR regions; and 3) SNPs with acceptable design scores according to the Illumina Assay Design Tool for genotyping on the Illumina™ GoldenGate platform. Priority for selecting SNPs included: 1) regulatory and coding SNPs; 2) highly polymorphic SNPs, preferably ≥10% minor allele frequency; 3) validated SNPs; and 4) SNPs at intron/exon boundaries.
Figure 1. Linkage Disequilibrium for 5 SNPs in the AQP5 Gene in European American participants with COPD.
Pairwise LD in subjects is represented as red squares for strong LD, blue squares for non-significant LD, and white squares for little or no LD. LD blocks are identified as noted.
Statistical Analysis
Mean annual decline in post-bronchodilator FEV1 % predicted (%predFEV1)(11) was calculated as a linear regression slope using all available time points. Linear regression models were used to adjust for potential confounders, including 1) baseline characteristics: smoking history (pack-years), age, sex, %predFEV1 and airway reactivity (AR); 2) change in body mass index (BMI); and 3) smoking status at Year 5 of the observation period, defined as 1) ‘continuous smoker’ or 2) ‘intermittent or former smokers’. Forward and backward selection was used to develop a parsimonious model. Age, baseline lung function, AR, and change in BMI were included in the final regression analyses. Interaction term for smoking and genetic effect were tested using PLINK. There was significant interaction (P<0.05) for at least one of the five SNPs, so analyses were stratified by smoking status. Residuals from this regression were used in an additive genetic model for each SNP where the most common homozygote for each SNP served as the reference category. Each SNP locus was evaluated for Hardy-Weinberg equilibrium. Pairwise linkage disequilibrium (LD) based on the D' statistic was measured using Haploview.[15] LD blocks were defined using their default algorithm.[16] Sliding windows of 2–4 adjacent SNPs were used to test for association. All analyses were performed with StataSE, version 8.0 (Stata Corp, College Station, TX) and PLINK.[17]
Functional Analysis
SNP information
A non-synonymous, but non-validated SNP (rs41495048) was subsequently identified from dbSNP build 128 (www.ncbi.nlm.nih.gov). It did not meet acceptable design criteria for genotyping on the Illumina™ GoldenGate platform or the alternative TaqMan™ probe-based, 5′ nuclease assay with minor groove binder (MGB) chemistry (ABI, Foster City, CA). However, given that this SNP is flanked by SNPs 3 and 4 ( Table 1 and Figure 1 ), which were significantly associated with lung function decline, and the only non-synonymous SNP identified at the time of this analysis, it was selected for further functional studies.
Table 1. Location, minor allele frequency, and type of selected AQP5 SNPs.
| SNPID | SNP | Position | Region | European American | |
| Minor Allele | MAF | ||||
| 1 | rs3782322 | 48631567 | promoter | A | 0.09 |
| 2 | rs2878771 | 48638660 | promoter | C | 0.19 |
| 3 | rs3736309 | 48644321 | intron (boundary) | G | 0.17 |
| 4 | rs296763 | 48649281 | downstream | G | 0.24 |
| 5 | rs2849266 | 48652567 | downstream | C | 0.08 |
Plasmid Generation
Human AQP5 was inserted into the EcoR1 site as previously described.[18]. In order to create N228K AQP5, degenerate primers were created and N228K human AQP5 was amplified by PCR using QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer's recommendations. Therefore both the wild-type and N228K plasmid have the same backbone and both express the hemagglutinin tag.
Cell culture and transfection
Immortalized human epithelial cells, which are endogenously AQP5 null (16HBE cells, gift of Gary Curtting) were grown in chamber slides to 50–60% confluence, and transiently transfected (1 µg/well) with either wild – or SNP-expressing HA-tagged human AQP5 cDNA using FuGENE 6 (Roche) (1.5 µl) according to the manufacturer's recommendations.
