Abstract
The goal of this study is to characterize the potential effect of artifacts originating from physiological noise on statistical analysis of diffusion tensor MRI (DTI) data in a population. DTI derived quantities including mean diffusivity (Trace(D)), fractional anisotropy (FA), and principal eigenvector (ε1) are computed in the brain of 40 healthy subjects from tensors estimated using two different methods: conventional nonlinear least-squares, and robust fitting (RESTORE). RESTORE identifies artifactual data points as outliers and excludes them on a voxel-by-voxel basis. We found that outlier data points are localized in specific spatial clusters in the population, indicating a consistency in brain regions affected across subjects. In brain parenchyma RESTORE slightly reduces inter-subject variance of FA and Trace(D). The dominant effect of artifacts, however, is bias. Voxel-wise analysis indicates that inclusion of outlier data points results in clusters of under- and over-estimation of FA, while Trace(D) is always over-estimated. Removing outliers affects ε1 mostly in low anisotropy regions. It was found that brain regions known to be affected by cardiac pulsation – cerebellum and genu of the corpus callosum, as well as regions not previously reported – splenium of the corpus callosum, show significant effects in the population analysis. It is generally assumed that statistical properties of DTI data are homogenous across the brain. This assumption does not appear to be valid based on these results. The use of RESTORE can lead to a more accurate evaluation of a population, and help reduce spurious findings that may occur due to artifacts in DTI data.
INTRODUCTION
Diffusion tensor imaging (DTI) (Basser et al. 1994) is a magnetic resonance imaging (MRI) technique that allows non–invasive investigation of structural and architectural features of living tissues (Pierpaoli et al. 1996). DTI is increasingly being used for clinical investigations and, in particular, for brain studies. Diffusion tensor derived quantities of interest generally include an index of diffusion anisotropy (the most popular is the fractional anisotropy (FA) (Pierpaoli and Basser 1996)); the mean diffusivity, <D> which is equal to 1/3 of the trace of the diffusion tensor, Trace(D); and the orientation of highest diffusivity, which is collinear with the principal eigenvector ε1.
There are a number of image artifacts that can corrupt diffusion weighted images (DWIs) which in turn affects DTI derived quantities. These include, but are not limited to, bulk subject motion, eddy current distortions, B0 susceptibility induced EPI distortions, signal dropouts, local mis-registration caused by pulsatile motion due to the cardiac cycle, and system related artifacts such as spike noise and temporal instabilities of the scanner. Correction of many of these artifacts is possible (Skare and Andersson 2001; Pierpaoli et al. 2003; Rohde et al. 2004; Nunes et al. 2005; Wu et al. 2008); for example, distortions due to bulk motion and eddy current distortions are generally corrected in post-processing in DTI studies using image registration strategies.
Noise in MR imaging is another complicating factor which can affect DWIs and, consequently, tensor derived quantities. Thermal noise in MR images can be modeled by a Gaussian distribution (Henkelman 1985) as long as the images have sufficient SNR (Koay and Basser 2006; Koay et al. 2006). The originally proposed least-squares regression approach of tensor fitting (Basser et al. 1994) takes into account thermal noise by including the assumed signal variance as a weighting factor in the fitting. However, artifacts generally cannot be modeled by a simple distribution. Traditionally, individual images affected by severe outlier type artifacts were simply removed manually on an image-by-image basis. A more sophisticated approach is to use robust tensor fitting techniques applied on a voxel-by-voxel basis which have been proposed for mitigating the effects of artifactual data points that manifest themselves as outlier data (Mangin et al. 2002; Chang et al. 2005). One of these robust fitting algorithms, called RESTORE (Chang et al. 2005), identifies outlier data points and removes them regardless of their origin: subject (e.g. cardiac pulsation and respiration) or scanner dependent (e.g. spike noise). RESTORE has shown good performance in Monte Carlo simulations (Chang et al. 2005) but few quantitative studies of in vivo brain data exist and little is known about the regional distribution of data points that would be identified as outliers in the human brain, in particular, in a population of subjects.
We previously reported in a preliminary study of healthy subjects that outliers appear clustered in certain regions of the brain with a consistent distribution across subjects (Walker et al. 2008) and that this will likely have an effect on the outcome of a population analysis (Walker et al. 2009). Additionally, preliminary findings suggest that the results of a statistical group analysis of a patient and a control population can differ if tensor fitting is performed using RESTORE rather than a traditional linear least-squares approach (Peterson et al. 2008). Apart from these findings, the general question whether outlier rejection is an important consideration when performing a clinical population analysis using tensor-derived quantities remains largely unanswered. Logically, one would assume that there can be two possible effects of outliers: 1) their presence could increase the variance of tensor derived quantities, modulating the statistical power of measurements in a heterogeneous way across the brain, and/or 2) they could introduce a bias in the value of tensor derived quantities, causing a systematic increase or decrease associated with artifacts. It is not unreasonable to suspect that either or both of these effects could affect the statistical properties of tensor-derived quantities; and there is no guarantee that different quantities will be affected equivalently. For example, bias or increased variability could exist for anisotropy, while absent for Trace(D) in certain brain regions.
In this work we present an investigation into the statistical effects of outlier data points in a population of 40 healthy human volunteers that underwent a fairly typical DTI brain study. We perform a qualitative assessment of the mean and standard deviation of tensor derived quantities across the population, as well as a voxel-wise group analysis of these quantities, and assessed differences in the orientation of highest diffusivity in tensors estimated first with the nonlinear least-squares (NLS) tensor fitting algorithm (referred to as conventional tensor fitting) and secondly with the RESTORE robust tensor fitting algorithm. Our goals are to characterize the regional distribution of data points that are identified as artifactual (outliers) and to investigate the potential consequences of outlier rejection on the statistical outcome of DTI population analysis. Additionally, we make freely available our population maps for investigators who may be interested in assessing variability in the brain regions of interest for their studies.
