Abstract
BACKGROUND
Differentiating ulcerative colitis (UC) from Crohn's colitis (CC) can be difficult and may lead to inaccurate diagnoses in up to 30 percent of inflammatory bowel disease (IBD) patients. Much of the diagnostic uncertainty arises from the overlap of clinical and histologic features. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) permits a histology-directed cellular protein analysis of tissues. As a pilot study, we evaluated the ability of histology-directed MALDI-MS to determine the proteomic patterns for potential differences between CC and UC specimens.
METHODS
Mucosal and submucosal layers of CC and UC colon resection samples were analyzed after histologic assessment. To determine whether MALDI-MS would distinguish inflammation, the uninflamed [n=21] vs. inflamed submucosa [n=22] were compared in UC and the uninflamed [n=17] vs. inflamed submucosa [n=20] in CC. To determine whether there were proteomic differences between the colitides, the uninflamed UC submucosa [n=21] was compared vs. the uninflamed CC submucosa [n=17], the inflamed UC submucosa [n=22] was compared vs. the inflamed CC submucosa [n=20], and inflamed UC mucosa vs. inflamed CC mucosa. Pair-wise statistics comparisons of the subsets were performed.
RESULTS
Pair-wise comparative analyses of the clinical groups allowed indentifying subsets of features important for classification. Comparison of inflamed vs. uninflamed CC submucosa showed two significant peaks: m/z 6445 (p=0.0003) and 12692 (p=0.003). In the case of inflamed vs. uninflamed UC submucosa, several significant differentiating peaks were found, but classification was worse. Comparisons of the proteomic spectra of inflamed submucosa between UC and CC identified two discrete significant peaks: m/z 8773 (p=0.006) and 9245 (p=0.0009). Comparisons of the proteomic spectra of uninflamed submucosa between UC and CC identified three discrete significant peaks: m/z 2778 (p=0.005), 9232 (p=0.005), and 9519 (p=0.005). No significantly different features were found between UC and CC inflamed mucosa.
CONCLUSION
MALDI-MS was able to distinguish CC and UC specimens while profiling the colonic submucosa. Further analyses and protein identification of the differential protein peaks may aid in accurately diagnosing IBD and developing appropriate personalized therapies.
Keywords: Ulcerative colitis, Crohn's colitis, Colon tissue profiling, Proteomics
INTRODUCTION
Current therapeutic options for inflammatory colitis, including ulcerative colitis (UC) and Crohn's colitis (CC), involve pharmacologic therapy1 or surgical resection or both.2,3 The distinction between CC and UC is of the utmost importance especially when determining the candidacy of a patient for restorative proctocolectomy (RPC). Differentiating CC and UC can be difficult at times even with a combination of clinical, radiologic, endoscopic and histopathology examinations. Up to 15 percent of colitis cases are labeled as “indeterminate colitis” (IC) when non-definitive evaluations have been made. In addition, up to 15 percent of UC patients that undergo RPC will have their diagnosis changed to CC based on final pathology or development of Crohn's disease in the ileal pouch.2,4,5 This diagnostic dilemma and the potential morbidity from an incorrect diagnosis and inappropriate operative therapy underscores the necessity of research efforts aimed at accurate colitis diagnosis.6–9 Early diagnostic accuracy of inflammatory colitis will lead to appropriate therapeutic medical options and will aid in determining appropriate candidates for RPC.
Cellular dysfunction leading to biopathophysiology may result in alterations of the protein microenvironment in the affected tissues.10 This generalized premise is applicable to the inflammatory colitides and includes colonic mucosal and submucosal protein microenviroments. Identification of any differentially expressed proteins in colonic tissues has significant potential to supplement known clinicopathologic variables currently used to differentiate the colitides.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) permits a histology-directed cellular protein analysis of tissues. Utilizing MALDI-MS, we evaluated the protein microenvironments in colonic mucosal and submucosal tissues to determine if molecular differences between the two inflammatory colitides exist. We hypothesized that MALDI-MS would effectively distinguish the presence of inflammation in the colonic mucosa and submucosa in CC and UC specimens. We also hypothesized that CC and UC are distinct pathologies that can be differentiated based on protein expression using specific proteomic peaks.
METHODS AND METHODS
PATIENT AND SAMPLE SELECTION
Snap frozen (−80°C) colon samples from 51 patients with inflammatory colitis (n=24 CC and n=27 UC) were available from the Vanderbilt Gastrointestinal Biospecimen Repository or from the Cooperative Human Tissue Network. The diagnosis for each patient was determined based on standard clinical and pathologic features11–13 and represented a consensus among treating physicians. A gastrointestinal pathologist blinded to clinical diagnosis confirmed the colitis diagnoses for each patient using de-identified final surgical pathology reports and hematoxylin and eosin (H & E) slides. For each sample, the mucosal and submucosal layers were analyzed.
