Abstract
Background & Aims
The Notch receptor family regulates cell fate through cell-cell communication. CSL (CBF-1/RBP-jκ, Su(H), Lag-1) drives canonical Notch-mediated gene transcription during cell lineage specification, differentiation and proliferation in the hematopoietic system, the intestine, the pancreas and the skin. However, the functional roles of Notch in esophageal squamous epithelial biology remain unknown.
Methods
Normal esophageal keratinocytes were stimulated with calcium chloride to induce terminal differentiation. The squamous epithelia were reconstituted in organotypic three-dimensional culture, a form of human tissue engineering. Notch was inhibited in culture with a γ-secretase inhibitor or dominant negative mastermind-like1 (DNMAML1). The roles of Notch receptors were evaluated by in vitro gain-of-function and loss-of-function experiments. Additionally, DNMAML1 was targeted to the mouse esophagus by cytokeratin K14 promoter-driven Cre (K14Cre) recombination of Lox-STOP-Lox-DNMAML1. Notch-regulated gene expression was determined by reporter transfection, chromatin immunoprecipitation (ChIP) assays, quantitative reverse-transcription polymerase chain reactions (RT-PCR), Western blotting, immunofluorescence and immunohistochemistry.
Results
NOTCH1 (N1) was activated at the onset of squamous differentiation in the esophagus. Intracellular domain of N1 (ICN1) directly activated NOTCH3 (N3) transcription, inducing HES5 and early differentiation markers such as involucrin (IVL) and cytokeratin CK13 in a CSL-dependent fashion. N3 enhanced ICN1 activity and was required for squamous differentiation. Loss of Notch signaling in K14Cre;DNMAML1 mice perturbed esophageal squamous differentiation and resulted in N3 loss and basal cell hyperplasia.
Conclusions
Notch signaling is important for esophageal epithelial homeostasis. In particular, the crosstalk of N3 with N1 during differentiation provides novel, mechanistic insights into Notch signaling and squamous epithelial biology.
Keywords: NOTCH1, NOTCH3, esophageal epithelium, squamous differentiation
Introduction
The stratified squamous epithelia are regulated at an exquisite level. The esophageal epithelium consists of keratinocytes that migrate in an outward fashion towards the luminal surface. During migration, the cells undergo terminal differentiation in the suprabasal layer. Then, the cells are desquamated into the lumen and the epithelium is renewed. Cytokeratins CK5 and CK14 are expressed in undifferentiated and proliferative keratinocytes in the basal cell layer. Cytokeratins CK4 and CK13 are specifically found in the suprabasal layer of the esophagus and other non-cornifying squamous epithelia1. Involucrin (IVL) is also expressed in the suprabasal, but not basal keratinocytes of the esophagus2. Filaggrin (FLG) is a late differentiation marker localized to keratohyalin granules3.
The Notch pathway regulates cell fate and differentiation through cell-cell communication4. The Notch family comprises four structurally related single transmembrane receptor proteins Notch1–4 (N1–4). Ligand (Jagged and Delta-like) binding triggers a series of enzymatic cleavages of the receptor proteins mediated by metalloprotease and γ-secretase, thereby resulting in nuclear translocation of the intracellular domain of Notch (ICN)4. ICN forms a transcriptional activation complex containing a DNA binding transcription factor CSL (CBF-1/RBP-jκ, Su(H), Lag-1) and the coactivator Mastermind-like (MAML)5. Notch target genes include the hairy and enhancer of split (HES) and related (HEY) family of transcription factors.
Notch signaling in the squamous epithelium has been investigated in the mouse epidermis6. Prenatal CSL deletion impairs squamous differentiation with loss of early and late differentiation markers such as CK1/CK10 and filaggrin, causing barrier defects and death at birth due to severe dehydration7. N1 has been proposed to have a central role in suppressing keratinocyte proliferation and promoting terminal differentiation6. In fact, the N1 deficient epidermis displays progressive hyperplasia as mice age8 and becomes prone to chemical induced carcinogenesis6. While N2 or N3 cannot compensate for N1 loss, compound loss of these Notch paralogues results in barrier defects and tumorigenicity even in the presence of N1, and that causes a more severe phenotype than N1 deficiency alone when all N1, N2 and N3 are knocked out9, indicating the presence of interplay amongst the Notch family members in squamous epithelial homeostasis. N3 shows a 0r0elatively weak CSL-dependent transcriptional activity compared to the other Notch family members10. Although N3 is expressed in the epidermis7, 11, no skin phenotype has been observed in N3 knockout mice11. Thus, N3 regulation and its role in squamous epithelial biology remain to be elucidated.
In this study, we have investigated how Notch signaling contributes to esophageal epithelial homeostasis in vitro and in vivo to find that N1 can trigger a robust induction and activation of N3 as a unique target for canonical CSL-dependent transcription at the onset of squamous differentiation, and that a novel functional interplay between ICN1 and ICN3 has a critical role in transcriptional activation of cellular components essential in terminal differentiation. Notch inhibition not only impairs squamous differentiation, but leads to basal cell hyperplasia and dysplasia in the mouse esophagus, providing novel insight into the role of Notch signaling in squamous epithelial biology.
Materials and Methods
Mice
DNMAML1f/f mice carrying Lox-STOP-Lox dominant negative mastermind-like1 (DNMAML1) were described previously12. DNMAML1f/f mice were intercrossed with K14Cre transgenic mice13, targeting DNMAML1 into the basal cell layer of the esophageal epithelium. The K14Cre;DNMAML1 mice were compared with age-matched DNMAML1f/f and K14Cre mice as controls. All experiments were done under approved protocols from the University of Pennsylvania Institutional Animal Care and Use Committee and NIH guidelines.