METHODS
Subjects
40 healthy volunteers aged 22 to 32 years (average 26 ± 3 years), 20 male, 20 female, right handed, self-reported as Caucasian, and with a minimum education of a college degree were scanned on a 3.0T GE Excite scanner using an eight channel coil (GE Medical Systems, Milwaukee, WI). All participants were fully collected under a Human Cortical Physiology and Stroke Neurorehabilitation Section protocol, and provided written informed consent before taking part in the study, which was approved by the Institutional Review Board of the National Institute of Neurological Disorders and Stroke, NIH. Whole brain single-shot echo-planar (EPI) DWI datasets were acquired with the following parameters: TE/TR = 76.4/18277.2ms, 2.5×2.5mm2 in-plane resolution, zero-filled by the scanner to 1.875×1.875mm2, with 60 slices at 2.5mm thickness, b-value of 1100s/mm2 in 50 non-collinear directions, and 10 images at a b-value of 0s/mm2, for a total of 60 brain volumes, SENSE acceleration (ASSET) factor = 2, no cardiac gating was performed. Structural T2-weighted (T2W) FSE images were acquired for each subject with TE/TR = 122/8333ms, FOV = 240mm2, acquisition matrix 512×512, 1.5mm slice thickness.
Image Processing
Preprocessing of the DWIs was performed with algorithms included in the TORTOISE software package (www.tortoisedti.org)(Pierpaoli et al. 2010). DWIs were first corrected for motion and eddy current distortions according to Rohde et al. including proper re-orientation of the b-matrix to account for the rotational component of the subject rigid body motion (Rohde et al. 2004; Leemans and Jones 2009). In addition, B0 susceptibility induced EPI distortions were corrected using an image registration based approach using B-Splines (Wu et al. 2008). All corrections were performed in the native space of the DWI images. For consistency, all images were reoriented into a common space defined by the mid-sagittal plane, the anterior commissure, and the posterior commissure (Bazin et al. 2007) also with appropriate rotations to the b-matrix.
Tensor fitting was performed twice on the corrected images; first using conventional fitting and second using RESTORE. The RESTORE algorithm uses iteratively reweighted least-squares fitting to identify outlier data points on a voxel-by-voxel basis. It then removes these outlier data points from consideration in the final tensor fitting, and performs conventional fitting on the remaining data points. An appropriate estimate of the “artifact free” signal standard deviation is required for accurate results with RESTORE; otherwise the algorithm could reject too many or too few data points. This signal standard deviation (σ) was initially estimated from the standard deviation of the signal measured in an ROI in the background using the classic Henkelman formula (Henkelman 1985). However, Henkelman's formula does not account for the many confounding effects on thermal noise in imaging, which includes the use of multiple coils, parallel imaging acquisition and reconstruction (Kellman and McVeigh 2005), apodization strategies, padding of background regions done by the proprietary software of the manufacturer, image interpolations for correction of non-linearity of the gradients (Wang et al. 2004), and subsequent image interpolations to correct for eddy distortion, motion (Rohde et al. 2005), and B0 susceptibility induced EPI distortion. Therefore, this approach resulted in an under-estimation of the true signal variability in the data. This conclusion was reinforced by the finding that the resulting reduced chi-square values () (Bevington 1969) of the non-linear tensor fitting in all voxels in the brain parenchyma were systematically greater than 1. If the expected signal variability is correctly estimated in artifact free regions the resulting values should be close to 1 by definition (Bevington 1969). Assuming that less than 50% of brain voxels are affected by outliers, we decided to extract a reasonable estimate the “artifact free” signal standard deviation to be used by the RESTORE algorithm directly from the residuals of the tensor fitting. The details of the procedure we employed are shown in Appendix A.
The RESTORE algorithm used in this work is a modified version (Chang et al. 2009) of the previously published algorithm (Chang et al. 2005), with an additional constraint to remove computational instabilities. This added constraint uses the condition number (Skare et al. 2000) to avoid too many data points from the same direction being excluded as outliers, which can yield an ill conditioning in the design matrix. If this situation arises RESTORE is not performed and NLS fitting is used with all data points included.
During the tensor fitting, maps of the outlier data points are created in order to investigate the regional distribution of outliers in each subject. An outlier rejection map was computed using the following formula on a voxel-by-voxel basis:
| (1) |
where the number of degrees of freedom is equal to the total number of images minus the number of fitted parameters. For this acquisition the number of degrees of freedom is equal to 50 DWI volumes − 6 fitted parameters = 44. For example, 8 corrupted data points identified by RESTORE and eliminated from the tensor fitting will result in an intensity value of 18 on the outlier map (8/44 = 18%).