This study was conducted in accordance with the Second International Helsinki Declaration14 and approved by the Vanderbilt Institutional Ethical Review Board Committee (file number 080898).
TISSUE PREPARATION
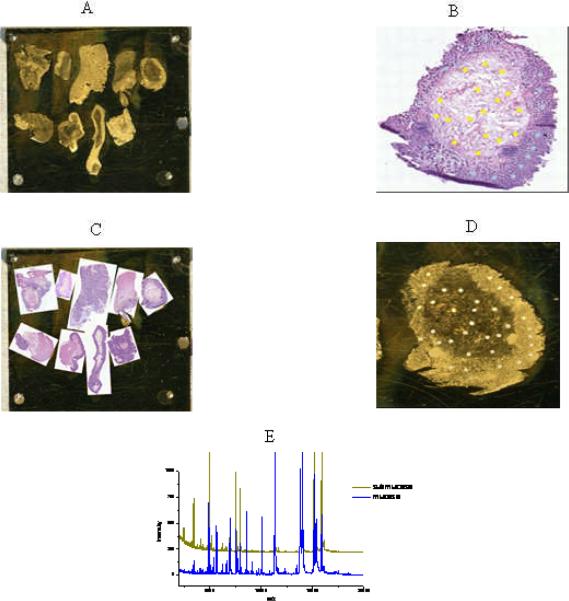
Histology-directed protein profiling6,7,15–17 was utilized to prepare tissues for analysis. This method of mass spectral data acquisition allows for spectra to be acquired from areas of mucosa or submucosa within a single tissue section and without the need for added sample preparation such as cell sorting or laser capture micro-dissection. The methods of histology-directed profiling for MALDI-MS are outlined in Figure 1. Briefly, after UC and CC specimens were identified, two twelve-micron thick sections of each sample were collected using a Leica CM1900 cryostat (Leica Microsystems, Bannockburn, IL). One section was placed on a conductive MALDI-MS target plate and fixed by submersion in graded ethanol (70 percent, 90 percent, and 95 percent) for 30 seconds each. The second contiguous section was placed on a standard microscope slide and stained with H & E and a digital image was acquired using a Mirax Desk Scan (Mirax, Budapest, Hungary) digital microscope at 40× magnification. The pathologist reviewed the digital images and identified areas of interest. These areas were annotated and marked (200 μm) on each tissue preparation. The marks corresponded to inflamed and uninflamed areas of mucosa and inflamed and uninflamed areas of submucosa. The classification of “inflamed” was based on the presence of neutrophils in the mucosa or submucosa or increased mononuclear cells in the submucosa. The classification of “uninflamed” was assigned by the absence of neutrophils and the presence of normal numbers of mononuclear cells. Each tissue sample preparation might have multiple marks indicating mucosal and submucosal inflamed or uninflamed areas of interest. The annotated histology images were exported and overlaid on a digital image of the MALDI target using PhotoShop.13 Coordinates of the annotated areas of interest were then determined and transferred to an acoustic robotic microspotter (LabCyte, Sunnyvale, CA) through the use of fiducial points visible both in the digital images and directly on the plate. The robotic microspotter deposited sinapinic acid (20 mg/ml in 50 percent acetonitrile, 0.1 percent trifluoroacetic acid) at the desired locations. Matrix spot coordinates were then transferred to a Bruker Autoflex II (Bruker Daltonik, Billerica, MA) mass spectrometer, equipped with a SmartBeam™ laser (Nd:YAG, 355 nm). Spectra were collected in linear positive ion mode as a sum of 400 laser spots from each matrix spot. The spectra were calibrated externally and the peak list was exported as ASCII files for further processing. A spectral peak is a unique fingerprint (or wave) which corresponds to a protein mass intensity and labeled as an m/z (mass-to-charge ratio) value.
Figure 1.
Workflow for histology-directed profiling for MALDI MS for colonic mucosal and submucosal tissue compartments.
A: Tissue section placed on a MALDI target
B: Areas marked on an H&E photomicrograph
C: The H&E picture is overlaid on a picture of the MALDI plate
D: Matrix is applied on the MALDI plate at the designated H&E locations
E: Spectra are generated from the matrix spots
To determine whether MALDI-MS would distinguish inflammation in the submucosa, the uninflamed areas of interest vs. inflamed areas of interest were compared in UC as well as the uninflamed vs. inflamed submucosa in CC. To determine whether there were proteomic differences between the inflammatory colitides, the uninflamed UC submucosa was compared to the uninflamed CC submucosa and the inflamed UC submucosa was compared to the inflamed CC submucosa. Additionally to assess any proteomic mucosal differences, the inflamed UC mucosa was compared to the inflamed CC mucosa. Both inflamed and uninflamed tissues were not observed in all patient samples. These comparisons are depicted in Table 1. The uninflamed mucosal layers were not compared between CC and UC due to insufficient numbers of uninflamed specimens for comparison.
