Abstract
Purpose
Mistakes in DNA repair can result in sustained damage and genetic instability. We comprehensively evaluated common variants in DNA repair pathway genes for their association with postmenopausal breast cancer risk with and without respect to estrogen receptor (ER) and progesterone receptor (PR) subtypes.
Methods
In this nested case-control study of 1,145 prospectively ascertained breast cancer cases and 1,142 matched controls within the Nurses’ Health Study Cancer Genetic Markers of Susceptibility project, we evaluated 1,314 common genetic variants in 68 candidate genes. These variants were chosen to represent five DNA repair pathways including base excision repair, nucleotide excision repair, double strand break repair (homologous recombination and non-homologous end-joining), direct reversal repair, and mismatch repair, along with candidate DNA polymerases, Fanconi Anemia complementation groups, and other genes relevant to DNA damage recognition and response. Main effects, pathway effects and pair-wise interactions were evaluated using Logistic Regression, and the Admixture Maximum Likelihood (AML) and Kernel Machine tests.
Results
Eight loci in linkage disequilibrium within the XRCC4 gene were associated with susceptibility to PR− breast cancer in main effect analyses (p-values corrected for multiple testing at the within-gene level <0.04). These loci drove the association between the non-homologous end-joining pathway, containing XRCC4, and PR− breast cancer (Admixture Maximum Likelihood p-value for the full pathway=0.002; p-value when the eight loci were removed=0.86). We performed the Kernel machine analysis to test the hypothesis of no linear or quadratic effects for any of the tested SNPs, or any SNP-SNP interactions among them, including those SNPs in XRCC4, and yielded a p-value of 0.85.
Conclusions
These findings suggest that common variation alone in DNA repair genes plays at most a small role in determining postmenopausal breast cancer risk among women of European ancestry, and support the theory that redundancies in DNA repair mechanisms may be compensatory.
Keywords: Polymorphism, DNA repair, Breast cancer, Postmenopausal women, Pathway
1 Introduction
Breast cancer is the most common type of cancer and the second leading cause of cancer death among women in the United States. While family history of breast cancer is an established predisposing factor, epidemiological studies suggest that only 5–10% of breast cancer cases are familial and the remaining proportion is sporadic [1]. The majority of genetic variants that influence susceptibility to sporadic breast cancer are unknown [2]. Common variants may explain a greater proportion of breast cancer morbidity and mortality than rare highly penetrant mutations, such as those in BRCA1 and BRCA2 which account for only 15–20% of familial breast cancer cases[1].
DNA repair plays an essential role in the maintenance of DNA integrity. Failure of DNA repair mechanisms can lead to sustained damage, potentially resulting in the malfunction of cellular systems and checkpoints, and the ability of a cell to over-proliferate or evade apoptosis. Deficient DNA repair capacity has been suggested as a predisposing factor in familial and sporadic breast cancer [3–5]. Substantial correlations have been found between DNA repair gene variants and DNA repair capacity[6]. Several studies have observed a low nucleotide excision repair capacity and direct reversion repair capacity of breast tissue [7–9] and suggest that the breast epithelium may uniquely lack redundant systems of double-strand break repair that are present in other tissues [10, 11]. If true, this suggests common genetic variation in DNA repair genes would have greater impact in breast tissues than other tissues with more extensive DNA repair redundancy.
Despite the relevance of DNA repair to carcinogenesis, the impact of common genetic variation on postmenopausal breast cancer susceptibility is not fully understood. In this case-control study of 2,287 postmenopausal women of European ancestry (1,145 cases and 1,142 controls matched on age and postmenopausal hormone use) nested within the Nurses’ Health Study (NHS), we comprehensively and systematically evaluated genetic variation in the coding and non-coding regions of 68 DNA repair genes in relation to invasive postmenopausal breast cancer risk. The association between breast cancer risk and each of these markers individually was assessed as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Project, although none reached conventional genome-wide significance in the initial scan [12] and only one reached genome-wide significance (rs999737 within RAD51L1) after extensive follow-up [13]. Here we present the results for these markers adjusted for multiple testing at the gene, rather than the genome-wide level. We also explore the possibility that markers in DNA repair pathways may be collectively associated with risk of breast cancer even though the association between breast cancer and any particular marker is too weak to detect. To this end, we conduct a test that aggregates evidence for association across multiple markers (the Admixture Maximum Likelihood test (AML)) and a test that explicitly allows for non-additive interactions among markers in the same pathway (the Kernel Machine test).