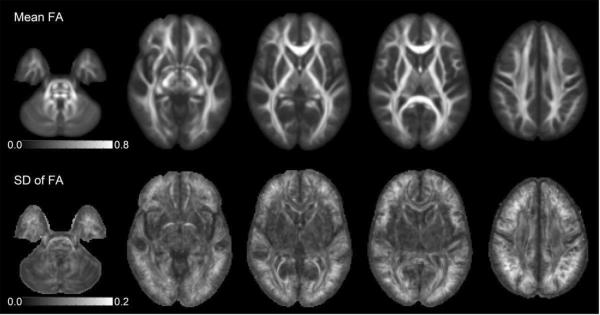
Following tensor estimation, spatial normalization was performed in order to do a voxel-wise analysis of the population. Ideally, local inter-subject variability due to mis-registration should be eliminated to study the effects of variability due to noise and artifacts. We used a non-parametric, diffeomorphic deformable image registration technique implemented in DTI-TK (Zhang et al. 2007), that incrementally estimates its displacement field using a tensor-based registration formulation (Zhang et al. 2006). It is designed to take advantage of similarity measures comparing tensors as a whole via explicit optimization of tensor reorientation and includes appropriate reorientation of the tensors following deformation (Zhang et al. 2006). Figure 1 shows the mean and standard deviation of FA of the population after registration. In general, the amount of residual mis-registration appears to be quite low in the major white matter areas, as well as in the cerebellum. Relatively high standard deviation is still observable at the periphery of the brain, which is to be expected because of the anatomical variability of the folding pattern in different individuals. The optic radiations, fornix, and anterior commissure also show some variability, possibly due to residual mis-registration.
Figure 1.
Mean and standard deviation of the population (N=40) for FA after diffeomorphic tensor registration. The contribution of mis-registration to the voxelwise variability is improved compared to affine registration (not shown), particularly in the major white matter tracts and the cerebellum. Higher levels of variability can be observed in peripheral region, particularly at the top of the brain.
The fully deformable registration was applied to all of the subjects' tensor images, and the resulting deformation field was then applied to the outlier maps, and to the FA and Trace(D) maps from both RESTORE and conventional fittings. An outlier rejection probability (ORP) map for the population was created by taking the mean of the 40 subjects' registered outlier maps (Figure 2). The average tensors were used to calculate the difference in angle of the orientation of the principal eigenvector (ε1) between the conventional and RESTORE data, as well as the expected average covariance matrix of the tensor (Koay et al. 2007) and covariance matrix of the orientation of ε1 (Koay et al. 2008). From the covariance matrices we computed the normalized area of the cone of uncertainty at 95% confidence for both conventional and RESTORE fittings.
Figure 2.
a) mean FA map with ORP overlayed in pink (ORP > 2%); b) outlier rejection probability (ORP) map is the mean outlier map of the 40 subjects. Note the well defined clusters of outlier data points within the brain; c) standard deviation of the 40 subjects' outlier maps. Areas of high probability outlier rejection also have a higher standard deviation within the population than areas not affected by outliers.
Image Analysis
Mean and standard deviation (SD) maps of FA and Trace(D) were calculated across the population for both the RESTORE and the conventional data. Subtraction maps (conventional minus RESTORE) were calculated to qualitatively evaluate the differences in variance and in mean between the two fitting algorithms. To further test the difference in mean between the two groups, voxel-wise analysis was performed using Randomise version 2.5 (Nichols and Holmes 2002) from the FSL software package version 4.1.4 (Smith et al. 2004). For this analysis, we performed a paired t-test type statistical analysis with the conventional data and the RESTORE data as the two groups, using the threshold-free cluster enhancement (TFCE) (Smith and Nichols 2009) function of Randomise, with 5000 permutations. Cluster maps presented here use family-wise error (FWE) corrected statistics for appropriate correction for multiple comparisons. Finally, we investigated the effect of outlier data points on the orientation of highest diffusivity by calculating the angular difference between the average principal eigenvector of the population of the conventional and the RESTORE data, as well as calculating the difference in area of the cone of uncertainty of ε1 (Koay et al. 2008) between the two fitting algorithms.
Mean and SD of FA, the ORP, the FSL cluster maps, and the mean tensor image created by DTI-TK are freely available for download at: http://science.nichd.nih.gov/confluence/display/nihpd/Data+Downloads. Investigators are invited to use these maps to identify whether their clinical findings lie within parenchyma regions of high or low probability outlier rejection, which can indicate whether it may be prudent to apply robust tensor fitting to their data.
RESULTS
The mean FA map, computed from the spatially normalized FA maps of the conventional data, the corresponding ORP map, and the standard deviation of the outlier maps are shown in Figure 2. Voxels with ORP values greater than 2% are overlaid in pink on the mean FA map (Figure 2a), showing the anatomical location of the regions likely to be affected by outlier data points. In the ORP map (Figure 2b) the brightest value (white) corresponds to a 10% rejection of data points as outliers. The ORP map shows a distinct regional distribution of outliers in the population, with a very low probability of outliers occurring in the superior parts of the brain, and a higher percentage of outliers in the medial portions of the cerebellum, middle cerebellar peduncles, discrete regions in the temporal lobes including the ventral medial portion of the temporal lobes which may encompass the fimbria of the hippocampus, the hippocampus and the amygdala, periventricular regions, the midline portion of the genu and splenium of the corpus callosum, insular regions, ventral frontal areas, as well as at air-tissue interfaces and cerebrospinal fluid (CSF)-tissue interfaces. In regions of high ORP there is a correspondingly high standard deviation, while areas with near zero ORP have a correspondingly low standard deviation (Figure 2c). These findings indicate that areas not affected by outliers are consistent within the population, while regions that have a higher probability of outlier rejection are more variable within the population.