Cell exposures
Shear stress was applied to 16HBE cells transfected with either wildtype hAQP5 or N228K hAQP5 plasmid as described previously.[19] Briefly, low levels of flow was used to create shear forces of 0.5–1 dyne/cm2, to estimate the shear produced by airflow through the tracheobronchial tree. CSE was prepared as previously published.[20], [21] Smoke from 1 research cigarette, 1R5F (University of Kentucky) was bubbled through 10 ml of PBS at 3000 rpm. The CSE was filtered through a 0.2 µm sterile filter (Corning, NY) and pH adjusted to 7.0. The CSE was diluted to 10% of initial concentration and used within 20 min to avoid breakdown of substances.
Immunoblotting and immunofluorescence
Total protein concentrations of the samples were determined by the BCA assay with bovine serum albumin as the standard. Depending on the experiment, 10 to 50 µg of total protein in 1.5%(w/v) SDS was loaded per lane and separated on 12% SDS-PAGE gels prior to transfer to a nitrocellulose membrane. Immunoblotting and blot visualization were performed as described.[18] Affinity purified antibody against the carboxy-terminus of human AQP5 were generated in our lab as previously described.[22] This antibody was used to detect both the wild-type as well as the N228K protein, as the single nucleotide switch did not affect the affinity of the antibody to the protein. Equivalence of samples was confirmed by probing for actin (Upstate Biotechnology). The bands are shown from a total of three different repeated experiments, and the densitometry was collected by comparing the changes in all of the samples to their controls, therefore representing the sum total of 3 different experiments, with replicates within each experiment. Confocal laser microscopy (SP5, Leica) was performed on cells grown in uncoated chamber slides (Falcon) using antibodies against human AQP5 and goat anti-rabbit secondary antibodies (Alexa 488; Molecular Probes). Merged images for analysis of double labeling or z-planes were reconstructed by differential interference contrast images.
Results
Clinical characteristics of subjects have been published previously[23] and are presented in Table 2 . There were no statistical differences in baseline characteristics between those included in the final analyses (n = 414) compared to those excluded because they had >2 missing data points in lung function (n = 15, data not shown). Distributions of baseline characteristics and lung function measurements were also similar to those in the full LHS cohort from which the subset was randomly selected (n = 5,887, data not shown).
Table 2. Patient characteristics.
| European Americans(n = 414) | |
| Baseline Characteristics | |
| Mean age (SD) | 48.6 (7.0) |
| Male, % | 65.0 |
| BMI, kg/m2 | 25.4 (3.6) |
| Smoking (pack-years) | 40.5 (19.2) |
| Pre-BD FEV1 % predicted, % | 76.3 (6.4) |
| Post-BD FEV1 % predicted, % | 79.2 (6.2) |
| Longitudinal Characteristics at 5 years | |
| Smoking history, % | |
| Continuous | 50 |
| Intermittent | 31 |
| Sustained quitter | 19 |
| Post-BD ΔFEV1, %/yr | −0.74 (1.63) |
BD – bronchodilator.
BMI – body mass index.
SD – standard deviation.
Single-marker analyses
All SNPs were in Hardy-Weinberg Equilibrium (HWE). All results of the two-point tests for association between AQP5 markers and %predFEV1 decline are presented in Table 3 , highlighting significant associations (P<0.05) between rate of lung function decline and three of the five SNPs tested in continuous smokers. There was no association between tested SNPs and lung function decline in the group of intermittent or former smokers. Results were similar when intermittent and former smokers were analyzed separately (data not shown).
Table 3. Association of AQP5 polymorphisms with lung function decline in continuous smokers and intermittent or former smokers.
