Abstract
C57BL/6J (B6) mice are susceptible to in utero growth retardation and a number of morphological malformations following prenatal alcohol exposure, while DBA/2J (D2) mice are relatively resistant. We have previously shown that genomic imprinting may play a role in differential sensitivity between B6 and D2 (Downing and Gilliam 1999). The best characterized mechanism mediating genomic imprinting is differential DNA methylation. In the present study we examined DNA methylation and gene expression, in both embryonic and placental tissue, at the mouse Igf2 locus following in utero ethanol exposure. We also examined the effects of a methyl-supplemented diet on methylation and ethanol teratogenesis. In embryos from susceptible B6 mice, we found small decreases in DNA methylation at four CpG sites in one of the differentially methylated regions of the Igf2 locus; only one of the four sites showed a statistically significant decrease. We observed no significant decreases in methylation in placentae. All Igf2 transcripts showed approximately 1.5 fold decreases following intrauterine alcohol exposure. Placing dams on a methyl-supplemented diet before pregnancy and throughout gestation brought methylation back up to control levels. Methyl-supplementation also resulted in lower prenatal mortality, greater prenatal growth, and decreased digit malformations; it dramatically reduced vertebral malformations. Thus, while prenatal alcohol had only small effects on DNA methylation at the Igf2 locus, placing dams on a methyl-supplemented diet partially ameliorated ethanol teratogenesis.
Keywords: Fetal Alcohol Syndrome, ethanol teratogenesis, Igf2, DNA methylation, methyl supplement
Introduction
Exposure to alcohol can be harmful to a developing fetus. The most severe cases are diagnosed with Fetal Alcohol Syndrome (FAS), a disorder defined by prenatal and/or postnatal growth retardation, a characteristic pattern of craniofacial abnormalities and central nervous system dysfunction (Jones and Smith, 1973, 1975; Sokol et al., 2003). Because not all offspring exposed to alcohol prenatally display the full spectrum of FAS symptoms, particularly the facial dysmorphology, the term Fetal Alcohol Spectrum Disorders (FASD; Koren et al., 2003; Sokol et al., 2003) has been coined to describe varying degrees of ethanol teratogenesis, including FAS. The estimated incidence of FASD in the United States is 1% of live births (May and Gossage, 2001; Sampson et al., 1997). However, not all women who consume alcohol during pregnancy give birth to children with deficits, which demonstrates individual differences in susceptibility to ethanol teratogenesis.
Many factors play a role in the development of FASD, including genetics. Human studies have shown that monozygotic twins are more similarly affected than dizygotic twins following prenatal alcohol exposure (Chasnoff, 1985; Christoffel and Salafsky, 1975; Palmer et al., 1974 Riikonen, 1994; Streissguth and Dehaene, 1993). Other studies have shown that different alleles of the alcohol dehydrogenase gene (Adh), an enzyme involved in ethanol metabolism, can influence the severity of teratogenesis in different ethnic populations (for review see Warren & Li, 2005). While these studies have shown a role for genetics in the development of FASD, they are few in number, and the range of genetic variation is unknown. Animal models, particularly mice, have proven to be an invaluable resource for investigating genetic influences on many phenotypes, including several prenatal alcohol traits. Both inbred and selectively bred mice can differ in susceptibility to many of the detrimental effects of in utero ethanol exposure, which provides additional support for the importance of genetics in the development of FASD (Boehm et al., 1997; Downing et al., 2009; Giknis et al., 1980; Gilliam et al., 1989, 1997; Webster et al., 1980).
Work from our laboratory and others has shown that C57BL/6J (B6) mice are susceptible to growth retardation and a number of morphological malformations following in utero ethanol exposure, while DBA/2J (D2) mice are relatively resistant (Boehm et al., 1997; Downing and Gilliam, 1999; Downing et al., 2009; Gilliam et al., 1997; Webster et al., 1980).In a reciprocal cross between B6 and D2 we identified a maternal effect on skeletal malformations following intrauterine ethanol exposure (Downing and Gilliam 1999). One source of variation that can account for a maternal effect is genomic imprinting, an epigenetic phenomenon whereby only one copy of an allele is expressed, either maternal or paternal. The best-characterized mechanism mediating genomic imprinting is differential DNA methylation. All known imprinted genes/gene clusters contain a differentially methylated region (DMR), a region that is methylated on one parental allele but not the other (Paoloni-Giacobino et al., 2007; Sha, 2008). Methylation occurs at cytosine bases within the context of CpG dinucleotides. In general, methylation of CpGs in promoter regions is associated with gene silencing, although there are exceptions, particularly for imprinted genes.