Figures 3a and 3b show the subtraction of SD maps for FA and Trace(D) respectively (conventional minus RESTORE). Bright areas indicate that processing with RESTORE reduces the variance, while dark areas indicate that processing with RESTORE increases the variance. If using RESTORE fitting is an improvement over conventional fitting, then we would expect a reduction in variance, and more bright areas than dark areas on the subtraction map. There are only small differences in the superior parts of the brain, mainly in the air-tissue interfaces, which do not show a systematic trend. In cerebellar regions (thick white arrows) there is a reduction of variance using the RESTORE fitting for both Trace(D) and FA. There is an increase in Trace(D) variance using RESTORE in the ventricular regions (black arrows). There appears to be a reduction in variance of Trace(D) in the insular region, which transitions to an increase in variance at the periphery of the insula where there is partial voluming with CSF (thin white arrows). There is also a small decrease in variance in the genu of the corpus callosum. Overall, the magnitude of the changes in variance is small, but regionally varying.
Figure 3.
Subtraction maps of a) FA and b) Trace(D) standard deviation across 40 subjects. Bright areas indicate that processing with RESTORE reduces the variance, while dark areas indicate that processing with RESTORE increases the variance. Largest decreases in variance are seen in the cerebellum (large white arrows) and insular regions (thin white arrows), largest increase in variance in CSF regions (black arrows).
Figures 4a and 4b show the subtraction of the mean values of FA and Trace(D) respectively (conventional minus RESTORE). Bright areas indicate that processing with RESTORE reduces the mean value, while dark areas indicate that processing with RESTORE increases the mean value. In the more superior parts of the brain, similar to the subtraction of SD, there is very little difference between the two fitting algorithms with the exception of the very peripheral regions at the tissue-air interfaces. In cerebellar regions, higher values of FA and Trace(D) in the population are found using conventional fitting (white arrows), which agrees with our previous findings in a different population (Walker et al. 2009). FA and Trace(D) values are slightly higher in the population in the ventricular regions using the RESTORE method. Additionally of note is a dark area on the midline of the genu (black arrows) and splenium (not pictured) of the corpus callosum in FA and the opposite on Trace(D), and bright areas in the insular regions for both FA and Trace(D) (thin white arrows). Overall, the presence of bias in FA and Trace(D) is more clearly demarcated than the effect on the variance and it is also regionally varying.
Figure 4.
Subtraction maps of a) FA and b) Trace(D) mean value across 40 subjects. Bright areas indicate that processing with RESTORE reduces the mean value, while dark areas indicate that processing with RESTORE increases the mean value.
Cluster maps in the brain parenchyma from the Randomise TFCE analysis for differences in the mean are presented in Figure 5. Voxels with high CSF contamination are not included in the analysis. These voxels were masked out by excluding voxels with mean Trace(D) value higher than 3000 μm2/s. Figure 5a shows the significant clusters of FA overlaid on the mean FA map. Blue clusters represent areas of lower FA with conventional fitting; red clusters represent areas of higher FA with conventional fitting (p ≤ 0.05). Red clusters are found only in the cerebellum. Blue clusters are more widespread, including clusters in the anterior portion of the pons, cerebellar peduncles, thalamic regions, corpus callosum, right anterior limb of the internal capsule, and the ventral medial portion of the temporal lobes, which may encompass the fimbria of the hippocampus, the hippocampus, and the amygdala. Figure 5b shows the significant clusters of Trace(D) overlaid on the mean Trace(D) map. There are no statistically significant clusters of lower Trace(D) with conventional fitting. Red clusters represent areas of higher Trace(D) with conventional fitting (p ≤ 0.05). These clusters are present throughout the parenchyma, and include the cerebellum, brainstem, thalamic regions, cingulum, corpus callosum, insular areas, the ventral medial portion of the temporal lobes, which may encompass the fimbria of the hippocampus, the hippocampus and the amygdala, and along the interhemispheric midline.
Figure 5.
Cluster maps from the Randomise TFCE analysis overlaid on a) mean FA and b) mean Trace(D) images. All clusters shown are p<0.05. Blue clusters indicate areas of significantly lower mean value with conventional fitting. Red clusters indicate areas of significantly higher mean value with conventional fitting.
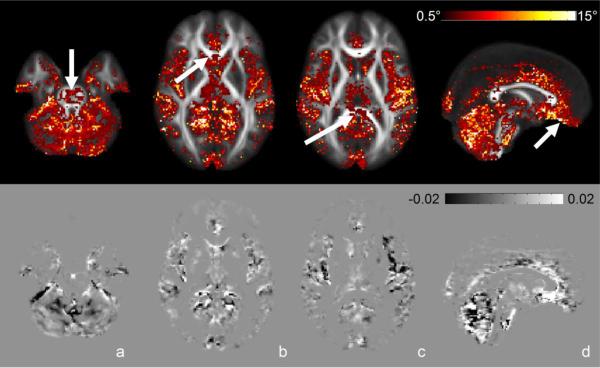
Figure 6 (top row) shows the angular difference between conventional and RESTORE data for the average principal eigenvector overlaid on the average FA map of the population. This angle reflects the orientational bias in ε1 introduced by outliers. The affected areas are consistent with areas highlighted by the ORP map, however, larger angular differences are found in low anisotropy regions; grey matter and CSF, and smaller differences in white matter. Most of the major white matter tracts have negligible angular differences except for the midline of the genu of the corpus callosum, the splenium of the corpus callosum and the transverse pontine fibers (arrows). In addition, there are large differences in the cerebellum, insula, and ventral frontal areas. Figure 6 (bottom row) shows the difference map of the normalized area of the cone of uncertainty of the average tensors (conventional minus RESTORE). In this map, dark voxels are where the area of the cone of uncertainty is larger when using RESTORE, while bright voxels are where the area of the cone is smaller when using RESTORE. There are dark voxels in the cerebellum and in the CSF spaces lateral to the insula, and bright areas in the pons, and in ventral frontal areas. While not clearly seen in the figure due to the small magnitude of the effect, there are slight positive values in the genu and splenium of the corpus callosum.