Table 1.
| Layer | Crohn's colitis | Ulcerative colitis | ||
|---|---|---|---|---|
| Inflamed (number of matrix spots) | Uninflamed (number of matrix spots) | Inflamed (number of matrix spots) | Uninflamed (number of matrix spots) | |
| Mucosa | A (18) | N=6 | B (24) | N=3 |
| Submucosa | C (20) | D (17) | E (22) | F (21) |
SPECTRA PRE-PROCESSING
Raw spectra were sent to Biodesix (Steamboat Springs, CO) for analysis. Pre-processing was performed using proprietary analysis tools developed by Biodesix Inc, Colorado. In brief, MALDI-MS spectra were baseline-corrected, normalized, and aligned using ProTS'-Marker (Biodesix).
FEATURE DEFINITION AND SELECTION
Each MALDI-MS spectrum was analyzed as integrated, background-subtracted and normalized spectral intensities over a chosen m/z range containing a peak. A combination of a selected subset of features and the classification algorithm (and its parameters), which assigns a clinical label to a spectrum, constitutes a classifier. The classification algorithm used was a straightforward implementation of a k-nearest-neighbor (KNN) algorithm.20 To classify a new spectrum, the KNN algorithm first calculates the Euclidean distance of the feature values of the new spectrum to those of the training set spectra. This calculation yields a list of distances from the test spectrum to each representative spectrum. For the KNN (those with the k smallest distances) the labels are compared. The assigned label is a simple majority vote over the KNN labels. The number “k” of the nearest neighbors was the only classifier parameter used. Candidate features for the classification algorithms were identified as differentially expressed m/z values from the groups of interest. The initial selection of significant features for the classifier was based on a calculation of univariate p-values from Mann-Whitney U-tests (Wilcoxon rank sum test, WIP), then a variant of a floating search method was applied.21 As an optimization criterion we used the leave-one-out cross validation (LOOCV) error on the training set. Finally, a visual inspection of features was carried out using the graphs of the group-averaged spectra.
CROSS-VALIDATION OF A CLASSIFIER
Due to the limited number of samples, the separation of the samples into independent and training test sets was not possible, and the evaluation of the classification was limited to LOOCV or leave-N-out cross validation (LNOCV). A fixed number (one for LOOCV or a prescribed number N for LNOCV) of spectra were removed from the training set, a classifier was generated using the remaining instances, and the performance of this classifier was evaluated by applying it to the left out instances and comparing classifier and true labels. Classifier parameters and the set of selected features were optimized using these cross-validations procedures.
RESULTS
To assess the ability of MALDI-MS to differentiate inflammation, spectra from inflamed vs. uninflamed CC submucosa were compared, as well as comparisons made between the inflamed vs. uninflamed UC submucosa. Several signals in the mass spectra at discrete m/z values were identified in the inflamed and uninflamed mucosal and submucosal layers of CC and UC specimens. The optimal classification between inflamed vs. uninflamed CC submucosa was achieved with two differentiating signals at m/z 6445 and 12692, WIP p=0.0003 and p=0.003, respectively (Table 2). These peaks showed a stable LNOCV classification error rate at 14 percent, providing an accuracy of 86 percent. When analyses were performed on inflamed vs. uninflamed UC submucosa, four differentiating m/z signatures were identified: 4627, 4024, 27848 and 25289. P-values ranged from p=0.001 to p=0.005 and permitted classifications with accuracy of 75 percent (Table 3).
Table 2.
JIBD
| M/Z | CROHN'S COLITIS SUBMUCOSA | WIP | |||
|---|---|---|---|---|---|
| UNINFLAMED | INFLAMED | ||||
| AVG | CV | AVG | CV | ||
| 6445 | 0.274 | 0.539 | 0.13 | 0.461 | P= 0.0003 |
| 12692 | 0.085 | 1.955 | 0.187 | 1.741 | P= 0.003 |
Table 3.
JIBD
| M/Z | ULCERATIVE COLITIS SUBMUCOSA | WIP | |||
|---|---|---|---|---|---|
| UNINFLAMED | INFLAMED | ||||
| AVG | CV | AVG | CV | ||
| 4627 | 0.269 | 0.683 | 0.122 | 1.148 | P= 0.002 |
| 4024 | 0.25 | 0.65 | 0.122 | 0.912 | P= 0.001 |
| 27848 | 0.104 | 0.602 | 0.185 | 0.501 | P= 0.003 |
| 25289 | 0.076 | 0.683 | 0.138 | 0.558 | P= 0.005 |
The MALDI-MS proteomic spectra of the inflamed and uninflamed submucosa of UC vs. CC were then respectively compared. Comparisons of the proteomic spectra of inflamed submucosa between UC and CC identified two significant differentiating peaks: m/z 8773 (p=0.006) and 9245 (p=0.0009) (Table 4). When proteomic spectra of uninflamed submucosa in UC and CC were compared, three discrete significant differentiating peaks were identified: m/z 2778 (p=0.005), 9232 (p=0.005), and 9519 (p=0.005) (Table 5). Cross-validation analysis showed an error rate of 20 percent and therefore an accuracy rate of 80 percent for each of the five peaks. Features of interest are presented in Figures 2, 3, and 4.