One of the first and best characterized loci is the reciprocally imprinted Igf2/H19 transcriptional unit, which plays a crucial role in placental and embryonic growth and development. This locus is complex and contains four DMRs, DMR0-DMR2 in the Igf2 region and an H19 DMR (Lopes et al., 2003). In the present study we examined DNA methylation at DMR1 of the mouse Igf2 locus following prenatal alcohol exposure. Our hypothesis was that in utero exposure to alcohol in B6 mice, which are susceptible to several different measures of ethanol teratogenesis, would result in a decrease in DNA methylation at DMR1, with a subsequent decrease in mRNA level. We also examined the effects of placing dams on a methyl-supplemented diet. Previous studies have shown that placing dams on a methyl-supplemented diet before mating and maintaining them on this diet throughout gestation can alter offspring DNA methylation and phenotype at epigenetically mediated loci (Cooney et al., 2002; Cropley et al., 2006; Waterland et al., 2006a; Wolff et al., 1998). We wanted to investigate if this was true for ethanol teratogenesis.
Methods
Mice
B6 mice were purchased from the Jackson Laboratory, Bar Harbor, Maine and housed in the animal facility at the Institute for Behavioral Genetics, Boulder, CO. Mice were maintained on a 12-hour light/dark cycle with lights on at 7:00 am; they were given access to food and water ad libitum. The animal colony was maintained at 22 ± 2°C. All procedures were approved by the University of Colorado Animal Care and Use Committee, in accordance with National Institutes of Health guidelines.
DNA Methylation
To generate tissue for methylation analyses, we placed two females with one male for two hours each morning and examined them for a seminal plug as evidence of mating. The morning of plug detection was designated gestational day 0 (GD 0). Plugged females were weighed and single-housed. At noon on GD 9, females were weighed to ascertain a 2 g minimum weight gain as evidence of pregnancy. Females were then intragastrically intubated with either 5.8 g/kg ethanol (20% v:v) or an isocaloric amount of maltose-dextrin. We included an additional group of dams that were placed on a high dose methyl-supplemented diet (3SZM; Wolff et al. 1998) two weeks before we began mating them, and maintained on this diet throughout pregnancy. These dams were given 5.8 g/kg ethanol on GD 9. Male mice used in the study and female mice on the regular (control) diet were given ad libitum access to NIH-31 diet. The 3SZM diet was designed to provide substantially increased amounts of cofactors and methyl donors for methyl metabolism. This diet was prepared by fortifying the control NIH-31 diet, as described by Wolff et al. (1998; Table 1).
Table 1.
The following components were added to the standard NIH-31 diet to give 1000 g of the 3SZM diet.