Figure 6.
Top Row; Angular difference between NLS and RESTORE fitting of the average principal eigenvector (ε1) for the population, overlaid on the mean FA map of the population. This reflects the orientational bias in ε1 introduced by outliers. Bottom Row; Subtraction of the normalized area of the cone of uncertainty of the average ε1 for the population. Bright regions indicate lower dispersion of the individual principal eigenvectors about the mean when outliers are removed by RESTORE and vice-versa for dark regions.
DISCUSSION
The first goal of our study was to characterize the regional distribution of artifactual DWI data points in the brain of a population of healthy subjects in a typical DTI study. We found that regions of high percentage of artifactual data points form very well defined clusters on the outlier rejection probability map (ORP) of the population (Figure 2). The anatomical location of the clusters in this study is consistent with findings of our previous preliminary study in a different group of subjects (Walker et al. 2008). Areas of the brain with low probability of outliers are extremely consistent within the population, indicating that areas that are not affected by artifacts in an individual are consistently not affected within the population. Areas with a higher percentage of outlier rejection show also higher variance of outlier rejection, indicating that areas affected by artifacts are spatially consistent, but the magnitude of the effect might vary across individuals.
The second goal of our study was to investigate the effect of artifactual data points on the statistical properties of tensor derived quantities in a population. The presence of artifacts in the DWIs per se does not necessarily imply measurable consequences in a population analysis of tensor derived quantities. Moreover, the effect could be in either or both the mean value of the metrics, increasing the risk of false positive findings, and/or their variance, thereby reducing the statistical power of DTI analysis in certain brain regions.
Subtraction of the mean and SD of FA and Trace(D) of the population processed with both RESTORE and conventional fitting shows differences in areas which are regionally consistent with the ORP map. These findings are also confirmed by voxel-by-voxel pair-wise statistical analysis, in which the clusters of highest significance are in brain regions that are generally consistent with the subtraction maps. Voxel-wise analysis also shows that effects on the mean of FA and Trace(D) are that outlier rejection using RESTORE nearly always results in an increased value of FA with the exception of the cerebellum and always results in a decreased value of Trace(D) in affected parenchyma (Figure 5).
If we assume that RESTORE identifies and rejects artifactual data points correctly, one can interpret a higher FA or Trace(D) value with conventional fitting as an over-estimation of the metric. This effect was observed in the cerebellum (thick white arrows on Figures 3 and 4), and is consistent with previous findings of artifacts associated with cardiac pulsation (Pierpaoli et al. 2003). Signal drops due to cardiac pulsation are likely to occur in particular directions of diffusion sensitization. In the cerebellum, for instance, the signal drop is more pronounced when the diffusion sensitizing gradient is applied with superior-inferior orientation. In relatively isotropic regions, signal drops will result in apparently increased anisotropy. In the cerebellum, cardiac pulsation artifacts, in fact, result in the spurious appearance of anisotropic tissue with superior-inferior apparent fiber orientations. In regions where anisotropy is already present, artifactual signal drops may cause a reduction in anisotropy. For example, cardiac pulsation is known to create a drop-out effect in the genu of the corpus callosum at the interhemispheric midline (Pierpaoli et al. 2003), resulting in reduced anisotropy in that area and a spurious “disconnection” of interhemispheric connectivity at each systole (Jones and Pierpaoli 2005). Our results are consistent with these previous findings (Figure 4 black arrows). Moreover, we identify an additional area of instability that has not been previously reported in a region encompassing the splenium of the corpus callosum. The effect on anisotropy in this area is the same as that which is observed in the genu, i.e. anisotropy is decreased by the presence of outliers. We hypothesize that vascular effects in the adjacent choroid plexi may contribute to the large presence of outliers in this region.
In addition to scalar tensor derived quantities, such as Trace(D) and FA, that are typically examined in population analysis of DTI data, we investigated the effects of outliers on the orientation of the principal eigenvector ε1, which is collinear with the orientation of highest diffusivity in each voxel. This directional information is used by diffusion based tractography algorithms. Not surprisingly, outliers induce ε1 perturbations that are larger in low anisotropy regions such as grey matter and CSF due to the fact that for more isotropic tensors the fiber orientation is poorly defined. This means that small perturbations may result in large apparent angular differences. Angular differences were found to be smaller in magnitude in areas of higher anisotropy. While the average orientation of ε1 in the population in the major white matter tracts is largely unaffected by outliers, there are a few exceptions, mainly the genu and splenium of the corpus callosum and the transverse pontine fibers. The magnitude of the angular difference is small in all major white matter tracts where anisotropy is high (maximum 1 to 2 degrees); however, these small errors may still have a significant impact on the results of both streamline and probabilistic tractography due to propagation of error effects (Lazar and Alexander 2003).
Differences in the angle and cone of uncertainty are apparent in the ventral frontal area, which has been previously identified as being affected by respiratory artifacts in EPI imaging by modulating the level of ghosting and distortion (Van de Moortele et al. 2002; Barry and Menon 2005; van Gelderen et al. 2007). Differences in orientation of highest diffusivity are also evident throughout the cerebellum, which is consistent with the aforementioned spurious appearance of anisotropic tissue with superior-inferior apparent fiber orientations. This is complemented by an increase in the dispersion of the individual eigenvectors around the mean (the normalized area of the cone of uncertainty is increased) when using RESTORE. Taken at face value this is a puzzling result. Why would RESTORE result in an increased directional variability in the population by removing artifactual data points? Remember that artifacts can manifest themselves by producing a bias more than increasing variability. In the low anisotropy regions of the cerebellum, artifacual data points create the presence of spurious anisotropy with consistent superior-inferior orientation. It has been shown that angular dispersion is higher in areas of low anisotropy (Jones 2003). Thus, when the spurious superior-inferior apparent fiber direction is removed by RESTORE, the resulting orientation of ε1 is intrinsically more variable because the tissue is more isotropic.