| Continuous smokers | Intermittent or former smokers | |||||
| SNPID | SNP | Minor Allele | Beta | p-val | Beta | P-val |
| 1 | rs3782322 | A | −0.072 | 0.782 | 0.075 | 0.807 |
| 2 | rs2878771 | C | 0.547 | 0.004 | −0.074 | 0.716 |
| 3 | rs3736309 | G | 0.502 | 0.024 | 0.119 | 0.544 |
| 4 | rs296763 | G | −0.430 | 0.012 | −0.077 | 0.679 |
| 5 | rs2849266 | C | 0.161 | 0.542 | 0.293 | 0.340 |
Haplotype Analyses
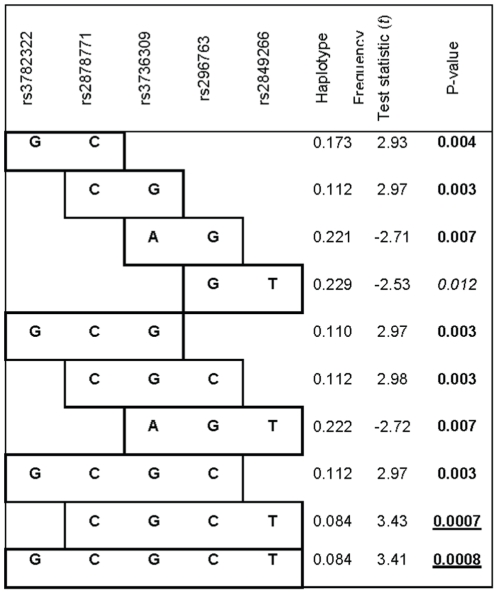
Strong linkage disequilibrium was observed for only one out of four pairings of contiguous SNPs and no LD blocks were identified in this gene under the Gabriel algorithm[16] ( Figure 1 ), therefore a systematic sliding window approach was implemented, considering haplotypes of 2–4 SNPs beginning with the first (5′) marker, and working across the gene. Haplotype tests further support the single SNP results with stronger P-values that appear to highlight the background haplotype likely carrying the potentially untyped but tagged causal SNP; multiple windows of signal that include the single SNPs with P<0.05 described above ( Figure 2 ). The haplotype CGCT (rs2878771, rs3736309, rs296763, rs2849266) showed the strongest association with rate of lung function decline of all haplotypes tested (P = 0.0007), and provides a stronger signal than any of the single SNPs, albeit with the same direction of effect (i.e., protective) on lung function decline.
Figure 2. Haplotype Analysis.
Using the sliding window approach of 2–4 SNPs/window beginning with the first (5′) marker, and working across the gene, one marker at a time, the results of significant (defined as P<0.05) haplotype signals in continuous smokers are shown.
Functional Analysis
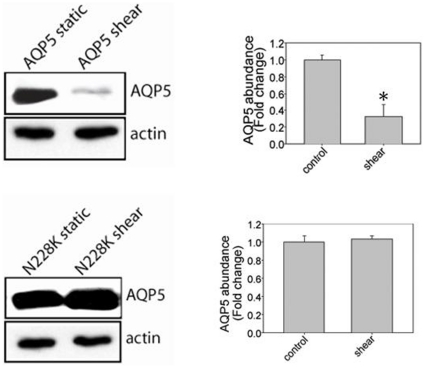
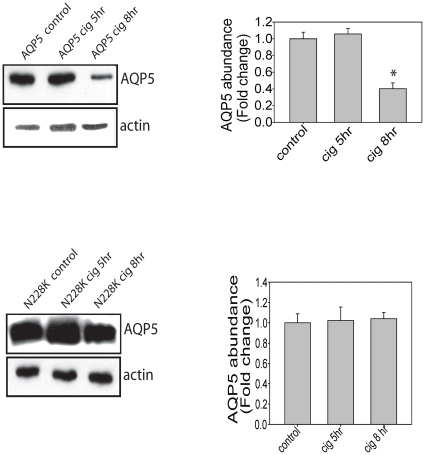
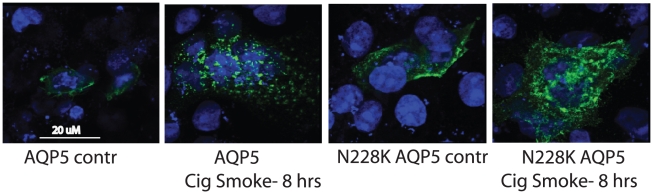
Under control conditions, there was similar abundance in both the wild-type AQP5 plasmid and N228K AQP5 plasmid. Shear stress led to a decrease in AQP5 abundance after two hours; however, similar changes were not observed in N228K AQP5, which showed no change in abundance after shear stress ( Figure 3 ). To test if the N228K AQP5 abundance was different from wildtype AQP5 in response to 10% CSE, 16HBE cells transfected with wildtype hAQP5 or N228K hAQP5 were exposed to either PBS or 10% CSE. CSE exposure led to decreased wild-type hAQP5 abundance at eight hours, and again there was no change in the N228K hAQP5 plasmid abundance ( Figure 4 ). Localization in response to these stimuli was not altered in response to either shear (data not shown) or CSE ( Figure 5 ), indicating that in 16HBE cells, the N228K AQP5 altered AQP5 abundance in response to physiologic stimuli (such as shear stress) as well as pathologic stimuli (such as cigarette smoke extract) without changes in distribution.