Human esophageal tissues
Normal human esophageal tissues (n=7) used herein were described previously14.
Cell lines and treatment
Primary and telomerase-immortalized non-transformed diploid human esophageal keratinocytes EPC2, EPC2-hTERT, EPC1-hTERT are described elsewhere15–16. Cells were kept undifferentiated in Keratinocyte-serum free medium (Invitrogen, Carlsbad, CA), containing a low concentration (0.09 mM) of CaCl2. Compound E (Calbiochem, La Jolla, CA), a γ-secretase inhibitor (GSI) was reconstituted in DMSO.
Retrovirus and lentivirus-mediated gene transfer and RNA interference (RNAi)
Recombinant DNA work is described in supplementary Materials and Methods. Replication-incompetent retroviruses and lentiviruses were produced and transduced as described previously17. Retroviral MigRI vector was used to express constitutively ICN118 as well as a green fluorescent protein (GFP)-DNMAML1 fusion gene19. pTOF-DNMAML1 retroviral vector was created to express DNMAML1 in a tetracycline-regulatable (Tet-Off) manner. Lentiviral pGIPZ vector (Open Biosystems, Huntsville, AL) was used to express short hairpin RNA (shRNA) directed against N3 (N3-A, V2LHS_229748 and N3-B, V2LHS_93017), or a non-silencing scramble control sequence (RHS4346). Cells transduced with MigRI or pGIPZ were selected, except for ICN1 transduction experiments, by flow sorting for the cells expressing GFP at the brightest level (top 20%). Cells transduced with pTOF-DNMAML1 were selected with 1 μg/ml of puromycin for 7 days. Small interfering RNA (siRNA) directed against N1 (N1-A, HSS181550 and N1-B, HSS107249), or a non-silencing scramble control sequence (12935-300)(Invitrogen) was transfected transiently using the Lipofectamine™ RNAiMAX reagent (Invitrogen), following manufacturer's instructions.
Luciferase assays
Transient transfection and luciferase assays were done as described previously17. Briefly, 400 ng of 8xCBF1-luc (designated as 8xCSL-luc)20, a reporter plasmid containing eight copies of the CSL DNA binding consensus sequence or INV2.5-pGL321 containing a 2.5 kb IVL promoter (−2466 to +41)(IVL-luc) was transfected along with or without ICN expression plasmids22 or a control empty vector (p3XFLAG-CMV-7). Cells were incubated for 48 hrs before cell lysis in ICN co-transfection experiments. Alternatively, CaCl2 was added to the final concentration of 0.6 mM at 24 hrs after transfection and for additional 48 hrs before cell lysis. The mean of firefly luciferase activity was normalized with the co-transfected renilla luciferase activity.
Chromatin immunoprecipitation (ChIP) assays
ChIP was performed as described previously23. Briefly, chromatin samples were prepared from fixed 2.5 million cells and immunoprecipitated with rabbit IgG (#sc-3888, Santa Cruz), anti-N118, anti-CSL/RBP-jκ (#AB5790, Millipore), or anti-Histone H3 (#ab1791-100 Abcam) antibody. Purified DNA was subjected to real-time PCR with primers flanking potential CSL-binding sites at −12 kilobases (kb)(TTCCCACa) and −2.3 kbp (agCATGGGAGa) upstream the translation start and within intron 2 (nt TTCCCACg-TTCCCACa) of N3 (GenBank NG_009819). The N3 3' end region was examined as an off target control. The DNA quantity recovered from each ChIP sample is shown as the relative value to anti-Histone H3 ChIP samples. Supplementary Table 1 lists primer sequences. Data represent three independent experiments.
RNA isolation, cDNA synthesis and real-time reverse-transcription polymerase chain reactions (RT-PCR)
RNA isolation, cDNA synthesis and real-time RT-PCR were done as described previously17. Supplementary Table 2 lists used TaqMan® Gene Expression Assays (Applied Biosystems). Relative gene expression was determined based upon corresponding threshold cycle values and normalized to β-actin as an internal control.
Western blot analysis
Western blotting was done as described previously16–17. Supplementary Table 3 lists primary antibodies and the titers used for Western blotting.
Immunofluorescence (IF) and Immunohistochemistry (IHC)
IF and IHC were performed as described previously14, 16. Supplementary Table 4 summarizes antibodies, titers and specific conditions. Stained objects were examined with a Nikon Microphot microscope and imaged with a digital camera. Ki67 labeling index was determined by counting at least 600 cells per group.
Statistical analyses
Data from triplicate and hexaduplicate experiments in real-time RT-PCR, luciferase assays and Ki67 labeling index were presented as mean ± SE and were analyzed by two-tailed Student's t test. Fisher's exact test was done to assess genotype-phenotype association in mice. P <0.05 was considered to be statistically significant.
Results
CSL-mediated canonical Notch signaling is required at the onset of esophageal squamous differentiation
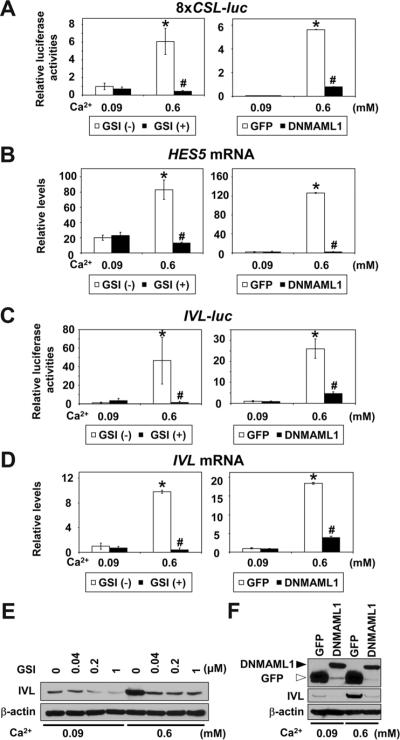
Calcium (Ca2+) induces keratinocyte differentiation24. Notch was activated in EPC2, EPC2-hTERT and EPC1-hTERT cells upon Ca2+ stimulation, which promotes cell-cell contact (Supplementary Figure 1), CSL-reporter activation and induction of HES5 and early differentiation markers IVL and CK13 in a temporal and dose-dependent manner (Figure 1 and Supplementary Figure 2). These Ca2+-induced responses were blocked effectively at the transcriptional, mRNA and protein levels by a GSI as well as DNMAML119, the latter antagonizing the MAML family of CSL-dependent transcriptional coactivator expressed in esophageal epithelial cells (Figure 1 and Supplementary Figure 3).