Table 4.
JIBD
| M/Z | INFLAMED SUBMUCOSA | WIP | |||
|---|---|---|---|---|---|
| ULCERATIVE COLITIS | CROHN'S COLITIS | ||||
| AVG | CV | AVG | CV | ||
| 8773 | 0.045 | 0.569 | 0.26 | 0.55 | P= 0.006 |
| 9245 | 0.041 | 0.407 | 0.23 | 0.612 | P= 0.0009 |
Table 5.
JIBD
| M/Z | UNINFLAMED SUBMUCOSA | WIP | |||
|---|---|---|---|---|---|
| ULCERATIVE COLITIS | CROHN'S COLITIS | ||||
| AVG | CV | AVG | CV | ||
| 2778 | 0.274 | 0.783 | 0.117 | 1.153 | P= 0.005 |
| 9232 | 0.1 | 0.711 | 0.047 | 0.42 | P= 0.005 |
| 9519 | 0.041 | 0.618 | 0.018 | 0.962 | P= 0.005 |
Figure 2.
Two individual protein peaks (m/z 6445 and 12692) that are discriminatory between uninflamed vs. inflamed submucosa in CC. Red (top) shows the inflamed group and Blue shows uninflamed group. The median of all spectra per group is shown in the first column on the left and individual spectra in the second column on the right. The LNOCV statistical analysis showed a stable error rate of 14 percent (The lighter traces in the mean spectra indicate 25 and 75 percent percentiles).
Figure 3.
Two individual protein peaks (m/z 8773 and 9245) that are discriminatory between inflamed submucosa in UC and CC. Red line (bottom) shows the UC group and Blue line (top) shows CC group. The median of spectra per group are shown in the first column on the left and individual spectra in the second column on the right. The LNOCV statistical analysis showed an accuracy of 80 percent. (The lighter traces in the mean spectra indicate 25 and 75 percent percentiles).
Figure 4.
Three individual protein peaks (m/z 2778, 9232 and 9519) that are discriminatory between uninflamed submucosa in UC and CC. Red line (bottom) shows the UC group and Blue line (top) shows CC group. The median of spectra per group is shown in the first column on the left and individual spectra in the second column on the right. The LNOCV statistical analysis showed an accuracy of 80 percent (The lighter traces in the mean spectra indicate 25 and 75 percent percentiles).
MALDI-MS profiling of inflamed mucosa identified three discriminating signals between CC vs. UC (m/z 31752, 4939, and 5677) with the p-values of 0.016, 0.046 and 0.054, respectively. Cross-validation analysis on these peaks yielded an accuracy of 70 percent.
DISCUSSION
The delineation of UC and CC has to date been based on the use of a combination of potentially clinical, endoscopic, histologic, and/or radiographic assessments that are utilized to guide both medical and surgical therapy. Despite advances in our understanding of the genetic,22,23 immunologic,23,24 and environmental25 factors that may contribute to the pathogenesis of IBD, there is no pathognomonic indicator sensitive enough to accurately and consistently discriminate CC from UC. MALDI-MS has proven to be an effective means to assess the protein microenvironment of tissue layers.15–17 MALDI-MS methodology used in a histology-directed manner permits us to target highly specific areas in colonic tissue for analysis without the need for extensive sample preparation, such as that needed for laser capture microdissection.15 This targeting allowed us to specifically assess the colonic mucosa and submucosa with MALDI-MS since the structural integrity of the tissue was maintained (as opposed to proteomic assessments using full thickness or morselated colonic tissue). In addition, the histology-directed approach permitted specific pathological assessments of inflamed and uninflamed areas within the mucosal and submucosal layers. Our study is the first report using histology-directed MALDI-MS to compare proteomic patterns in the colonic tissue layers in order to distinguish differences between the two inflammatory colitides.
Histology-directed mass spectral protein analysis was able to identify colonic protein signatures that statistically distinguished inflammation in the submucosal layer of CC with a classification (LNOCV and LOOCV) accuracy of 86 percent, and to a lesser extent, in the UC specimens with an accuracy of 75 percent. This lower accuracy may be due to a lesser degree of inflammation in UC versus CC, different types of disease activities or inflammation, or due to the low sample numbers in this study. Another possibility could be because of the statistical high dimensional variable selection based on molecular features of disease on cross-validation results.26 Further validation studies are ongoing with greater numbers of samples to better assess the ability of MALDI-MS to distinguish inflammation in the submucosa of UC as well as CC specimens. The addition of normal colonic submucosa as controls in our comparisons may better assess proteomic differences to truly see if MALDI-MS can distinguish inflamed and uninflamed submucosa.