Similarly, we surprisingly found an increased variability of both FA and Trace(D) with RESTORE fitting in the population in areas occupied by CSF. Potential instabilities with RESTORE have been reported, mainly that this algorithm removes outlier data points from consideration in tensor fitting, but, if too many points are identified as outliers, then removing those points from the fitting will reduce the accuracy of the results (Chang et al. 2009). Moreover, the performance of RESTORE is expected to vary depending on the underlying noise and artifact distribution in the data. In general, removal of artifacts by outlier rejection is ideally suited for a situation in which a relatively small number of severely artifactual data points corrupt an otherwise relatively good set of data, creating a well identifiable bimodal distribution. On the contrary, when the number of artifactual data points is large and the effect of the artifacts is a broadening of the distribution, RESTORE type methods will be progressively less effective in correcting the data. It is easy to see that in the extreme case in which the effect of the artifacts is so random to result in a perfectly Gaussian but broader distribution of errors, RESTORE will reject points in a random fashion, de facto rejecting good and bad data points with equal probability. We believe that differences in the distribution of errors between brain parenchyma and CSF spaces could be a contributing factor in the different performance of RESTORE in these regions. For example, in the brain tissue cardiac related artifacts are large for a limited period of about 200 ms in the systolic phase of the cardiac cycle (Pierpaoli et al. 2003). Essentially, cardiac pulsation artifacts, and similarly respiratory artifacts, occur only occasionally creating the basis for a bimodal distribution of errors for which RESTORE excels, allowing us to capture these outliers very well in the ORP. In CSF areas, velocity gradients of flowing CSF spins are temporally more dispersed throughout the entire cardiac cycle resulting in an expanded temporal window in which artifactual data can be collected. Moreover, signal attenuation due to CSF flow can be compounded with other signal instabilities caused by blood flow in the vessels and choroid plexi.
Regardless of the explanation, an important practical take home message of our study is that RESTORE improves the quality of the DTI results in the population only in the brain parenchyma not in the ventricular areas. Estimating diffusion parameters in the ventricles is usually not of interest; however, we recommend not using RESTORE if the ventricles are of interest. One other cautionary note is that RESTORE needs adequate data redundancy to work properly, i.e. a sufficiently large number of diffusion encoding directions, or independent replicates of DWIs. We believe that the results of our study are representative of what should be expected with a reasonably redundant dataset, but different results would be found if the number of gradient directions and/or independent replicates of DWIs are insufficient for RESTORE to work properly.
In brain parenchyma, RESTORE robust tensor fitting generally reduces variance and normalizes the mean value of metrics which would otherwise be under- or over-estimated by the presence of outlier data points in a population. The magnitude of reduction in variance is small, but is heterogeneous across the brain, which has implications for the statistical analysis of DTI data. In general the variance of the measurement directly affects the statistical power of the experiment. The statistical power of the experiment is rarely considered in DTI studies, in part due to the difficulty of characterizing the expected variance in the population. Our results indicate that variance in the population is heterogeneous throughout the brain, which implies that the statistical power is also regionally varying in the brain. Moreover, the variance modulation when RESTORE is used also has a specific regional distribution, which implies that using RESTORE will affect the statistical power in some brain regions but not in others. Using the standard equation for statistical power, and the assumption that the diffusion quantity of interest is normally distributed across the population, it is found that the population size is proportional to the variance of the population over the square of the effect size. It is therefore possible to estimate the population size increase required if one chooses to use conventional fitting in place of using the RESTORE method while maintaining the same level of statistical power as with the RESTORE algorithm. We investigated this in 4 regions; the cerebellum and genu of the corpus callosum where there are a large number of outliers on the ORP map, and in the cingulum, and the posterior limb of the internal capsule where there are few outliers on the ORP. As expected, the difference in population size requirements is regionally varying in the brain, and also differs for FA and Trace(D). The cerebellum requires a population size increase of approximately 11% for FA and 4% for Trace(D), the genu of the corpus callosum requires an increase of approximately 4% for FA and 2% for Trace(D), while the regions with few outliers require no population size increase in order to maintain the same statistical power due to the differences of variance between conventional and RESTORE fitting. While the magnitude of the difference in variance is small between the two fitting algorithms, there is a regionally varying effect on the statistical power in the brain.
We found however that the strongest statistical effect of artifacts is to produce a bias in the values of tensor derived quantities rather than increasing their variance in the population. Voxel-wise analysis confirms this, with the result that inclusion of outlier data points will generally under-estimate FA and over-estimate Trace(D). Therefore, when considering a statistical analysis of a population, one needs to consider that the effect of outliers may not be the same for both patient and control populations. If one population contains more physiological noise than the other, one could find statistically significant results that are attributable to the presence of outlier data points as opposed to the presence of a disease or pathology. We recommend that investigators use the ORP, which can be downloaded from http://science.nichd.nih.gov/confluence/display/nihpd/Data+Downloads, to determine whether their statistically significant results are in regions which are affected by outlier data points. This can be done by using the provided mean FA image for registering their data into the space of the ORP and overlaying their results with the ORP. If the areas of interest coincide with regions highlighted by the ORP, it is recommended to investigate the source of the difference between the populations, and/or to use RESTORE to reevaluate the data to be confident that the significant results are due to disease, and not due to the presence of artifacts.