Figure 3. Exposure to shear stress leads to a decrease in wild-type AQP5 but no change in N228K AQP5 (n = 3; * p<0.05 with ANOVA one-way).
Figure 4. Eight hours of 10% CSE leads to a decrease in wild-type AQP5 but no change in N228K AQP5 (n = 3; * p<0.05 with ANOVA one-way).
Figure 5. Both wild-type and N228K AQP5 are present on the membrane under control conditions. CSE leads to internalization of both.
Discussion
We identified three SNPs in the AQP5 gene significantly associated with rate of lung function decline in a European American population of continuous smokers with COPD, which was further supported by haplotype analysis. Furthermore, a single non-synonymous SNP, flanked by SNPs which were significantly associated with rate of lung function decline, has been identified in AQP5; with this information, we created a human-AQP5 plasmid using site-directed mutagenesis. Wildtype hAQP5 and N228K hAQP5 responded differently to both the physiologic stimulus of low level shear stress, as well as pathologic exposure of cigarette smoke exposure (CSE) in human bronchial epithelial cells. Collectively our results implicate AQP5 as a novel candidate gene for rate of lung function decline and COPD.
Our strongest single-marker association was for a marker in the promoter region, wherein the C allele at rs2878771 was associated with a 0.55% per year attenuation (i.e., protection) in loss of FEV1 % predicted in continuous smokers, a considerable effect size. In other words, over a 40 year period, a person homozygous for the C variant will be predicted to experience a decline in FEV1 % predicted of 44% less compared to a homozygote for the major allele. Thus over time, this genetic variant could substantially impact disease progression. For example, given a MAF of 0.19, as seen in our cohort, we might extrapolate 4% of the population to be homozygote for the minor allele (CC), 31% to be heterozygote and 66% to be homozygote for the major allele. Even after adjusting for smoking status, this genetic variant could explain the difference between mild and severe COPD under current GOLD criteria.[24]
We identified two additional SNPs in the AQP5 gene significantly associated with rate of lung function decline, including a SNP in intron 3 (rs3736309), where carriers of the G allele had a lower prevalence of COPD in a Chinese population.[9] Our results confirm that the G allele may confer some protection from COPD as the G allele in rs3736309 was associated with a significant attenuation in lung function decline among smokers.
Though our strongest haplotype signals span the gene (∼3 kb) rendering it difficult to narrow the association to a causal variant, to facilitate identifying variants/mutations in AQP5 that may alter expression of AQP5, the non-synonymous SNP in exon 4 of AQP5, which results in an amino acid change (Asn to Lys) was created in a human-AQP5 plasmid using site-directed mutagenesis. This SNP was selected because it was the only non-synonymous SNP identified in AQP5 at the time of the study and was flanked by two intronic SNPs, including rs3736309, used in our analysis and found to be associated with rate of lung function decline. Our results show that this functional mutation alters hAQP5 abundance in HBE-16 cells treated with CSE and low level shear stress. The wildtype hAQP5 abundance decreases; however, no similar reduction was observed in the N228K hAQP5 treated with low level shear stress and CSE at two and eight hours, respectively. This differential regulation of protein expression and protein stability to these stimuli could contribute to functional differences in lung function seen in individuals with COPD.