Figure 1.
Ca2+ activates Notch signaling to induce squamous differentiation in esophageal keratinocytes
EPC2-hTERT cells were exposed to Ca2+ at indicated concentrations in the presence or absence of GSI or DNMAML1 for 48 hrs after transfection in (A) and (C), or 72 hrs in (B), (D)–(F). (A) 8×CSL-luc reporter activities, (B) HES5 mRNA levels, (C) IVL-luc reporter activities, (D) IVL mRNA levels, (E) IVL protein levels, (F) DNMAML1 and IVL protein levels. Luciferase assays, real-time RT-PCR and Western blotting determined reporter activities, mRNA and protein, respectively. β-actin served as an internal or loading control in (B), (D), (E) and (F). GSI (−), DMSO only; GSI (+), 1 μM compound E; GFP, control vector in (A)–(D) and (F). *, P< 0.001 vs. 0.09 mM Ca2+ + GSI (−) or GFP; #, P< 0.001 vs. 0.6 mM Ca2+ + GSI (−) or GFP; n=6 in (A) and (C); n=3 in (B) and (D).
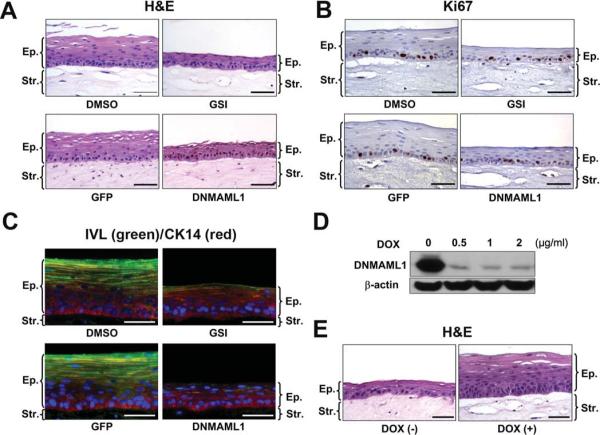
In the esophageal epithelium reconstituted in organotypic 3D culture, Notch inhibition suppressed sharply epithelial stratification as well as early (i.e. IVL and CK13) and late (FLG) differentiation markers without affecting basal cell marker (i.e. CK14) expression and proliferation (Figure 2 and supplementary Figure 4). These data suggest there is canonical Notch signaling in the early stages of esophageal squamous epithelial differentiation.
Figure 2.
Notch inhibition impairs squamous differentiation of esophageal epithelia reconstituted in organotypic 3D culture
EPC2-hTERT cells were grown in 3D (A–C and E) and monolayer (D) cultures in the presence or absence of 1 μM compound E (GSI) or DNMAML1, and subjected to H&E staining in (A) and (E), IHC for Ki67 in (B) and IF for CK14 (red) and IVL (green) in (C). Ki67 labeling index in (B) was 36.7 ± 9.7 for DMSO; 32.8 ± 4.2 for GSI; 33.9 ± 5.4 for GFP and 36.8 ± 3.8 for DNMAML1 without significant difference (n=6). In (D), Western blotting determined DNMAML1 repression by doxycycline (DOX) at indicated concentrations in the Tet-Off system. DOX (−), 0 μg/ml DOX; DOX (+), 2 μg/ml DOX in (E). Scale bar, 50 μm.; Ep., epithelium; Str., stroma.
The lack of squamous differentiation in the presence of Notch inhibitors can be accounted for by depletion of committed cells that are capable of undergoing differentiation in response to Ca2+ stimulation. When DNMAML1 was repressed by doxycycline in the Tet-Off system, Notch functions and squamous differentiation were restored in both monolayer and 3D cultures (Figure 2D and E, and Supplementary Figure 5), consistent with Notch-mediated direct transcriptional regulation of squamous differentiation.
N1 and N3 are induced and activated during esophageal squamous differentiation
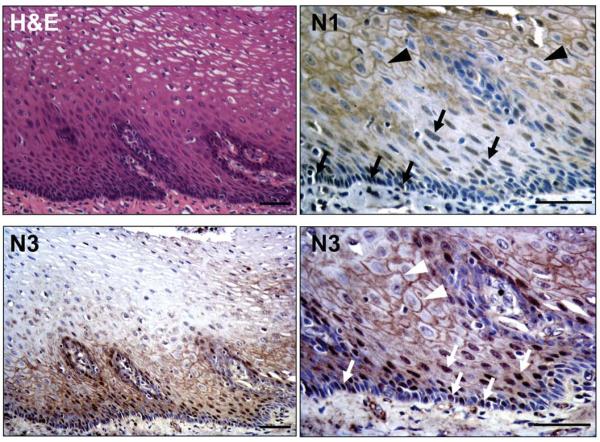
We next explored whether the Notch receptor family members are involved in the regulation of squamous differentiation in the esophagus. N1 and N3 were expressed on the cell membrane of the suprabasal cells within the normal human esophageal epithelium where nuclear localization of both N1 and N3 was also observed in the cells within the first 3–4 layers above the basal cell layer (Figure 3). Additionally, a subset of basal cells showed nuclear N1 and N3 expression (Figure 3).