These pathways/genes included direct reversion repair (MGMT), base excision repair (BER) (ADPRT, APEX1, FEN1, LIG1, LIG3, NEIL1, NEIL2, OGG1, PCNA, UNG2, XRCC1), nucleotide excision repair (NER) (CKN1, ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, ERCC6, RAD23a, RAD23b, RPA1, RPA2, RPA3, XPA, XPC), double-strand break (DSB) repair via (a) homologous recombination (HR) (BRCA1, BRCA2, MRE11A, NBS1, RAD50, RAD51, RAD51c, RAD51L1, RAD51L3, RAD52, RAD54L, XRCC2, XRCC3), or (b) non-homologous end-joining (NH) (DCLRE1C, G22P1, LIG4, PRKDC, XRCC4, XRCC5), mismatch repair (MMR) (MLH1, MLH3, MSH2, MSH3, MSH6, PMS1, PMS2), DNA polymerases (POL) (POLB, POLD1, POLE, POLI, POLK), Fanconi Anemia complementation groups (FAN) (FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG), and DNA damage recognition and response (REG)(ATM, ATR, CHEK1, CHEK2, TP53).
Breast cancer tumors are heterogeneous across ER and PR status with respect to tumor characteristics, response to treatment, and risk profiles [14–17]. In this study we investigate overall breast cancer susceptibility as well as ER and PR subtype specific susceptibility. Preliminary reports suggest that categorization into ER and PR subtypes may be particularly useful when studying the etiology of breast cancer with respect to DNA repair [18–22].
2 Materials and Methods
2.1 Population
This study population is nested within the Nurses’ Health Study (NHS) Cancer Genetic Markers of Susceptibility (CGEMS) breast cancer case-control study (described elsewhere [12]). Participants provided a blood sample (between 1989 and 1990), were free of diagnosed breast cancer at blood collection, and followed for incident disease until May 2004. Controls were matched to cases based on age, blood collection variables (time, date, and year of blood collection, as well as recent (<3 months) use of postmenopausal hormones (PMH)). The analysis was restricted to women of self-reported European ancestry (with no substantial genetic evidence of non-European ancestry) who were menopausal at blood draw. Informed consent was obtained from all participants. The study was approved by the Institutional Review Board of the Brigham and Women’s Hospital, Boston, MA, USA.
Here we investigate the 1,314 CGEMS tag SNPs which had a minor allele frequency (MAF)>0.01 among controls and were located within 30Kb upstream or downstream of 68 genes chosen to represent the above-mentioned five DNA repair pathways, candidate DNA polymerases, Fanconi Anemia complementation groups, and genes relevant to DNA damage recognition and response. These SNPs were selected from the 528,173 genome-wide-tagging SNPs genotyped successfully among 1,145 CGEMS cases and 1,142 controls. Detailed methods for the CGEMS genome wide association study have been previously reported [12].
2.2 Analysis of main effect
The main effect of each SNP on invasive breast cancer susceptibility was estimated using unconditional logistic regression, adjusted for matching factors (four indicators of age quartile, PMH use (Yes/No)) and three eigenvectors previously shown to be effective in reducing population stratification in this cohort [12]. All SNPs were modeled additively using their minor allele count. Two-sided p-values were calculated for each SNP. Measures of linkage disequilibrium (r2) presented for genotyped SNPs within a gene were calculated among the controls.
2.3 Correction for multiple testing
The Bonferroni correction, which divides the type I error rate (α) by the total number of tests performed, is overly conservative in the presence of linkage disequilibrium (LD); tests of SNPs in LD cannot be considered independent tests as their outcomes will be correlated. We apply the method proposed by Gao et al. [23] to estimate the effective number of independent tests (Meff,i) for each gene i. To obtain a gene specific corrected type I error rate we substitute this value into the standard Bonferroni correction in place of the total number of SNPs (α/Meff,i). P-values for main effects analyses are presented as both uncorrected and Meff,i corrected p-values (pcorrected=puncorrected*Meff,i). The sum of the Meff,i over the 68 genes studied was 862.