Research Highlights.
physiological noise artifacts in DTI show well defined regional patterns in the brain
these patterns are highly reproducible in a population of healthy subjects
artifacts bias diffusion tensor derived metrics rather than increasing their variance
robust tensor fitting is effective in reducing the effect of artifacts
ACKNOWLEDGEMENTS
We would like to thank Gary Hui Zhang for use of and help with the software DTI-TK (http://www.nitrc.org/projects/dtitk). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the NICHD Intramural Program, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke as part of the NIH MRI Study of Normal Pediatric Brain Development with supplemental funding from the NIH Neuroscience Blueprint.
APPENDIX A
In this appendix, we will describe the procedure of our noise estimation derived from a collection of the fitted sum of squares of the ordinary nonlinear least squares estimation of the diffusion tensor on a volumetric data. First, we shall denote the ratio of the residual sum of squares of the fit to the degrees of freedom ν = n−p, at voxel location (i,j,k). Note that n is the number of data points and p is 7 for the number of unknown parameters in the diffusion tensor estimation. It is known that follows the Chi-square distribution and follows the reduced Chi-square distribution (Bevington 1969) when the DW signals are of sufficiently high SNR and the data fit the model well (Koay and Basser 2006; Koay et al. 2006). In absence of artifactual data points, the sample mean of for all (i,j,k) from the region of interest should be close to 1. Therefore, one could estimate the noise level σ2, by imposing and get . However, the sample mean is not a robust measure and its value is biased by artifactual points, so we need to use a more robust metric such as the sample median. To use the sample median in place of the mean in theory a scaling factor α(ν) is needed (see Appendix B for the derivation) so that .
In our case the scaling factor was negligibly different from 1 so we did not apply it. Therefore, our procedure of noise estimation is as follows:
Step 1. Compute from the voxel locations (i,j,k) that are associated with a specific region of interest. In our case, the region of interest included the whole brain, masked to remove skull and background voxel values.
Step 2. Find the sample median of this collection of .
Step 3. Compute the estimated noise level as given by
APPENDIX B
In this appendix, we will outline the derivation of the scaling factor mentioned in Appendix A. Given that follows the reduced Chi-square distribution, we would like to compute the median of of the reduced Chi-square distribution so as to be able to use the sample median of more accurately.
For completeness, we give definitions of the probability densities needed in this work. The Chi-square probability density, gχ2, is given by
and the reduced Chi-square probability density, , can be expressed as:
To compute the median, μv, of the reduced Chi-square distribution, we use the following equation,
After some manipulations, it can be shown that (Koay et al. 2009)
where Q−1 is the inverse cumulative distribution function (CDF) of the gamma distribution. Specifically, the notation of Q−1 is exactly equivalent to the Mathematica function called InverseGammaRegularized. Listed here are some numerical values of μv from v=10 to v=100 at a step of 10, {0.934, 0.966, 0.977, 0.983, 0.986, 0.988, 0.99, 0.991, 0.992, 0.993}. It is clear from these numerical values that the median is lower in value than the mean, which is equal to 1. Therefore, , and
or
where the scaling factor, α(v), is related to μv, by α(v)=1/μv. Finally, we note that the scaling factor used in this study is α(53)=1.012 because μ53 is 0.987.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barry RL, Menon RS. Modeling and suppression of respiration-related physiological noise in echo-planar functional magnetic resonance imaging using global and one-dimensional navigator echo correction. Magn Reson Med. 2005;54(2):411–8. doi: 10.1002/mrm.20591. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 1994;103(3):247–54. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Bazin PL, Cuzzocreo JL, Yassa MA, Gandler W, McAuliffe MJ, Bassett SS, Pham DL. Volumetric neuroimage analysis extensions for the MIPAV software package. J Neurosci Methods. 2007;165(1):111–21. doi: 10.1016/j.jneumeth.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington P. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill Book Company; New York, NY: 1969. [Google Scholar]
- Chang L-C, Jones DK, Pierpaoli C. RESTORE: robust estimation of tensors by outlier rejection. Magn Reson Med. 2005;53(5):1088–95. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- Chang L-C, Walker L, Pierpaoli C. Making the Robust Tensor Estimation Approach: “RESTORE” more Robust. ISMRM 17th annual mtg; Honolulu, Hawaii. 2009. [Google Scholar]
- Henkelman RM. Measurement of signal intensities in the presence of noise in MR images. Medical Physics. 1985;12(2):232–233. doi: 10.1118/1.595711. [DOI] [PubMed] [Google Scholar]
- Jones DK. Determining and visualizing uncertainty in estimates of fiber orientation from diffusion tensor MRI. Magn Reson Med. 2003;49(1):7–12. doi: 10.1002/mrm.10331. [DOI] [PubMed] [Google Scholar]
- Jones DK, Pierpaoli C. Contribution of Cardiac Pulsation to Variability of Tractography Results. ISMRM 13th annual mtg; Miami Beach, Florida. 2005. [Google Scholar]