Figure 3.
N1 and N3 expression in normal human esophageal epithelium
Representative H&E and IHC. N1 and N3 were detected in the nucleus (arrows) and on the membrane (arrowheads). Scale bar, 50 μm.
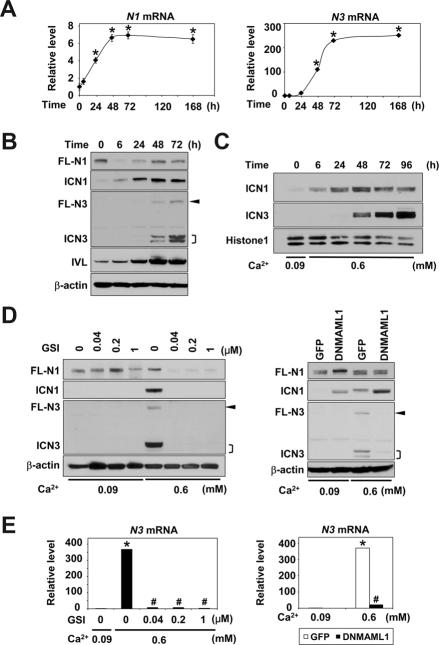
We determined also the expression of N1–4 in primary and immortalized esophageal cells by real-time RT-PCR. Threshold cycle values implied relatively high basal levels of N1 and N2 mRNA and very low to undetectable levels of N3 and N4 mRNA (data not shown). Ca2+ increased N1 and N3, but not N2 and N4 mRNA (Figure 4A and Supplementary Figure 6), and that was reflected by the HES5, IVL and CK13 mRNA levels (Figure 1, Supplementary Figures 2). Constitutive N1 mRNA expression was corroborated by full-length (FL)-N1 protein detected by Western blotting in unstimulated EPC2-hTERT cells (Figure 4B, time 0). Upon Ca2+ stimulation, FL-N1 protein was downregulated within 6 hrs while ICN1 emerged in a reciprocal fashion (Figure 4B), indicating N1 activation. Both FL-N1 and ICN1 levels were increased thereafter (Figure 4B). Since ICN1 is generated by enzymatic FL-N1 cleavage triggered by ligand binding, ICN1 upregulation (Figure 4B, 6h–48h) can be accounted for by an increase in the N1 mRNA level (Figure 4A), consistent with autoregulation of N1 transcription23, leading to de novo FL-N1 protein synthesis (Figure 4B).
Figure 4.
Sequential induction and activation of N1 and N3 during Ca2+-induced squamous differentiation in esophageal keratinocytes
EPC2-hTERT cells were exposed to 0.6 mM Ca2+ for indicated time periods in (A)–(C) or 72 hrs in (D) and (E), and subjected to real-time RT-PCR in (A) and (E) or Western blotting in (B)–(D). In (D) and (E), Ca2+ stimulation was done in the presence or absence of GSI or DNMAML1. Compound E (GSI) was used at indicated concentrations. β-actin served as an internal or loading control in (A), (B), (D) and (E). Histone H1 served as a loading control for nuclear extracts in (C). *, P< 0.001 vs. time 0 (n=3) in (A). *, P< 0.001 vs. 0.09 mM Ca2+ + 0 μM GSI or GFP; #, P< 0.001 vs. 0.6 mM Ca2+ + 0 μM GSI or GFP (n=3) in (E). FL-N, full-length Notch; ICN, intracellular Notch. Arrowhead, FL-N3;], doublets of ICN3.
Interestingly, N3 mRNA induction occurred more robustly and in a delayed fashion compared to N1 mRNA (Figure 4A). Western blotting detected neither FL-N3 nor ICN3 prior to Ca2+ stimulation in EPC2-hTERT cells (Figure 4B). Consistent with a late N3 mRNA induction, both FL-N3 and ICN3 levels increased dramatically at 48 hrs after Ca2+ stimulation (Figure 4A and B). Such a successive induction pattern of ICN3 preceded by ICN1 was documented in nuclear extracts (Figure 4C), implying sequential activation of N1 and N3.
Ligand-induced activation of N1 and N3 was suggested by the finding that the GSI blocked both ICN1 and ICN3 generation upon Ca2+ stimulation (Figure 4D). Interestingly, GSI suppressed FL-N3 and N3 mRNA (Figure 4D and E), suggesting that N3 per se may be subjected to regulation by Notch signaling. Consistent with this premise, DNMAML1 prevented Ca2+ from inducing N3 mRNA and FL-N3 protein (Figure 4D and E). Importantly, ICN3 was found in the nuclear fraction by 48 hrs after Ca2+ stimulation and that was suppressed by GSI or DNMAML1 (Figure 4C and Supplementary Figure 7A and B). Moreover, repression of DNMAML1 by doxycycline in the Tet-Off system augmented N3 mRNA and ICN3 induction (Supplementary Figure 7C and D), reinforcing the notion of direct CSL-mediated induction and nuclear translocation of the activated N3. N3 activation upon Ca2+-mediated squamous differentiation was observed also in primary mouse esophageal keratinocytes, and was antagonized by GSI (Supplementary Figure 8). Although GSI also suppressed N1 mRNA and ICN1 induction by Ca2+, DNMAML1 augmented N1 mRNA and ICN1 expression upon Ca2+ stimulation (Figure 4D and Supplementary Figure 9), suggesting that N1 mRNA induction may be CSL-independent. These observations imply dynamic changes in expression and activation of N1 and N3 during esophageal squamous differentiation.