2.4 Pathway analyses
2.4.1 Admixture Maximum Likelihood test
For each of the five DNA repair pathways and the three additional categories of genes important to DNA repair, we applied Admixture Maximum Likelihood (AML) [24]. AML estimates the proportion of associated SNPs and their typical effect size to test the global null hypothesis of no association between any SNP and breast cancer susceptibility within the pathway. 1,000 permutations were used to estimate the AML p-values for trend. Due to computational limitations AML analyses were not adjusted for covariates. However, the minimum estimated p-values obtained for each SNP when using this method were similar to those obtained in the above covariate-adjusted main effects analyses. The NH and HR double strand break repair pathways were analyzed separately.
2.4.2 Kernel machine test
Kernel machine analyses [25] were conducted using a quadratic kernel to test the hypothesis of no linear or quadratic effects for any of the tested SNPs, or any SNP-SNP interactions among them. This analysis is equivalent to fitting a mixed model and testing whether the variance of the random effects τ2=0. The mixed model is:
where pi is the probability individual i has breast cancer; X is a vector of observed covariates (including an intercept) and β a vector of their fixed effects; Gij is the genotype at SNP j for individual i, coded additively (0, 1, or 2 counts of the minor allele); γj and γij are independent random effects, each distributed as N(0,τ2). The kernel defines a similarity matrix among subjects based on their genotypes; the quadratic kernel used here was K(Gi,Gj)=(1+ Gi’Gj)2.
Due to computational intensity only those SNPs with a raw p-value less than 0.05 in the covariate-adjusted single SNP analyses were fed into the kernel machine. Because restricting the analysis to univariately significant SNPs can downwardly bias p-values, we used permutation to assess statistical significance (1,000 permutations). The complete case approach was used to handle missing data; to verify the appropriateness of the complete case approach, the kernel analysis was repeated after filling in missing genotypes with imputed CGEMS data, and a similar result was obtained. The imputation methods applied are described elsewhere [26].
2.5 Breast cancer subtype analyses
Main effect and pathway analyses were repeated classifying invasive breast cancer cases according to their ER or PR status, and comparing them to non-cases. ER and PR status were available from pathology reports for a subset of the 1,145 breast cancer cases (ER+ cases: 807, ER− cases: 181, PR+ cases: 666, PR− cases: 297). AML analyses were performed using AMLcalc [24]; all other analyses including the calculation of Meff,i were performed using R 2.8.0 [27].
3 Results
3.1 Analysis of main effect
Table 1 lists, by outcome of interest (any breast cancer, ER+, ER−, PR+, PR−), all common variants with a Meff,i corrected p-value less than 0.05 in the main effects analysis using unconditional logistic regression adjusted for matching factors (age, PMH use) and population structure. Given the total number of effective tests performed per outcome in the main effects analysis (ΣMeff,i = 862) one would expect 43 Meff,i corrected p-values to exceed this 0.05 threshold per outcome when the null hypothesis of no association is true (862 × 0.05 = 43.1). Meff,i adjusted p-values presented represent a gene-level correction and do not take into account the number of genes or outcomes tested.
Table 1.
Gene and SNP information for all SNPs with a Meff adjusted p-value (PMeff) less than 0.05 in a single SNP unconditional logistic regression analysis adjusted for matching factors (age, PMH use) and population structure. The outcome, breast cancer, was considered generally and with respect to estrogen receptor (ER) and progesterone receptor (PR) subtypes. (M: Total number of SNPs investigated within a gene. Meff: Effective number of tests given the linkage disequilibrium pattern within a gene. OR: Odds Ratio comparing heterozygous carriers of the minor allele to non-carriers.)