COPD is influenced by genetic and environmental factors and the interaction between genetic and environmental influences are likely to be important in the pathogenesis of COPD.[25] Cigarette smoking is the major environmental determinant of COPD and genetic and smoking interactions have been associated with lung function in COPD and other chronic lung diseases.[26]–[29] We found effects of genetic variants in AQP5 are stronger in continuous smokers than among those who have quit smoking or smoke intermittently. A specific mechanism by which this interaction could occur is suggested by in vitro studies showing smoking attenuates expression of AQP5 in submucosal glands of subjects with COPD.[7] This decreased expression of human AQP5 has been associated with mucus overproduction in the airways of subjects with COPD and lower lung function.[7] Our in vitro studies confirm wildtype hAQP5 abundance decreases after exposure to CSE and low level shear stress.
In addition to mucous hyper-secretion, another potential mechanism by which AQP5 may lead to lung function loss in subjects with COPD includes a predisposition to airway reactivity as aqp5 −/− mice have been shown to be hyperresponsive to bronchoconstriction by cholinergic stimulation.[30] Increased airway reactivity characterizes both asthma and COPD and has been previously associated with longitudinal changes in lung function in the LHS cohort.[14]
In recent years, there has been interest in the ‘common variant/multiple disease’ hypothesis and suggests certain disease genes may contribute to related clinical phenotypes.[31] For example, previous literature suggests that there are overlapping associations between several genes and chronic lung diseases, including asthma, cystic fibrosis and COPD.[32]–[36] In addition, AQP5 has been suggested to be implicated in asthma pathogenesis,[30], [37], [38] however, whether AQP5 polymorphisms contribute to rate of lung function decline in other lung diseases, such as asthma, remains to be determined.
In summary, our results demonstrate AQP5 polymorphisms are associated with rate of lung function decline among continuous smokers with COPD in the LHS. A missense mutation may be involved in regulation of AQP-5 expression in response to cigarette smoke extract and shear stress, however the functional variant in human AQP5 responsible for differential rate of lung function decline and its mechanism of action is yet to be identified. Moreover, the relative lack of information with regard to non-synonymous SNPs in the AQP5 gene points to the importance of deep re-sequencing. Collectively, these results identify AQP5 as a novel target in COPD and lung function loss.
Acknowledgments
We would like to thank Gary Cutting for providing the 16HBE cell line; Peter Chi, Monica Campbell and Pat Oldewurtel for technical assistance; Alan L Scott, PhD, Anne E. Jedlicka and Margaret V. Mintz of the Malaria Research Institute, Gene Array Core Facility, Johns Hopkins University for assistance with DNA isolation; Alan F. Scott, PhD, Kimberly Doheny, PhD, Roxann Ashworth and Corinne Boehm of the Center for Inherited Disease Research Institute for genotyping; and Helen Voelker and Kathy Farnell of the LHS Data Coordinating Center, University of Minnesota for assistance with the LHS database. The principal investigators and senior staff of the clinical and coordinating centers, the NHLBI, and members of the Safety and Data Monitoring Board of the Lung Health Study are as follows: Case Western Reserve University, Cleveland, OH: M.D. Altose, M.D. (Principal Investigator), C.D. Deitz, Ph.D. (Project Coordinator); Henry Ford Hospital, Detroit, MI: M.S. Eichenhorn, M.D. (Principal Investigator), K.J. Braden, A.A.S. (Project Coordinator), R.L. Jentons, M.A.L.L.P. (Project