N1 induces N3 through CSL-dependent transcription
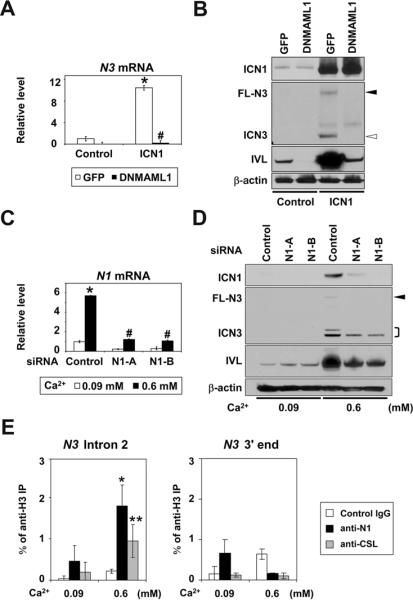
N1 activation preceded N3 induction (Figure 4A–C). This prompted us to investigate the possibility that N1 may regulate N3 directly. To this end, ICN1 was ectopically expressed in EPC2-hTERT cells. ICN1 not only activated the CSL-reporter but also induced HES5, CK13 and IVL whereas DNMAML1 blocked all these activities (supplementary Figure 10). ICN1 induced N3 mRNA in the absence of DNMAML1 (Figure 5A), indicating that N1 may induce N3 through CSL-dependent transcription. Moreover, Western blotting demonstrated that DNMAML1 blocked not only FL-N3 and ICN3, but also IVL proteins (Figure 5B).
Figure 5.
ICN1 directly induces and activates N3 through CSL-dependent transcription in esophageal keratinocytes
EPC2-hTERT cells expressing either DNMAML1 or GFP (control) were transduced with ICN1 to determine (A) N3 mRNA and (B) N3 protein levels 72 hrs after retrovirus infection. In (C) and (D), EPC2-hTERT cells were stimulated with Ca2+ for 72 hrs following N1 siRNA transfection to determine indicated molecules. Real-time RT-PCR and Western blotting determined mRNA and protein, respectively. β-actin served as an internal or loading control in (A)–(D). *, P< 0.001 vs. GFP + control or 0.09 mM Ca2+ + siRNA control; #, P< 0.001 vs. ICN1 + GFP or 0.6 mM Ca2+ + siRNA control; n=3 in (A) and (C). Closed arrowhead, FL-N3; open arrowhead, ICN3;], doublets of ICN3. In (E), cells were stimulated with Ca2+ for 48 hrs for ChIP assays with indicated antibodies. *, P< 0.01, **, P< 0.05 vs. 0.09 mM Ca2+; n=3.
We next addressed whether N1 is required for N3 induction by RNAi experiments. N1 knockdown compromised Ca2+-mediated induction and activation of N3 as well as Notch target genes (Figure 5C and D, Supplementary Figure 11), implying N1 in N3-mediated squamous differentiation.
Furthermore, ChIP assays documented Ca2+-induced binding of ICN1 and CSL to the intron 2 of the N3 gene containing a putative CSL binding cis-element (Figure 5E and supplementary Figure 12), providing evidence for direct transcriptional regulation of N3 by N1.
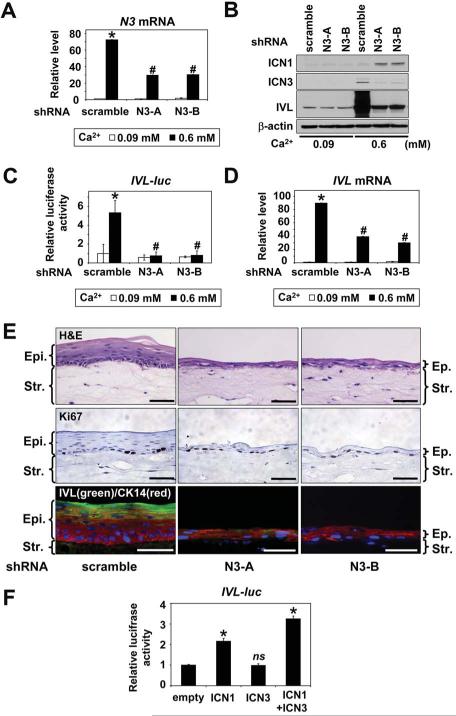
N3 cooperates with N1 for esophageal squamous differentiation
To elucidate the role of N3 in squamous differentiation, we established two independent lines of EPC2-hTERT cells of stable N3 knockdown. The RNAi effect was validated by inhibition of Ca2+- or ICN1-mediated N3 mRNA and ICN3 protein induction (Figure 6A and B, and supplementary Figure 13A and B). N3 knockdown resulted in suppression of the IVL promoter activation as well as IVL mRNA and IVL protein induction (Figure 6B–D, and supplementary Figure 13B and C). Additionally, N3 knockdown suppressed HES5 and CK13 induction (Supplementary Figure 13D and E), implying N3 is involved in their transcriptional regulation. Moreover, in organotypic 3D culture, N3 knockdown impaired squamous epithelial stratification and IVL expression without affecting basal cell proliferation (Figure 6E). Interestingly, ICN1 was induced upon Ca2+ stimulation in N3 knockdown cells without circumventing the N3 knockdown effect upon IVL expression (Figure 6B). Given the role of N3 in IVL transcriptional activation, we carried out transient transfection of ICN1 and ICN3, either alone or in combination. ICN1, but not ICN3, activated the IVL promoter. However, ICN3 enhanced significantly the ICN1-mediated IVL promoter transactivation in EPC2-hTERT cells (Figure 6F). These findings indicate cooperation between N1 and N3 in squamous differentiation.
Figure 6.