| Gene Name | Path | M | Meff | rs number | OR | Raw P | PMeff | SNP Type |
|---|---|---|---|---|---|---|---|---|
| Breast Cancer vs. No Breast Cancer | ||||||||
| RPA3 | NER | 65 | 42 | rs17136898 | 1.34 | 0.0011 | 0.046 | intronic |
| MSH3 | MMR | 46 | 20 | rs6151838 | 0.73 | 0.0024 | 0.048 | intronic |
| ER+ Breast Cancer vs. No Breast Cancer | ||||||||
| LIG1 | BER | 30 | 22 | rs274860 | 1.82 | 0.0003 | 0.006 | intronic |
| LIG1 | BER | 30 | 22 | rs2304136 | 1.63 | 0.0006 | 0.013 | intronic |
| ER− Breast Cancer vs. No Breast Cancer | ||||||||
| RPA3 | NER | 65 | 42 | rs13237260 | 1.60 | 0.0002 | 0.007 | intergenic |
| RPA3 | NER | 65 | 42 | rs17136898 | 1.73 | 0.0002 | 0.010 | intronic |
| RPA3 | NER | 65 | 42 | rs10952069 | 1.57 | 0.0005 | 0.019 | intergenic |
| POLE | POL | 18 | 10 | rs5745066 | 2.44 | 0.0027 | 0.027 | missense |
| APEX1 | BER | 25 | 21 | rs2275008 | 1.46 | 0.0016 | 0.033 | intronic |
| PR+ Breast Cancer vs. No Breast Cancer | ||||||||
| FANCF | FAN | 3 | 3 | rs12805507 | 0.76 | 0.0029 | 0.009 | intergenic |
| FANCF | FAN | 3 | 3 | rs408199 | 0.77 | 0.0029 | 0.009 | intergenic |
| LIG1 | BER | 30 | 22 | rs274860 | 1.80 | 0.0006 | 0.013 | intronic |
| RPA3 | NER | 65 | 42 | rs17136898 | 0.34 | 0.0009 | 0.038 | intronic |
| LIG1 | BER | 30 | 22 | rs2304136 | 1.40 | 0.0020 | 0.045 | intronic |
| PMS1 | MMR | 12 | 10 | rs5743030 | 0.61 | 0.0047 | 0.047 | intronic |
| PR− Breast Cancer vs. No Breast Cancer | ||||||||
| XRCC4 | NH | 34 | 23 | rs10080123 | 0.71 | 0.0005 | 0.012 | intronic |
| XRCC4 | NH | 34 | 23 | rs1193693 | 0.72 | 0.0005 | 0.012 | intronic |
| XRCC4 | NH | 34 | 23 | rs1017794 | 0.72 | 0.0006 | 0.014 | intronic |
| DCLRE1C | NH | 15 | 12 | rs12572872 | 1.38 | 0.0016 | 0.020 | intronic |
| XRCC4 | NH | 34 | 23 | rs10040363 | 0.73 | 0.0009 | 0.021 | intronic |
| XRCC4 | NH | 34 | 23 | rs1011981 | 0.72 | 0.0011 | 0.026 | intronic |
| XRCC4 | NH | 34 | 23 | rs10042854 | 0.73 | 0.0013 | 0.029 | intronic |
| XRCC4 | NH | 34 | 23 | rs4591730 | 0.73 | 0.0013 | 0.031 | intronic |
| XRCC4 | NH | 34 | 23 | rs11949301 | 0.73 | 0.0016 | 0.038 | intronic |
Two SNPs exceeded the corrected threshold with respect to Breast Cancer susceptibility. Rs6151838, an intronic variant within MSH3 (member of the MMR pathway), had a Meff,MSH3 adjusted p-value of 0.048. Rs17136898, an intronic variant within RPA3 (member of the NER pathway), had a Meff,RPA3 adjusted p-value of 0.046. This RPA3 variant was also associated with ER− breast cancer (Meff,RPA3 adjusted p-value = 0.007) and PR+ breast cancer (Meff,RPA3 adjusted p-value = 0.038).
Two intronic SNPs (rs274860 and rs2304136) within LIG1 (a member of the BER pathway) had Meff,LIG1 adjusted p-values <0.05 for association with both ER+ and PR+ breast cancer susceptibility. These two SNPs are in high LD (r2 =0.84). Rs274860 had the smaller set of p-values (ER+: Meff,LIG1 adjusted p-value = 0.006; PR+: Meff,LIG1 adjusted p-value =0.013).
In addition to the previously mentioned LIG1 and RPA3 associations, three SNPs exceeded their corrected gene specific threshold for association with PR+ breast cancer. Rs12805507 and rs408199, both had a Meff,FANCF adjusted p-value of 0.009. They are intergenic SNPs in high LD (r2=0.89) located near FAN gene FANCF. Rs5743030, an intronic SNP within PMS1 (a member of the MMR pathway had Meff,PMS1 adjusted p- value of 0.047).