Coordinator); Johns Hopkins University School of Medicine, Baltimore, MD: R.A. Wise, M.D. (Principal Investigator), C.S. Rand, Ph.D. (Co-Principal Investigator), K.A. Schiller (Project Coordinator); Mayo Clinic, Rochester, MN: P.D. Scanlon, M.D. (Principal Investigator), G.M. Caron (Project Coordinator), K.S. Mieras, L.C. Walters; Oregon Health Sciences University, Portland: A.S. Buist, M.D. (Principal Investigator), L.R. Johnson, Ph.D. (LHS Pulmonary Function Coordinator), V.J. Bortz (Project Coordinator); University of Alabama at Birmingham: W.C. Bailey, M.D. (Principal Investigator), L.B. Gerald, Ph.D., M.S.P.H. (Project Coordinator); University of California, Los Angeles: D.P. Tashkin, M.D. (Principal Investigator), I.P. Zuniga (Project Coordinator); University of Manitoba, Winnipeg: N.R. Anthonisen, M.D. (Principal Investigator, Steering Committee Chair), J. Manfreda, M.D. (Co-Principal Investigator), R.P. Murray, Ph.D. (Co-Principal Investigator), S.C. Rempel-Rossum (Project Coordinator); University of Minnesota Coordinating Center, Minneapolis: J.E. Connett, Ph.D. (Principal Investigator), P.L. Enright, M.D., P.G. Lindgren, M.S., P. O'Hara, Ph.D., (LHS Intervention Coordinator), M.A. Skeans, M.S., H.T. Voelker; University of Pittsburgh, Pittsburgh, PA: R.M. Rogers, M.D. (Principal Investigator), M.E. Pusateri (Project Coordinator); University of Utah, Salt Lake City: R.E. Kanner, M.D. (Principal Investigator), G.M. Villegas (Project Coordinator); Safety and Data Monitoring Board: M. Becklake, M.D., B. Burrows, M.D. (deceased), P. Cleary, Ph.D., P. Kimbel, M.D. (Chairperson; deceased), L. Nett, R.N., R.R.T. (former member), J.K. Ockene, Ph.D., R.M. Senior, M.D. (Chairperson), G.L. Snider, M.D., W. Spitzer, M.D. (former member), O.D. Williams, Ph.D.; Morbidity and Mortality Review Board: T.E. Cuddy, M.D., R.S. Fontana, M.D., R.E. Hyatt, M.D., C.T. Lambrew, M.D., B.A. Mason, M.D., D.M. Mintzer, M.D., R.B. Wray, M.D.; National Heart, Lung, and Blood Institute staff, Bethesda, MD: S.S. Hurd, Ph.D. (Former Director, Division of Lung Diseases), J.P. Kiley, Ph.D. (Former Project Officer and Director, Division of Lung Diseases), G. Weinmann, M.D. (Former Project Officer and Director, Airway Biology and Disease Program, DLD), M.C. Wu, Ph.D. (Division of Epidemiology and Clinical Applications).
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd, and its Licensees to permit this article (if accepted) to be published in Thorax and any other BMJPGL products and to exploit all subsidiary rights, as set out in our licence.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by National Heart, Lung and Blood Institute (HL076322, HL066583, HL010342), NIA (AG21057), Flight Attendant Medical Research Institute Young Clinician Scientist Award and HL085763 (VS). Genotyping services were provided by the Johns Hopkins University under U.S. Federal Government contract number N01-HV-48195 from the NHLBI. KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mannino DM, Homa DM, Akinbami LJ, Ford ES, Redd SC. Chronic obstructive pulmonary disease surveillance–nited States, 1971-2000. Respir Care. 2002;47(10):1184–1199. [PubMed] [Google Scholar]
- 2.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet. 1997;349(9064):1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 3.Bascom R. Differential susceptibility to tobacco smoke: possible mechanisms. Pharmacogenetics. 1991;1(2):102–106. doi: 10.1097/00008571-199111000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Hogg JC. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet. 2004;364(9435):709–721. doi: 10.1016/S0140-6736(04)16900-6. [DOI] [PubMed] [Google Scholar]