N3 is required and cooperates with N1 during esophageal squamous differentiation
EPC2-hTERT cells stably expressing two independent shRNA sequences directed against N3 (N3-A and N3-B) or control shRNA were stimulated with 0.6 mM Ca2+ for 72 hrs in (A), (B) and (D), or 48 hrs after IVL-luc transfection in (C). (A), N3 mRNA; (B), ICN1, ICN3 and IVL protein levels; (C), IVL-luc activities; and (D), IVL mRNA levels. Luciferase assays, real-time RT-PCR and Western blotting determined reporter activities, mRNA and protein, respectively. β-actin served as an internal or loading control. *, P< 0.001 vs. 0.09 mM Ca2+ + scramble; #, P< 0.001 vs. 0.6 mM Ca2+ + scramble; n=3 in (A) and (D); n=6 in (C). In (E), cells were grown in organotypic 3D culture and subjected to H&E staining (top panels), IHC for Ki67 (middle panels) and IF for CK14 (red) and IVL (green)(lower panels). Scale bar, 50 μm.; Ep., epithelium; Str., stroma. In (F), EPC2-hTERT cells were transfected with IVL-luc along with an empty vector (200 ng), ICN1 (200 ng), ICN3 (200 ng), or in combination (100 ng each) for luciferase assays. *, P< 0.001 vs. empty; ns, not significant vs. empty; n=6 in (F).
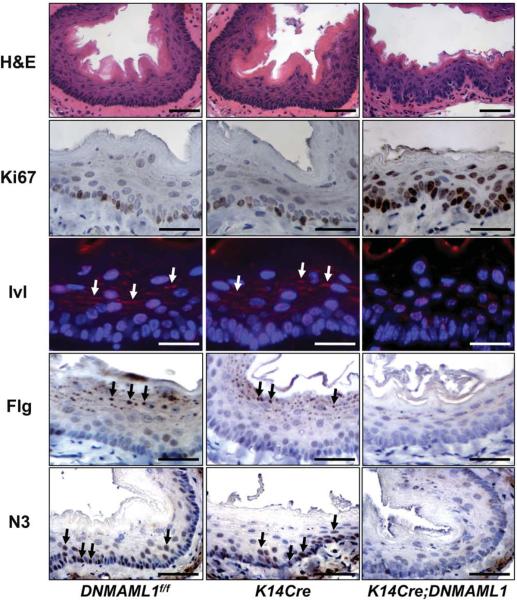
Notch inhibition in the mouse esophagus leads to deregulated squamous differentiation and aberrant basal cell proliferation with Notch3 downregulation
To determine the functional consequences of Notch inhibition in vivo, we targeted DNMAML1 in the mouse esophagus. Born in the anticipated Mendelian ratio, the K14Cre;DNMAML1 mice developed normally, yet four out of ten mice exhibited weight loss by 3 months of age. Histological examination revealed basal cell hyperplasia and/or dysplasia with disorganized nuclear polarity associated with an increase in Ki67 positive cells in the affected mouse esophagi. Neither the rest of K14Cre;DNMAML1 nor age-matched control mice presented histological changes (Figure 7). Nevertheless, there was a statistically significant genotype-phenotype association in the affected K14Cre;DNMAML1 mouse esophagi, compared to DNMAML1f/f or K14Cre controls (P=0.023, n=10). There was a marked reduction in the number and size of keratohyalin granules in the suprabasal stratum granulosum cell layer in the affected K14Cre;DNMAML1 esophagi, compared to unaffected K14Cre;DNMAML1 or control mouse esophagi, suggesting impaired terminal differentiation. This was corroborated by decreased Ivl and Flg expression (Figure 7). Finally, N3 expression was diminished greatly in the affected K14Cre;DNMAML1 esophagi (Figure 7), implicating CSL-dependent N3 expression in vivo. In aggregate, DNMAML1 perturbed Notch3 expression as well as the squamous differentiation program through inhibition of CSL-dependent transcription in the mouse esophagus.
Figure 7.
DNMAML1 induce basal cell hyperplasia/dysplasia impairing squamous differentiation and N3 expression in the mouse esophageal epithelium
Representative K14Cre;DNMAML1 and control (DNMAML1f/f and K14Cre) mouse esophagi were subjected to H&E staining, IHC for Ki67, Flg, and N3, or IF for Ivl. Ki67 labeling index, 19.9 ± 5.0% for DNMAML1f/f; 21.8 ± 4.8% for K14Cre (not significant vs. DNMAML1f/f); and 69.6 ± 5.5% for K14Cre;DNMAML1 (P< 0.001 vs. DNMAML1f/f and K14Cre)(n=6). Arrows denotes Ivl cytoplasmic staining. Arrowheads indicate Flg localized to keratohyalin granules. Scale bar, 50 μm.
Discussion
Our data indicate that the Notch pathway plays a critical role in esophageal squamous differentiation involving the canonical CSL-mediated transcriptional network (Figures 1–4). A novel functional interplay between N1 and N3 appears to be essential as N1 transcriptionally activates N3 to drive squamous differentiation (Figures 5 and 6). In mouse, Notch inhibition not only impaired esophageal squamous differentiation, but caused basal cell hyperplasia and dysplasia (Figure 7). These findings establish a novel model whereby crosstalk between N1 and N3 regulates squamous differentiation (Supplementary Figure 14).
To our knowledge, this is the first demonstration of functionally active Notch signaling in normal esophageal epithelial cell biology. Both primary and immortalized human esophageal keratinocytes underwent terminal differentiation and formed stratified squamous epithelium in 3D culture in a CSL-dependent manner. Notch inhibition abrogated early and late differentiation in culture as well as in mice. GSI induces goblet cell metaplasia in the rat intestine without affecting normal esophageal squamous epithelium after five daily intraperitoneal injections25. However, long-term GSI treatment may be required before GSI takes effect in the esophagus, given slower epithelial turnover (> 14 days)26. DNMAML1 did not affect basal cell proliferation in 3D culture (Figure 2). Thus, esophageal hyperplasia and dysplasia in mice may be explained by non-cell autonomous effects of Notch inhibition9.