Five SNPs exceeded their corrected gene-specific threshold for association with ER− breast cancer. RPA3 contained three associated SNPs including rs17136898 (previously mentioned) and two additional associated SNPs (rs13237260 and rs10952069; intergenic SNPs in high LD located downstream of RPA3 (r2=0.96)). Some LD exists between rs17136898 and rs13237260 (r2=0.69) and between rs17136898 and rs10952069 (r2=0.68). Rs13237260 had the lowest Meff,RPA3 adjusted p-value (0.007). POL gene POLE contained a missense SNP (rs5745066) with Meff,POLE adjusted p-value of 0.027. APEX1, of the BER pathway contained intronic SNP rs2275008 which had a Meff,APEX1 adjusted p-value of 0.033.
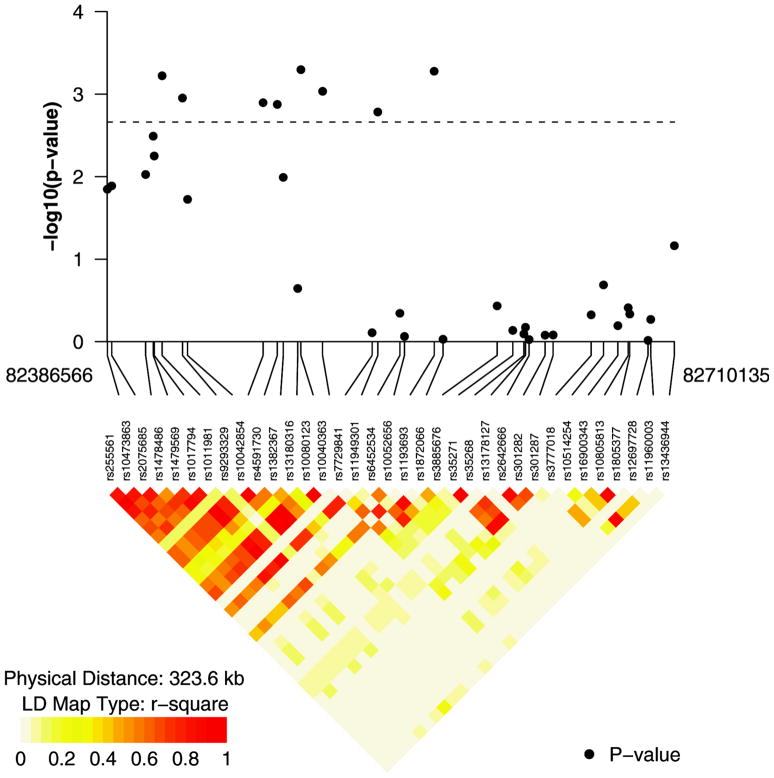
Eight of the nine SNPs with Meff adjusted p-values < 0.05 for their association with risk of PR− breast cancer reside on XRCC4, a key player in the NH pathway. Fig. 1 depicts r-square values or SNPs on XRCC4 and their corresponding p-values for the PR–analysis. It suggests that the large number of associated SNPs on XRCC4 may be due to high LD (the smallest Meff,XRCC4 adjusted p-value observed was 0.012). The 9th SNP to exceed the corrected threshold is rs12572872, and intronic SNP within DCLRE1C, also residing within the NH pathway (Meff,DCLRE1C adjusted p-value =0.020).
Fig. 1.
R2 values for SNPs on XRCC4 and their corresponding p-values from the progesterone receptor negative (PR–)breast cancer subtype analysis.
3.2 Pathway analyses
None of the pathways or groupings of DNA repair related genes, tested using AML, appeared associated with susceptibility to breast cancer independent of ER or PR status. Raw p-values for trend for all AML analyses are presented in Table 2, uncorrected for the number of pathways or outcomes tested. BER appeared mildly associated with ER+ and PR+ breast cancer (p-values of 0.032 and 0.049 respectively), as did NER with ER− breast cancer (p-value=0.048). However, none of these associations are strong enough to survive a multiple testing correction for the number of pathways investigated. The NH association with PR− breast cancer is strong enough to survive a multiple testing correction (p-value=0.002). To examine whether this NH pathway effect is driven by the multitude of highly linked SNPs on XRCC4, we repeated the analysis excluding the eight associated XRCC4 SNPs, and obtained a p-value of 0.86.
Table 2.