When using histology-directed MALDI-MS to compare proteomic profiles of CC and UC inflamed and uninflamed submucosal layers, five differentially expressed m/z values were identified that could distinguish the colitides. These proteomic peaks were noted to show greater intensity in the CC submucosa, perhaps indicative of the greater degree or different type of inflammation in the tissues underlying the mucosa. It is possible that these differing peaks may represent candidate biomarkers that could be utilized to delineate the inflammatory colitides. This pilot study is the first of its kind to utilize tissue assessment with MALDI-MS to delineate CC and UC.
There has been great interest in trying to identify protein biomarkers that might delineate the inflammatory colitides. These studies have been variably successful using serum,27,28 mucosal biopsy,29,30 and fecal31,32 biomarkers as prognostic indicators or in assessing whether the IBD is in a quiescent or active state. However, these biomarkers have not been shown to distinguish UC from CC, but rather as prognostic for the known diagnosed disease as being quiescent or active.27–34 Recently, Meuwis, et al.35 reported the first proteomic study on the sera of IBD patients using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry technology, and they identified four biomarkers (PF4, MRP8, FIBA and Hpα2) of interest. These biomarkers had been identified previously in the setting of IBD36,37 and only serve to confirm the presence of IBD rather than differentiating the disease phenotypes. The fact that these biomarkers were selected through a robust statistical process among a large range of potential serological markers highlights their potential importance in both IBD pathophysiology and diagnostic confirmation. The potential biomarkers they identified are small proteins or fragments of proteins (or small protein isoforms potentially with variable posttranslational modifications) that would not necessarily be easily detected by strategies other than proteomic analysis and mass spectrometry. Ansari, et al.,38 in another recent study showed significantly higher expression of serological RANTES (Regulated on Activation, Normal T cell Expressed and Secreted) in UC patients rather than CC patients. It is unclear, as of yet, whether the proteins from these reported studies correspond to the proteomic peaks that we are currently in the process of identifying in our study using MALDI-MS.
There are a number of limitations in our study. The major one is the small sample size of 51 patients (n=24 CC and n=27 UC) which renders the estimation of statistical significance of the differences between groups approximate. It also limits the evaluation of classification to LNOCV performance without the independent validation of the test set. While the data indicate statistically significant differences of m/z peaks used in classification, studies of larger numbers of patients are needed to validate these results on the independent sample cohorts.
The fact that inflammation in the UC submucosa in our study showed features with average p-values in the range from 1.5 to 5.5×10−3 (and accuracy of 75 percent) by MALDI-MS may be due to (apart from other reasons such as inadequate sample size) the extent of inflammation being limited to the mucosa38 although the submucosa is often also inflamed10–12 as we also noted in our studies6,7 and have confirmed that there is some degree of an inflammatory and immune response taking place in this layer that can be studied by mass spectrometry.6–9 Our study is also limited by how we determined the underlying diagnosis of UC and CC by the current gold standard of histology using H & E. As noted in the introduction, this method of diagnosis (in combination with other clinical and diagnostic techniques) may be incorrect in up to 15 percent of cases making CC appear to be UC. Even if up to 15 percent of the UC samples that we examined were in fact CC, this would actually bias the data toward the null hypothesis when comparing the proteomic patterns between the inflamed and uninflamed submucosa of the two colitides, but statistically significant differentiating peaks were still identified in our study. Another limitation is the lack of a normal colonic submucosal control for comparison. While we wanted to focus on the proteomic differences between the colitides (which did not require a normal control), our further validations of MALDI-MS as an effective technique for identifying tissue inflammation will include normal controls to better determine its effectiveness.
The distinctive m/z values that we identified showed statistical differences between CC and UC only when profiling the submucosal layers. Protein signatures obtained from inflamed mucosa showed discrimination between UC and CC, but the accuracy of classification estimated by LNOCV was rather low. However, there is a possibility that better results can be achieved on a larger sample set. This may limit the clinical usefulness of our results when assessing endoscopic biopsies of the colon given that these most abundantly include the mucosa. Submucosal layer biopsies, however, although limited, may as well be obtained endoscopically.39–41 The need to evaluate submucosa to differentiate UC and CC with MALDI-MS is therefore a potential limitation, except for assessment of colectomy specimens, given that submucosal tissue is abundantly available after surgery.