- 5.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 6.Verkman AS, Matthay MA, Song Y. Aquaporin water channels and lung physiology. Am J Physiol Lung Cell Mol Physiol. 2000;278(5):L867–L879. doi: 10.1152/ajplung.2000.278.5.L867. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Feng YL, Wen FQ, Chen XR, Ou XM, et al. Decreased expression of human aquaporin-5 correlated with mucus overproduction in airways of chronic obstructive pulmonary disease. Acta Pharmacol Sin. 2007;28(8):1166–1174. doi: 10.1111/j.1745-7254.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee MD, Bhakta KY, Raina S, Yonescu R, Griffin CA, et al. The human Aquaporin-5 gene. Molecular characterization and chromosomal localization. J Biol Chem. 1996;271(15):8599–8604. doi: 10.1074/jbc.271.15.8599. [DOI] [PubMed] [Google Scholar]
- 9.Ning Y, Ying B, Han S, Wang B, Wang X, et al. Polymorphisms of aquaporin5 gene in chronic obstructive pulmonary disease in a Chinese population. Swiss Med Wkly. 2008;138(39-40):573–578. doi: 10.4414/smw.2008.12240. [DOI] [PubMed] [Google Scholar]
- 10.Anthonisen NR. Lung Health Study. Am Rev Respir Dis. 1989;140(4):871–872. doi: 10.1164/ajrccm/140.4.871. [DOI] [PubMed] [Google Scholar]
- 11.Anthonisen NR, Connett JE, Kiley JP, Altose MD, Bailey WC, et al. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. JAMA. 1994;272(19):1497–1505. [PubMed] [Google Scholar]
- 12.Connett JE, Kusek JW, Bailey WC, O'Hara P, Wu M. Design of the Lung Health Study: a randomized clinical trial of early intervention for chronic obstructive pulmonary disease. Control Clin Trials. 1993;14(2) Suppl:3S–19S. doi: 10.1016/0197-2456(93)90021-5. [DOI] [PubMed] [Google Scholar]
- 13.Kanner RE, Connett JE, Williams DE, Buist AS. Effects of randomized assignment to a smoking cessation intervention and changes in smoking habits on respiratory symptoms in smokers with early chronic obstructive pulmonary disease: the Lung Health Study. Am J Med. 1999;106(4):410–416. doi: 10.1016/s0002-9343(99)00056-x. [DOI] [PubMed] [Google Scholar]
- 14.Tashkin DP, Altose MD, Connett JE, Kanner RE, Lee WW, et al. Methacholine reactivity predicts changes in lung function over time in smokers with early chronic obstructive pulmonary disease. The Lung Health Study Research Group. Am J Respir Crit Care Med. 1996;153(6 Pt 1):1802–1811. doi: 10.1164/ajrccm.153.6.8665038. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidhaye V, Hoffert JD, King LS. cAMP has distinct acute and chronic effects on aquaporin-5 in lung epithelial cells. J Biol Chem. 2005;280(5):3590–3596. doi: 10.1074/jbc.M411038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidhaye VK, Schweitzer KS, Caterina MJ, Shimoda L, King LS. Shear stress regulates aquaporin-5 and airway epithelial barrier function. Proc Natl Acad Sci U S A. 2008;105(9):3345–3350. doi: 10.1073/pnas.0712287105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glader P, Moller S, Lilja J, Wieslander E, Lofdahl CG, et al. Cigarette smoke extract modulates respiratory defence mechanisms through effects on T-cells and airway epithelial cells. Respir Med. 2006;100(5):818–827. doi: 10.1016/j.rmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Serikov VB, Leutenegger C, Krutilina R, Kropotov A, Pleskach N, et al. Cigarette smoke extract inhibits expression of peroxiredoxin V and increases airway epithelial permeability. Inhal Toxicol. 2006;18(1):79–92. doi: 10.1080/08958370500282506. [DOI] [PubMed] [Google Scholar]
- 22.Steinfeld S, Cogan E, King LS, Agre P, Kiss R, et al. Abnormal distribution of aquaporin-5 water channel protein in salivary glands from Sjogren's syndrome patients. Lab Invest. 2001;81(2):143–148. doi: 10.1038/labinvest.3780221. [DOI] [PubMed] [Google Scholar]