- Kellman P, McVeigh ER. Image reconstruction in SNR units: a general method for SNR measurement. Magn Reson Med. 2005;54(6):1439–47. doi: 10.1002/mrm.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay CG, Basser PJ. Analytically exact correction scheme for signal extraction from noisy magnitude MR signals. J Magn Reson. 2006;179(2):317–22. doi: 10.1016/j.jmr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Koay CG, Chang L-C, Carew JD, Pierpaoli C, Basser PJ. A unifying theoretical and algorithmic framework for least squares methods of estimation in diffusion tensor imaging. J Magn Reson. 2006;182(1):115–25. doi: 10.1016/j.jmr.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Koay CG, Chang LC, Pierpaoli C, Basser PJ. Error propagation framework for diffusion tensor imaging via diffusion tensor representations. IEEE Trans Med Imaging. 2007;26(8):1017–34. doi: 10.1109/TMI.2007.897415. [DOI] [PubMed] [Google Scholar]
- Koay CG, Nevo U, Chang LC, Pierpaoli C, Basser PJ. The elliptical cone of uncertainty and its normalized measures in diffusion tensor imaging. IEEE Trans Med Imaging. 2008;27(6):834–46. doi: 10.1109/TMI.2008.915663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay CG, Ozarslan E, Pierpaoli C. Probabilistic Identification and Estimation of Noise (PIESNO): a self-consistent approach and its applications in MRI. J Magn Reson. 2009;199(1):94–103. doi: 10.1016/j.jmr.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M, Alexander AL. An error analysis of white matter tractography methods: synthetic diffusion tensor field simulations. Neuroimage. 2003;20(2):1140–53. doi: 10.1016/S1053-8119(03)00277-5. [DOI] [PubMed] [Google Scholar]
- Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med. 2009;61(6):1336–49. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- Mangin JF, Poupon C, Clark C, Le Bihan D, Bloch I. Distortion correction and robust tensor estimation for MR diffusion imaging. Med Image Anal. 2002;6(3):191–8. doi: 10.1016/s1361-8415(02)00079-8. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes RG, Jezzard P, Clare S. Investigations on the efficiency of cardiac-gated methods for the acquisition of diffusion-weighted images. J Magn Reson. 2005;177(1):102–10. doi: 10.1016/j.jmr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Peterson DJ, Landman BA, Cutting LE. The impact of robust tensor estimation on voxel-wise analysis of DTI data. ISMRM 16th annual mtg; Toronto, Ontario, Canada. 2008. [Google Scholar]
- Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201(3):637–48. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Marenco S, Rohde GK, Jones DK, Barnett AS. Analyzing the contribution of cardiac pulsation to the variability of quantities derived from the diffusion tensor. ISMRM 11th annual mtg; Toronto, Ontario, Canada. 2003. [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett AS, Chang L-C, Koay CG, Pajevic S, Rohde GK, Sarlls J, Wu M. TORTOISE: an integrated software package for processing of diffusion MRI data. ISMRM 18th annual mtg; Stockholm, Sweden. 2010. [Google Scholar]
- Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004;51(1):103–14. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- Rohde GK, Barnett AS, Basser PJ, Pierpaoli C. Estimating intensity variance due to noise in registered images: applications to diffusion tensor MRI. Neuroimage. 2005;26(3):673–84. doi: 10.1016/j.neuroimage.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Skare S, Andersson JL. On the effects of gating in diffusion imaging of the brain using single shot EPI. Magn Reson Imaging. 2001;19(8):1125–8. doi: 10.1016/s0730-725x(01)00415-5. [DOI] [PubMed] [Google Scholar]
- Skare S, Hedehus M, Moseley ME, Li TQ. Condition number as a measure of noise performance of diffusion tensor data acquisition schemes with MRI. J Magn Reson. 2000;147(2):340–52. doi: 10.1006/jmre.2000.2209. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Van de Moortele PF, Pfeuffer J, Glover GH, Ugurbil K, Hu X. Respiration-induced B0 fluctuations and their spatial distribution in the human brain at 7 Tesla. Magn Reson Med. 2002;47(5):888–95. doi: 10.1002/mrm.10145. [DOI] [PubMed] [Google Scholar]
- van Gelderen P, de Zwart JA, Starewicz P, Hinks RS, Duyn JH. Real-time shimming to compensate for respiration-induced B0 fluctuations. Magn Reson Med. 2007;57(2):362–8. doi: 10.1002/mrm.21136. [DOI] [PubMed] [Google Scholar]
- Walker L, Chang L-C, Kanterakis E, Bloy L, Simonyan K, Verma R, Pierpaoli C. Regional Distribution of Outliers of Diffusion MRI in the Human Brain. ISMRM 16th annual mtg; Toronto, Ontario, Canada. 2008. [Google Scholar]
- Walker L, Chang L-C, Kanterakis E, Bloy L, Simonyan K, Verma R, Pierpaoli C. Statistical Assessment of the Effects of Physiological Noise and Artifacts in a Population Analysis of Diffusion Tensor MRI Data. ISMRM 17th annual mtg; Honolulu, Hawaii. 2009. [Google Scholar]
- Wang D, Strugnell W, Cowin G, Doddrell DM, Slaughter R. Geometric distortion in clinical MRI systems Part I: evaluation using a 3D phantom. Magn Reson Imaging. 2004;22(9):1211–21. doi: 10.1016/j.mri.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Wu M, Chang L-C, Walker L, Lemaitre H, Barnett AS, Marenco S, Pierpaoli C. Comparison of EPI distortion correction methods in diffusion tensor MRI using a novel framework. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 2):321–9. doi: 10.1007/978-3-540-85990-1_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Avants BB, Yushkevich PA, Woo JH, Wang S, McCluskey LF, Elman LB, Melhem ER, Gee JC. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE Trans Med Imaging. 2007;26(11):1585–97. doi: 10.1109/TMI.2007.906784. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Alexander DC, Gee JC. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med Image Anal. 2006;10(5):764–85. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]