Admixture maximum likelihood (AML) p-values for trend corresponding to nine pathways involved in DNA repair. The outcome, breast cancer, was considered generally and with respect to estrogen receptor (ER) and progesterone receptor (PR) subtypes.
| Pathway | Genes | SNPs | BC P trend | ER+ P trend | ER− P trend | PR+ P trend | PR− P trend |
|---|---|---|---|---|---|---|---|
| BER | 11 | 178 | 0.151 | 0.032 | 0.451 | 0.049 | 0.656 |
| DIRECT | 1 | 58 | 0.924 | 0.865 | 0.143 | 0.958 | 0.256 |
| HR | 13 | 352 | 0.606 | 0.582 | 0.36 | 0.702 | 0.331 |
| FAN | 6 | 74 | 0.177 | 0.209 | 0.901 | 0.191 | 0.524 |
| MMR | 7 | 124 | 0.416 | 0.288 | 0.92 | 0.416 | 0.724 |
| NER | 14 | 291 | 0.105 | 0.446 | 0.048 | 0.436 | 0.155 |
| NH | 6 | 109 | 0.93 | 0.946 | 0.43 | 0.553 | 0.002 |
| POL | 5 | 64 | 0.568 | 0.953 | 0.212 | 0.983 | 0.245 |
| REG | 5 | 64 | 0.883 | 0.99 | 0.502 | 0.944 | 0.598 |
3.2.1 SNP*SNP interaction
916 of the 1,000 kernel permutations had a p-value more extreme than that observed in our data, therefore we cannot reject the null hypothesis that there are no main effects or pair-wise SNP-SNP interactions (p=0.92). To assess whether it was appropriate to apply a complete case approach, the same analysis was repeated using the imputed CGEMS data, and a similar result was obtained (permutation p-value=0.85).
4 Discussion
Specific types of DNA damage trigger a repair response from specific DNA repair pathways. For example, the BER pathway responds to a wide variety of non-bulky exogenous and endogenous oxidative DNA damage and single strand breaks [28] while the NER pathway is a versatile repair system to remove a wide variety of bulky, helix-distorting lesions and adducts [29, 30]. Therefore, considering common genetic variants individually may mask biologically relevant pathway effects. We applied AML and Kernel machine analyses to examine the effect of common genetic variation at the pathway level.
In addition to five traditional DNA repair pathways, we considered Fanconi Anemia complementation group genes and DNA polymerases for their role in DNA repair. FAN genes interact with DNA-damage response proteins and other proteins related to cellular responses to carcinogenic stress and to caretaker and gatekeeper functions. The products of POL genes are specialized for operation in distinct DNA repair pathways, or for bypass of specific classes of adducts in DNA [31]. To our knowledge this study is the most exhaustive investigation to date into pathway effects of DNA repair related genes with respect to breast cancer.
In this study we have presented a comprehensive screen of the effect of common variation within a broader range of DNA repair genes than has, to our knowledge, been previously studied on breast cancer and specifically invasive postmenopausal breast cancer. Additionally this study builds on previous work [32, 33] by considering the potential impact of full pathways and pairwise interaction. We have also examined the relevance of each SNP and pathway with and without respect to ER and PR subtypes -acknowledging the heterogeneity of breast cancer and allowing detection for subtype specific effects that may not be noticeable when pooling all breast cancer cases together.
We observed no more independent associations than would be expected by chance. Thus it is probable that most if not all of our observed nominally significant associations are likely to be false positive results. However, the identity of false positive and the false negative results cannot be discerned from our data. Replication is essential in studies of large numbers of common variants to identify the true nature of each SNP’s association.
SNP rs999737 within RAD51L1, a gene on the NH pathway, achieved genome-wide significance in the third stage of the CGEMS initiative [13] (p-value = 1.74×10(−7); ORheterozygote = 0.94(95% CI: 0.88–0.99)). In this study we observed rs999737 to have an uncorrected p-value of 0.012. We have not found evidence of common variants with large effects; however, our study is underpowered to detect modest effects and we may therefore lack the ability to detect individual SNPs with small effects.
Due to the number of tests performed in our analysis, one ought to anticipate a high degree of false positive test results in the absence of appropriate statistical controls. We applied a correction for multiple testing to reduce false positive associations that takes into account LD structure in determining the number of effective tests. Many of the SNPs which exceeded our corrected threshold at the gene level were in high LD with one another and therefore each cluster of LD was likely explaining a single underlying haplotype.