Our study provides potential opportunities for future analyses. While the pattern of proteomic expression and/or the protein localization patterns may hold diagnostic potential, individual proteins may not conclusively be identified by molecular weight (m/z) alone. Analyses are underway to identify the signals to see if they represent discriminative proteins. Identified proteins may be used in studies on endoscopic biopsies or, if confirmed, as serum biomarkers for discrimination between CC and UC (and even as differentiators of indeterminate colitis). Protein identification and validation may provide differentiating biomarkers that would not only improve diagnostic accuracy but may aid in more appropriate, personalized treatment of UC and CC patients and may also provide the basis for future studies on the underlying pathophysiology of IBD.
In conclusion, we found that MALDI MS tissue profiling as described significantly distinguished the inflammatory colitides. Significant discriminatory m/z peaks were identified in these pilot proteomic analyses of both inflamed and uninflamed colonic submucosa from UC and CC. Histology-directed MALDI MS profiling allows for the analysis of high enriched cell types and has the power to predict protein differentiation that can allow inflammatory colitis delineation via submucosal analyses. The methodology revealed 8 m/z peaks of interest, and of these, 5 peaks were individually considered as “good classifiers”. Further analyses and protein identification may provide differentiating biomarkers that would not only improve diagnostic accuracy but may aid in the personalized treatment of UC and CC.
ACKNOWLEDGEMENTS
We are grateful to Kerry R. Wiles and Anthony L. Frazier for sample collection coordination and guidance. We thank Jamie L. Allen for assistance with sample preparation, Wael El-Rifai, MD, PhD, and Phillip E. Williams for scientific guidance, and Deming Mi and Julia Grigorieva, PhD, for assistance with the statistical analyses. This paper was presented, in parts, at the 7th NIH-Network of Research Investigators, Bethesda, MD, April 23 – 24, 2009; at The American Gastroenterological Association Academic Skills Workshop, Phoenix, AZ, March 13 – 14, 2009; at The Annual Congress of The American Society of Colon and Rectal Surgeons, Hollywood, FL, May 2 – 6, 2009 6 (receiving the “New Jersey Society of Colon and Rectal Surgeons” award for the best basic science presentation); at Annual Congress of The Digestive Disease Week, Chicago, IL, May 30 – June 4, 2009;7 at Annual Congress of The Digestive Disease Week, New Orleans, LA, May 2 – 5, 2010,8 and at The Annual Congress of The American Society of Colon and Rectal Surgeons, Minneapolis, MN, May 15 – 19, 2010.9
Source of support: This study is supported by Research Foundation, American Society of Colon and Rectal Surgeons, Limited Project Grant (LPG-086 to A. E. M'Koma), 3U54 CA091408-09S1(to MMC-VICC partnership), Vanderbilt CTSA grant 1 UL1 RR024975 from NCRR/NIH, (to A. E. M'Koma), Core Support Grant [(NIH/NCI-CA068485), DOD (W81XWH-05-1-0179 and 5RO1-GM58008-11 (to R. M. Caprioli)], National Foundation for Cancer Research - NFCR Center for Proteomics and Drug Action (to R. M. Caprioli), and 5P30 DK58404-08 Silvio O. Conte Digestive Diseases Research Core Centers.
ABBREVIATIONS
- m/z
Mass-to-charge ratio. Each m/z peak corresponds to a protein. With a mass spectrometer, we measure them as mass-to-charge (m/z) values. Without further work, we cannot identify the protein
- AVG
Average value
- WIP
P value from Wilcoxon rank sum test, otherwise known as Mann-Whitney U-test
- CV
Coefficient of Variation
- MALDI-MS
Matrix-assisted laser desorption/ionization mass spectrometry
- UC
Ulcerative colitis
- CC
Crohn's colitis
- IBD
Inflammatory Bowel Disease
- NS
Not significant
- H & E
Hematoxylin and eosin
- LOOCV
Leave One Out Cross-validation
- LNOCV
Leave N Out Cross-validation
- WFCCM
Weight-Flexible Compound Covariate
- [n=21] etc.