Loss of Notch signaling in the skin leads to barrier defects and squamous cell carcinoma9 as well as atopic dermatitis-like disease27. Thus, our mouse model may have implication in esophageal diseases such as cancer and eosinophilic esophagitis. Interestingly, focal inflammation was noted in a few K14Cre;DNMAML1 esophagi (Shinya Ohashi et al. unpublished data). Only forty percent of the K14Cre;DNMAML1 mice displayed esophageal phenotypes at three months after birth although hair loss occurred with 100% penetrance rate, implying hair follicle dysfunction (John T. Seykora, manuscript in preparation). We suspect mosaicism in the K14Cre;DNMAML1 mouse esophagus as a possible explanation. Such a difference in Cre-mediated recombination efficiency between the esophagus and the epidermis may be accounted for by the fact that CK14 is induced postnatally in the esophagus28.
Induction of CK13, an esophageal-specific cytokeratin, implies tissue-specific differentiation while other markers such as IVL and FLG are shared with other squamous epithelia including that in the epidermis. Lack of Ivl and Flg expression in the K14Cre;DNMAML1 mouse is in concordance with impaired differentiation by embryonic K14Cre-mediated Csl loss in the mouse epidermis7. Similar to CK13 expression in esophageal cells, Ca2+ induces skin-specific suprabasal cytokeratins CK1 and CK10 in a Csl-dependent fashion7. However, there may be certain differences in the modes of Notch signaling during squamous differentiation between the esophagus and the skin, which includes hair follicle and interfollicular keratinocytes that are regulated differentially.
First, Ivl can be induced in a Csl-independent manner in mouse skin keratinocytes29. By contrast, our data point to CSL-dependent IVL transcription in esophageal keratinocytes. Second, Hes1 is induced most predominantly amongst the HES/HEY family of transcription factors upon Notch activation in epidermal keratinocytes7 where Hes1 regulates Hey1/Hey2 and other Notch downstream target molecules such as p21 and Wnt430. In human esophageal cells, Ca2+ induced HES5 most robustly (Figures 1 and Supplementary Figure 2) with HES1 induction only to a modest extent (data not shown). Functional redundancy of Hes1 and Hes5 has been implicated in neuronal differentiation31. However, Hes5 determines T-cell fate depending upon the ligand density while Hes1 is indispensible for T-cell differentiation32. Our data imply N3 serving an important functional role in HES5 induction.
ICN3, a weak transcriptional activator10, failed to stimulate the IVL promoter by itself, but enhanced ICN1-mediated IVL transactivation (Figure 6F). Our data suggest a novel modulatory role of N3 upon N1-mediated IVL induction. Although ICN and CSL binding was not validated in this study, four potential CSL-binding cis-elements exist within the 2.5 kb promoter sequence in IVL-luc construct. It is tempting to speculate that ICN1-ICN3 heterodimers may be formed on the IVL promoter to recruit histone acetyltransferases, chromatin remodeling factors and other transcription factors as proposed previously33. AP-1 and SP1 also regulate IVL34. Thus, further study will be needed to identify factors cooperating with ICN1, ICN3 and CSL to regulate IVL.
N1-mediated CSL-dependent N3 transcription agrees with N3 mRNA induction by ICN135. Our ChIP assay data establish a direct link between ICN1 and N3. N3 downregulation in the K14Cre;DNMAML1 mouse esophagus supports this notion. What is the biological role of N3 in the esophagus? Reminiscent of our K14Cre;DNMAML1 mouse phenotypes, esophageal hyperplasia has been noted when N3 was deleted in the N1N2 compound knockout mice, phenocopying the γ-secretase deficient skin11. We did not observe apoptosis upon N3 knockdown or DNMAML1 expression (data not shown). However, N3 may inhibit apoptosis through AKT36 and the mitogen-activated protein kinase pathways37. AKT activation by epidermal growth factor receptor (EGFR) leads to esophageal hyperplasia suppressing late differentiation in organotypic 3D culture38. Since EGFR signaling negatively regulates N139, it will be of great interest to explore the role of N3 in this context.
In conclusions, our innovative approaches reveal novel functional crosstalk between N1 and N3 during differentiation, providing mechanistic insights into the role of Notch signaling in esophageal squamous epithelial biology.
Supplementary Material
Acknowledgments
The authors thank Mr. Ben Rhoades, Ms. Daniela Budo, Dr. Gary P. Swain and Dr. Xiaoping Yang for technical assistance, members of the Nakagawa lab for helpful discussions as well as Drs. Tao Wang (University of Manchester), Anthony J. Capobianco (University of Miami), Raphael Kopan (Washington University), Marjorie A. Phillips and Robert H. Rice (University of California) for reagents.
Funding: This study was supported in part by NIH Grants R01DK077005 (to SO, MN, HN), P01-CA-098101 (Mechanisms of Esophageal Carcinogenesis to HN, AJK, MH, JAD), P30-DK050306 (to RK and Momo N), AGA Foundation Student Research Fellowship Award (to Momo N), and P30-DK050306 (to the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases).
Abbreviations used in this paper
- ChIP
chromatin immunoprecipitation
- CSL
CBF-1/RBP-jκ, Su(H), Lag-1
- DAPI
4',6-diamidino-2-phenylindole
- DNMAML1
dominant negative mastermind-like1
- EGFR
epidermal growth factor receptor
- DOX
doxycycline
- FLG
Filaggrin
- FL
full-length
- GSI
γ-secretase inhibitor
- GFP
green fluorescent protein
- HES
hairy and enhancer of split
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- ICN
Intracellular domain
- IVL
Involucrin
- kb
kilobases
- K14Cre
K14 promoter-driven Cre recombinase
- MAML
Mastermind-like
- N
NOTCH
- RT-PCR
reverse-transcription polymerase chain reactions
- RNAi
RNA interference
- shRNA
short hairpin RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors disclose no conflicts.