We have previously investigated the association between 1,050 common variants in 60 DNA repair genes and pre-menopausal breast cancer risk among 239 cases and 477 matched controls within the Nurses’ Health Study II [33]. Within these premenopausal women, we found suggestive evidence that common variants in XPF and XRCC3 genes may influence risk, as well as common variation in the NH pathway.
Haiman et al. [32] investigated common genetic variation in 60 genes from within the same set of pathways using the Multiethnic Cohort Study (MEC). This analysis utilized the genotypes for 1367 tag SNPs from 2,093 breast cancer cases and 2,303 controls stemming from five ethnic sub-populations to identify candidate loci. The study sought replication of 15 tag SNPs in three populations of varying ethnic composition including the NHS to identify “pan-ethnic” alleles. The authors observed a lack of replication when comparing United States and United Kingdom study populations.
Both of the above studies investigated pathway effects by studying whether an increased count of variant alleles within a pathway was correlated with increased breast cancer susceptibility. Our study utilizes a more inclusive set of tag SNPs, and more comprehensive pathway approach to investigate this question in greater detail.
Our observations support the general conclusions and lack of replication between the majority of the previous studies into common variation within DNA repair genes. They suggest that when not taking into account environmental interaction, common variation in DNA repair genes plays at most a small role in determining postmenopausal invasive breast cancer risk among American women of European ancestry.
While mutations in key DNA repair genes (for example BRCA1, BRCA2, ATM) have been found to have a major impact on breast cancer risk, it is understandable that the tangible impact of common variation in these genes could be low due to redundant repair systems compensating for one another. Breast tissue is thought to lack redundancy relative to other tissues; this study is consistent with the hypothesis that residual redundancies may be sufficient or that variants that markedly degrade DNA repair capacity are under selective pressure.
It is possible that environmental factors, such as antioxidant or folate deficiency, that insult the DNA or impair DNA repair capacity, may interact with common variants in DNA repair genes in a way such that the meaningful impact of the variant is only detectable when stratifying by the presence or absence of the environmental factor. Such interaction would make replication across diverse populations, as was attempted in the multiethnic study by Haiman et al. [32], unlikely if the populations geographically or temporally differ greatly with respect to environment. Further research is needed to investigate the effect of environmental interaction on the relationship between common variation in DNA repair genes and breast cancer susceptibility.
Acknowledgments
This project was funded by NCI grant CA118447. Genevieve Monsees was supported by PHS T32-ES016645-01. The authors would like to thank Constance Chen for preparing the data and for producing Fig. 1. We also thank the participants of the Nurses’ Health Study for their dedication and commitment.
Abbreviations
- BER
Base excision repair
- CGEMS
Cancer Genetic Markers of Susceptibility
- DIRECT
Direct reversal repair
- DSB
Double strand break repair
- ER
Estrogen receptor
- FAN
Fanconi anemia complementation groups
- HR
Homologous recombination
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- MMR
Mismatch repair
- NER
Nucleotide excision repair
- NH
Non-homologous end joining
- NHS
Nurses’ Health Study
- PMH
Postmenopausal hormone
- POL
Candidate DNA polymerases
- PR
Progesterone receptor
- REG
DNA damage recognition and response
- SNP
Single nucleotide polymorphism
References
- 1.Ponder BA. Cancer genetics. Nature. 2001;411:336–41. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 2.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33(Suppl):238–44. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 3.Helzlsouer KJ, Harris EL, Parshad R, Fogel S, Bigbee WL, et al. Familial clustering of breast cancer: possible interaction between DNA repair proficiency and radiation exposure in the development of breast cancer. Int J Cancer. 1995;64:14–7. doi: 10.1002/ijc.2910640105. [DOI] [PubMed] [Google Scholar]
- 4.Helzlsouer KJ, Harris EL, Parshad R, Perry HR, Price FM, et al. DNA repair proficiency: potential susceptiblity factor for breast cancer. J Natl Cancer Inst. 1996;88:754–5. doi: 10.1093/jnci/88.11.754. [DOI] [PubMed] [Google Scholar]
- 5.Parshad R, Price FM, Bohr VA, Cowans KH, Zujewski JA, et al. Deficient DNA repair capacity, a predisposing factor in breast cancer. Br J Cancer. 1996;74:1–5. doi: 10.1038/bjc.1996.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12:689–98. [PubMed] [Google Scholar]