number of patients analyzed
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
REFERENCES
- 1.Lakatos PL. Prevalence, predictors, and clinical consequences of medical adherence in IBD: how to improve it? World J Gastroenterol. 2009;15:4234–9. doi: 10.3748/wjg.15.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen B. Crohn's disease of the ileal pouch: Reality, diagnosis, and management. Inflamm Bowel Dis. 2009;15:284–94. doi: 10.1002/ibd.20661. [DOI] [PubMed] [Google Scholar]
- 3.Wagner-Bartak NA, Levine MS, Rubesin SE, Laufer I, Rombeau JL, Lichtenstein GR. Crohn's disease in the ileal pouch after total colectomy for ulcerative colitis: findings on pouch enemas in six patients. AJR Am J Roentgenol. 2005;184:1843–7. doi: 10.2214/ajr.184.6.01841843. [DOI] [PubMed] [Google Scholar]
- 4.Shen B, Remzi FH, Brzezinski A, Lopez R, Bennett AE, Lavery IC, Queener E, Fazio VW. Risk factors for pouch failure in patients with different phenotypes of Crohn's disease of the pouch. Inflamm Bowel Dis. 2008;14:942–8. doi: 10.1002/ibd.20409. [DOI] [PubMed] [Google Scholar]
- 5.Keighley MR. The final diagnosis in pouch patients for presumed ulcerative colitis may change to Crohn's disease: patients should be warned of the consequences. Acta Chir Iugosl. 2000;47:27–31. [PubMed] [Google Scholar]
- 6.M'Koma AE, Wise PE, Seeley EH, Washington MK, Schwartz DA, Herline AJ, Muldoon RL, Caprioli RM. Proteomic Patterns of Colonic Submucosa Discriminates Inflammatory Colitides. P47. Annual Congress - The America Society of Colon and Rectal Surgeons; Hollywood, Florida: 2009. p. 146. [Google Scholar]
- 7.M'Koma AE, Seeley EH, Wise PE, Washington MK, Schwartz DA, Herline AJ, Muldoon RL, Caprioli RM. Proteomic analysis of colonic submucosa differentiates Crohn's and ulcerative colitis. M1096. Annual Congress - Digestive Disease week; Chicago, Illinois: 2009. p. 600. [Google Scholar]
- 8.M'Koma AE, Wise PE, Seeley EH, Washington MK, Schwartz DA, Muldoon RL, Herline AJ, Caprioli RM. Human Alpha Defensins are Differentially Expressed Between the Inflammatory Colitides. T1270. Annual Congress - Digestive Disease Week; New Orleans, LA: 2010. p. 720. [Google Scholar]
- 9.M'Koma AE, Wise PE, Schwartz DA, Washington MK, Muldoon RL, El-Rifai WM, Herline AJ. Gene Expression of Colonic Submucosa Differs Between the Inflammatory Colitides. RF6. Annual Congress - The America Society of Colon and Rectal Surgeons; Minneapolis, MN: 2010. p. 117. [Google Scholar]
- 10.Rodrigues-Lisoni FC, Peitl P, Jr, Vidotto A, Polachini GM, Maniglia JV, Carmona-Raphe J, Cunha BR, Henrique T, Souza CF, Teixeira RA, Fukuyama EE, Michaluart P, Jr, de Carvalho MB, Oliani SM, Gencapo HA, Tajara EH. Genomics and proteomics approaches to the study of cancer-stroma interactions. BMC Med Genomics. 2010;3:14. doi: 10.1186/1755-8794-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulubova MV, Manolova IM, Vlaykova TI, Prodanova M, Jovchev JP. Adhesion molecules in chronic ulcerative colitis. Int J Colorectal Dis. 2007;22:581–9. doi: 10.1007/s00384-006-0236-0. [DOI] [PubMed] [Google Scholar]
- 12.Buckell NA, Williams GT, Bartram CI, Lennard-Jones JE. Depth of ulceration in acute colitis: correlation with outcome and clinical and radiologic features. Gastroenterology. 1980;79:19–25. [PubMed] [Google Scholar]
- 13.Cook MG, Dixon MF. An analysis of the reliability of detection and diagnostic value of various pathological features in Crohn's disease and ulcerative colitis. Gut. 1973;14:255–62. doi: 10.1136/gut.14.4.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri KS, Suresh KR, Gogtay NJ, Thatte UM. Declaration of Helsinki, 2008: implications for stakeholders in research. J Postgrad Med. 2009;55:131–4. doi: 10.4103/0022-3859.52846. [DOI] [PubMed] [Google Scholar]
- 15.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145–55. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 16.Chaurand P, Schwartz SA, Capriolo RM. Profiling and imaging proteins in tissue sections by MS. Anal Chem. 2004;76:87A–93A. [PubMed] [Google Scholar]
- 17.Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, Caprioli RM. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5:1975–83. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S, Moore JH, Caprioli RM, Carbone DP. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 19.Bauer JA, Chakravarthy AB, Rosenbluth JM, Mi D, Seeley EH, De Matos Granja-Ingram N, Olivares MG, Kelley MC, Mayer IA, Meszoely IM, Means-Powell JA, Johnson KN, Tsai CJ, Ayers GD, Sanders ME, Schneider RJ, Formenti SC, Caprioli RM, Pietenpol JA. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res. 2010;15:681–90. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theodoritis S, Koutroumbas K. Pattern Recognition. 3rd Edition Academic Press; 2006. [Google Scholar]