References
- 1.Viaene AI, Baert JH. Expression of cytokeratin-mRNAs in squamous-cell carcinoma and balloon-cell formation of human oesophageal epithelium. Histochem J. 1995;27:69–78. doi: 10.1007/BF00164174. [DOI] [PubMed] [Google Scholar]
- 2.Banks-Schlegel S, Green H. Involucrin synthesis and tissue assembly by keratinocytes in natural and cultured human epithelia. J Cell Biol. 1981;90:732–7. doi: 10.1083/jcb.90.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale BA, Scofield JA, Hennings H, et al. Identification of filaggrin in cultured mouse keratinocytes and its regulation by calcium. J Invest Dermatol. 1983;81:90s–5s. doi: 10.1111/1523-1747.ep12540769. [DOI] [PubMed] [Google Scholar]
- 4.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27:5138–47. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- 6.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–23. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanpain C, Lowry WE, Pasolli HA, et al. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–35. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Basak JM, Demehri S, et al. Bi-compartmental communication contributes to the opposite proliferative behavior of Notch1-deficient hair follicle and epidermal keratinocytes. Development. 2007;134:2795–806. doi: 10.1242/dev.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellavia D, Checquolo S, Campese AF, et al. Notch3: from subtle structural differences to functional diversity. Oncogene. 2008;27:5092–8. doi: 10.1038/onc.2008.230. [DOI] [PubMed] [Google Scholar]
- 11.Pan Y, Lin MH, Tian X, et al. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–43. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Tu L, Fang TC, Artis D, et al. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–42. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasioukhin V, Degenstein L, Wise B, et al. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–6. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ, Natsuizaka M, Ohashi S, et al. Hypoxia activates the cyclooxygenase-2-prostaglandin E synthase axis. Carcinogenesis. 2010;31:427–34. doi: 10.1093/carcin/bgp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada H, Nakagawa H, Oyama K, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–38. [PubMed] [Google Scholar]
- 16.Andl CD, Mizushima T, Nakagawa H, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–30. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi S, Natsuizaka M, Wong GS, et al. EGFR and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–84. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aster JC, Xu L, Karnell FG, et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–15. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maillard I, Weng AP, Carpenter AC, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 20.Jeffries S, Capobianco AJ. Neoplastic transformation by Notch requires nuclear localization. Mol Cell Biol. 2000;20:3928–41. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips MA, Qin Q, Rice RH. Identification of an involucrin promoter transcriptional response element with activity restricted to keratinocytes. Biochem J. 2000;348(Pt 1):45–53. [PMC free article] [PubMed] [Google Scholar]
- 22.Ong CT, Cheng HT, Chang LW, et al. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–19. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 23.Yashiro-Ohtani Y, He Y, Ohtani T, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–76. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuspa SH, Kilkenny AE, Steinert PM, et al. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J Cell Biol. 1989;109:1207–17. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menke V, van Es JH, de Lau W, et al. Conversion of metaplastic Barrett's epithelium into post-mitotic goblet cells by gamma-secretase inhibition. Dis Model Mech. 2010;3:104–10. doi: 10.1242/dmm.003012. [DOI] [PubMed] [Google Scholar]
- 26.Squier CA, Kremer MJ. Biology of oral mucosa and esophagus. J Natl Cancer Inst Monogr. 2001:7–15. doi: 10.1093/oxfordjournals.jncimonographs.a003443. [DOI] [PubMed] [Google Scholar]
- 27.Dumortier A, Durham AD, Di Piazza M, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of notch signaling in the murine skin. PLoS One. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd C, Yu QC, Cheng J, et al. The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J Cell Biol. 1995;129:1329–44. doi: 10.1083/jcb.129.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangarajan A, Talora C, Okuyama R, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen BC, Lefort K, Mandinova A, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–42. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtsuka T, Ishibashi M, Gradwohl G, et al. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varnum-Finney B, Dallas MH, Kato K, et al. Notch target Hes5 ensures appropriate Notch induced T- versus B-cell choices in the thymus. Blood. 2008;111:2615–20. doi: 10.1182/blood-2007-03-079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam Y, Sliz P, Pear WS, et al. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–8. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckert RL, Crish JF, Efimova T, et al. Regulation of involucrin gene expression. J Invest Dermatol. 2004;123:13–22. doi: 10.1111/j.0022-202X.2004.22723.x. [DOI] [PubMed] [Google Scholar]
- 35.Luo B, Aster JC, Hasserjian RP, et al. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–67. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Prince CZ, Hu X, et al. HRT1 modulates vascular smooth muscle cell proliferation and apoptosis. Biochem Biophys Res Commun. 2003;308:596–601. doi: 10.1016/s0006-291x(03)01453-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Prince CZ, Mou Y, et al. Notch3 signaling in vascular smooth muscle cells induces c-FLIP expression via ERK/MAPK activation. Resistance to Fas ligand-induced apoptosis. J Biol Chem. 2002;277:21723–9. doi: 10.1074/jbc.M202224200. [DOI] [PubMed] [Google Scholar]
- 38.Oyama K, Okawa T, Nakagawa H, et al. AKT induces senescence in primary esophageal epithelial cells but is permissive for differentiation as revealed in organotypic culture. Oncogene. 2006 doi: 10.1038/sj.onc.1210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolev V, Mandinova A, Guinea-Viniegra J, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–11. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.