- 7.Fong LY, Jensen DE, Magee PN. DNA methyl-adduct dosimetry and O6-alkylguanine-DNA alkyl transferase activity determinations in rat mammary carcinogenesis by procarbazine and N-methylnitrosourea. Carcinogenesis. 1990;11:411–7. doi: 10.1093/carcin/11.3.411. [DOI] [PubMed] [Google Scholar]
- 8.Hamdan MA, Guzman RC, Yang J, Beattie C, Mascharack P, et al. Depletion of O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine in three-dimensional collagen cultures of normal human breast epithelial cells. Carcinogenesis. 1992;13:1743–9. doi: 10.1093/carcin/13.10.1743. [DOI] [PubMed] [Google Scholar]
- 9.Latimer JJ, Nazir T, Flowers LC, Forlenza MJ, Beaudry-Rodgers K, et al. Unique tissue-specific level of DNA nucleotide excision repair in primary human mammary epithelial cultures. Exp Cell Res. 2003;291:111–21. doi: 10.1016/s0014-4827(03)00368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welcsh PL, Schubert EL, King MC. Inherited breast cancer: an emerging picture. Clin Genet. 1998;54:447–58. doi: 10.1111/j.1399-0004.1998.tb03764.x. [DOI] [PubMed] [Google Scholar]
- 11.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potter JD, Cerhan JR, Sellers TA, McGovern PG, Drinkard C, et al. Progesterone and estrogen receptors and mammary neoplasia in the Iowa Women’s Health Study: how many kinds of breast cancer are there? Cancer Epidemiol Biomarkers Prev. 1995;4:319–26. [PubMed] [Google Scholar]
- 15.Robertson JF. Oestrogen receptor: a stable phenotype in breast cancer. Br J Cancer. 1996;73:5–12. doi: 10.1038/bjc.1996.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson WF, Chatterjee N, Ershler WB, Brawley OW. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat. 2002;76:27–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 17.Colditz GA, Rosner BA, Chen WY, Holmes MD, Hankinson SE. Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst. 2004;96:218–28. doi: 10.1093/jnci/djh025. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez R, Silva JM, Dominguez G, Garcia JM, Martinez G, et al. Detection of loss of heterozygosity at RAD51, RAD52, RAD54 and BRCA1 and BRCA2 loci in breast cancer: pathological correlations. Br J Cancer. 1999;81:503–9. doi: 10.1038/sj.bjc.6690722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skog S, He Q, Khoshnoud R, Fornander T, Rutqvist LE. Genes related to growth regulation, DNA repair and apoptosis in an oestrogen receptor-negative (MDA-231) versus an oestrogen receptor-positive (MCF-7) breast tumour cell line. Tumour Biol. 2004;25:41–7. doi: 10.1159/000077722. [DOI] [PubMed] [Google Scholar]
- 20.Lee KM, Choi JY, Kang C, Kang CP, Park SK, et al. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res. 2005;11:4620–6. doi: 10.1158/1078-0432.CCR-04-2534. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SM, Hankinson SE, Hunter DJ, Giovannucci EL, Colditz GA, et al. Folate intake and risk of breast cancer characterized by hormone receptor status. Cancer Epidemiol Biomarkers Prev. 2005;14:2004–8. doi: 10.1158/1055-9965.EPI-05-0083. [DOI] [PubMed] [Google Scholar]
- 22.Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER. Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell. 2009;20:3374–89. doi: 10.1091/mbc.E09-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–9. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 24.Tyrer J, Pharoah PD, Easton DF. The admixture maximum likelihood test: a novel experiment-wise test of association between disease and multiple SNPs. Genet Epidemiol. 2006;30:636–43. doi: 10.1002/gepi.20175. [DOI] [PubMed] [Google Scholar]
- 25.Wu MC, Zhang L, Wang Z, Christiani DC, Lin X. Sparse linear discriminant analysis for simultaneous testing for the significance of a gene set/pathway and gene selection. Bioinformatics. 2009;25:1145–51. doi: 10.1093/bioinformatics/btp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 28.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 29.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–8. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 30.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–60. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 31.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–40. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 32.Haiman CA, Hsu C, de Bakker PI, Frasco M, Sheng X, et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17:825–34. doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 33.Han J, Haiman C, Niu T, Guo Q, Cox DG, et al. Genetic variation in DNA repair pathway genes and premenopausal breast cancer risk. Breast Cancer Res Treat. 2009;115:613–22. doi: 10.1007/s10549-008-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]