- 21.Webb A. Statistical pattern recognition. Wiley; Chichester, UK: 2000. [Google Scholar]
- 22.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 23.Pallone F, Blanco Gdel V, Vavassori P, Monteleone I, Fina D, Monteleone G. Genetic and pathogenetic insights into inflammatory bowel disease. Curr Gastroenterol Rep. 2003;5:487–92. doi: 10.1007/s11894-003-0038-2. [DOI] [PubMed] [Google Scholar]
- 24.Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002;17:629–38. doi: 10.1016/s1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan A, Korzenik JR. Inflammatory bowel disease and environmental influences. Gastroenterol Clin North Am. 2002;31:21–39. doi: 10.1016/s0889-8553(01)00003-6. [DOI] [PubMed] [Google Scholar]
- 26.Wasserman L, Roeder K. High dimensional variable selection. Ann Stat. 2009;37:2178–2201. doi: 10.1214/08-aos646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kader HA, Tchernev VT, Satyaraj E, Lejnine S, Kotler G, Kingsmore SF, Patel DD. Protein microarray analysis of disease activity in pediatric inflammatory bowel disease demonstrates elevated serum PLGF, IL-7, TGF-beta1, and IL-12p40 levels in Crohn's disease and ulcerative colitis patients in remission versus active disease. Am J Gastroenterol. 2005;100:414–23. doi: 10.1111/j.1572-0241.2004.40819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bossuyt X. Serologic markers in inflammatory bowel disease. Clin Chem. 2006;52:171–81. doi: 10.1373/clinchem.2005.058560. [DOI] [PubMed] [Google Scholar]
- 29.Fukushima K, Yonezawa H, Fiocchi C. Inflammatory bowel disease-associated gene expression in intestinal epithelial cells by differential cDNA screening and mRNA display. Inflamm Bowel Dis. 2003;9:290–301. doi: 10.1097/00054725-200309000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Shkoda A, Werner T, Daniel H, Gunckel M, Rogler G, Haller D. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–25. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 31.Langhorst J, Elsenbruch S, Koelzer J, Rueffer A, Michalsen A, Dobos GJ. Noninvasive markers in the assessment of intestinal inflammation in inflammatory bowel diseases: performance of fecal lactoferrin, calprotectin, and PMN-elastase, CRP, and clinical indices. Am J Gastroenterol. 2008;103:162–9. doi: 10.1111/j.1572-0241.2007.01556.x. [DOI] [PubMed] [Google Scholar]
- 32.Costa F, Mumolo MG, Ceccarelli L, Bellini M, Romano MR, Sterpi C, Ricchiuti A, Marchi S, Bottai M. Calprotectin is a stronger predictive marker of relapse in ulcerative colitis than in Crohn's disease. Gut. 2005;54:364–8. doi: 10.1136/gut.2004.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, Casciotti L, Maganti V, Reddy PS, Strahs A, Immermann F, Spinelli W, Schwertschlag U, Slager AM, Cotreau MM, Dorner AJ. Molecular classification of Crohn's disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8:51–61. doi: 10.2353/jmoldx.2006.050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felley-Bosco E, Andre M. Proteomics and chronic inflammatory bowel diseases. Pathol Res Pract. 2004;200:129–33. doi: 10.1016/j.prp.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Meuwis MA, Fillet M, Geurts P, de Seny D, Lutteri L, Chapelle JP, Bours V, Wehenkel L, Belaiche J, Malaise M, Louis E, Merville MP. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. 2007;73:1422–33. doi: 10.1016/j.bcp.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Simi M, Leardi S, Tebano MT, Castelli M, Costantini FM, Speranza V. Raised plasma concentrations of platelet factor 4 (PF4) in Crohn's disease. Gut. 1987;28:336–8. doi: 10.1136/gut.28.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meuwis MA, Fillet M, Lutteri L, Maree R, Geurts P, de Seny D, Malaise M, Chapelle JP, Wehenkel L, Belaiche J, Merville MP, Louis E. Proteomics for prediction and characterization of response to infliximab in Crohn's disease: a pilot study. Clin Biochem. 2008;41:960–7. doi: 10.1016/j.clinbiochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Ansari N, Abdulla J, Zayyani N, Brahmi U, Taha S, Satir AA. Comparison of RANTES expression in Crohn's disease and ulcerative colitis: an aid in the differential diagnosis? J Clin Pathol. 2006;59:1066–72. doi: 10.1136/jcp.2005.034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley E, Jenkins D, Campbell F, Warren B. How could pathologists improve the initial diagnosis of colitis? Evidence from an international workshop. J Clin Pathol. 2002;55:955–60. doi: 10.1136/jcp.55.12.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wytock DH, Baybick J. Depth of colorectal biopsies with proctoscopic forceps. Gastrointest Endosc. 1987;33:15–7. doi: 10.1016/s0016-5107(87)71477-1. [DOI] [PubMed] [Google Scholar]
- 41.Yardley JH, Donowitz M. Colo-rectal biopsy in inflammatory bowel disease. Monogr Pathol. 1977;18:50–94. [PubMed] [Google Scholar]