- 23.Hansel NN, Gao L, Rafaels NM, Mathias RA, Neptune ER, et al. Leptin receptor polymorphisms and lung function decline in COPD. Eur Respir J. 2009;34(1):103–110. doi: 10.1183/09031936.00120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 25.Sandford AJ, Silverman EK. Chronic obstructive pulmonary disease. 1: Susceptibility factors for COPD the genotype-environment interaction. Thorax. 2002;57(8):736–741. doi: 10.1136/thorax.57.8.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He JQ, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Glutathione S-transferase variants and their interaction with smoking on lung function. Am J Respir Crit Care Med. 2004;170(4):388–394. doi: 10.1164/rccm.200312-1763OC. [DOI] [PubMed] [Google Scholar]
- 27.He JQ, Burkett K, Connett JE, Anthonisen NR, Pare PD, et al. Interferon gamma polymorphisms and their interaction with smoking are associated with lung function. Hum Genet. 2006;119(4):365–375. doi: 10.1007/s00439-006-0143-z. [DOI] [PubMed] [Google Scholar]
- 28.Seo T, Pahwa P, McDuffie HH, Nakada N, Goto S, et al. Interactive effect of paraoxonase-1 Q192R polymorphism and smoking history on the lung function decline in grain workers. Ann Epidemiol. 2008;18(4):330–334. doi: 10.1016/j.annepidem.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Collaco JM, Vanscoy L, Bremer L, McDougal K, Blackman SM, et al. Interactions between secondhand smoke and genes that affect cystic fibrosis lung disease. JAMA. 2008;299(4):417–424. doi: 10.1001/jama.299.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krane CM, Fortner CN, Hand AR, McGraw DW, Lorenz JN, et al. Aquaporin 5-deficient mouse lungs are hyperresponsive to cholinergic stimulation. Proc Natl Acad Sci U S A. 2001;98(24):14114–14119. doi: 10.1073/pnas.231273398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker KG. The common variants/multiple disease hypothesis of common complex genetic disorders. Med Hypotheses. 2004;62(2):309–317. doi: 10.1016/S0306-9877(03)00332-3. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa E, Ruan J, Connett JE, Anthonisen NR, Pare PD, et al. Transforming growth factor-beta1 polymorphisms, airway responsiveness and lung function decline in smokers. Respir Med. 2006;101(5):938–943. doi: 10.1016/j.rmed.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Arkwright PD, Laurie S, Super M, Pravica V, Schwarz MJ, et al. TGF-beta(1) genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax. 2000;55(6):459–462. doi: 10.1136/thorax.55.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eden E. Asthma and COPD in Alpha-1 Antitrypsin Deficiency. Evidence for the Dutch Hypothesis. COPD. 2010;7(5):366–374. doi: 10.3109/15412555.2010.510159. [DOI] [PubMed] [Google Scholar]
- 35.Tzetis M, Efthymiadou A, Strofalis S, Psychou P, Dimakou A, et al. CFTR gene mutations–including three novel nucleotide substitutions–and haplotype background in patients with asthma, disseminated bronchiectasis and chronic obstructive pulmonary disease. Hum Genet. 2001;108(3):216–221. doi: 10.1007/s004390100467. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang J, Huang J, Li X, He C, et al. Polymorphisms in the transforming growth factor-beta1 gene and the risk of asthma: A meta-analysis. Respirology. 2010;15(4):643–650. doi: 10.1111/j.1440-1843.2010.01748.x. [DOI] [PubMed] [Google Scholar]
- 37.Krane CM, Deng B, Mutyam V, McDonald CA, Pazdziorko S, et al. Altered regulation of aquaporin gene expression in allergen and IL-13-induced mouse models of asthma. Cytokine. 2009;46(1):111–118. doi: 10.1016/j.cyto.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y, Wang Y, Chen Z, Wang D, Wang X, et al. Role of Aquaporin 5 in antigen-induced airway inflammation and mucous hyperproduction in mice. J Cell Mol Med [Epub ahead of print] 2010. Available: http://onlinelibrary.wiley.com/doi/10.1111/j.1582-4934.2010.01103.x/pdf. Accessed November 15, 2010. [DOI] [PMC free article] [PubMed]