| Supplement | Amount |
|---|---|
| Choline | 15 g |
| Betaine | 15 g |
| Folic Acid | 15 mg |
| Vitamin B12 | 1.5 mg |
| L-methionine | 7.5 g |
| Zinc | 150 mg |
Four hours after intubation, dams were sacrificed. Embryonic and placental tissues were excised and immediately placed in a −80° freezer. Tissue was sent to the laboratory of Dr. Craig Cooney at the University of Arkansas for Medical Sciences (UAMS) for subsequent DNA extraction, bisulfite conversion and sequencing. DNA was extracted from tissues using an Epicentre MasterPure DNA purification kit (Epicentre Biotechnologies, Madison, WI) according to manufacturer’s instructions with minor modifications. Phenol (Amresco, Solon, OH) and 1-bromo-3chloropropane (Molecular Research Center, Inc., Cincinnati, OH) extraction steps were added prior to isopropanol precipitation. Purified DNA was washed with Tris-EDTA buffer in Montage centrifugal filters (Millipore, Bedford, MA). DNA was then sodium-bisulfite modified with an Epitect Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. For each bisulfite modification 300 ng of DNA was used; bisulfite-treated DNA was stored at −20°C..The Igf2 DMR1 was PCR amplified, producing a 265 bp product, using primers taken from Waterland et al. (2006b). PCR products were then sequenced at the UAMS DNA sequencing core facility using a Model 3100 Genetic Analyzer (Applied Biosystems [ABI], Foster City, CA) and a Big Dye terminator sequencing kit (ABI). Quantification of DNA methylation levels was done using the Mquant algorithm, with minor modifications, as described previously (Leakey et al., 2008). Each methylation level determination represents the mean of at least 3 independent PCR amplifications and bisulfite genomic sequencings. The number of litters per diet/treatment was as follows: 8 control, maltose; 5 control, ethanol, 3 methyl-supplemented, ethanol. The Igf2 DMR1 contains 4 CpG sites. Data were analyzed with MANOVA, using the average methylation level at each CpG site as dependent variables and diet/treatment as the independent variable. Post hoc analyses consisted of Bonferroni-corrected t-tests.
Quantitative PCR
DNA methylation is often correlated with gene expression, particularly for imprinted genes. Therefore, we examined mRNA levels of several Igf2 transcripts following prenatal alcohol exposure, using quantitative, real-time PCR (q-RT-PCR). Mice were mated and treated, and tissue was excised, as described above. Three litters were generated per treatment; within a litter, all placental and all embryonic tissue was pooled for RNA extraction. Tissue was homogenized and RNA extracted using a Qiagen RNeasy Tissue kit, following manufacturer’s instructions. Total RNA was then reverse transcribed to yield single-strand cDNA using a Promega ImPromII Reverse Transcription System, following manufacturer’s instructions. Primers were designed using Primer Express software (ABI). Quantitative RT-PCR was performed on cDNAs using SYBR green chemistry and an ABI Prism 7000 system. Relative quantification of mRNA levels was determined by normalizing against a control gene, Gapdh, using the comparative CT method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Each time a sample was assayed, it was run in triplicate; each sample was assayed three times.
Ethanol Teratogenesis
In addition to examining the effects of a 3SZM diet on DNA methylation at the Igf2 locus, we tested the hypothesis that placing dams on a 3SZM diet would ameliorate ethanol teratogenesis. Female B6 mice were placed on either the control NIH-31 diet or the 3SZM diet two weeks prior to mating and were maintained on the diet throughout pregnancy. At noon on GD9, dams were intubated with maltose-dextrin or 5.8 g/kg alcohol. At 2:00 pm on GD 18, dams were sacrificed; uterine horns were exposed and a count made of live, dead and resorbed fetuses. Live fetuses were weighed, sexed and examined for gross morphological malformations. Gross morphological malformations consisted primarily of forepaw adactyly (missing digits) or syndactyly (fused digits). Every other fetus within a litter was then placed in Bouin’s fixative a minimum of 4 weeks for subsequent soft-tissue examination. Soft-tissue malformations included hydronephrosis and dilated brain ventricles. The remaining fetuses were placed in ethanol a minimum of 2 weeks. They were then macerated for 72 hours in a 1% KOH solution, followed by 6–9 hours in a 1% KOH solution containing alizarin red. Stained fetuses/skeletons were then placed in 25% glycerin for 24 hours and stored in 75% glycerin for subsequent skeletal examinations. Skeletal malformations included missing, fused, wavy or bifurcated ribs and asymmetrical, fused or missing vertebral arches and centra. All teratological examinations were done blind, without knowledge of diet or treatment. Data were analyzed using ANOVA, with diet (control or 3SZM) and treatment (maltose or ethanol) as the independent variables. For all malformations, litter means were the unit of analysis.
Results
DNA Methylation
We observed only small decreases in methylation at the 4 CpG sites of DMR1 following in utero ethanol exposure. In embryos, MANOVA showed main effects of treatment at site 2 (p < .05) and site 3 (p < .02). Post-hoc analysis showed that the only significant difference was at site 3, where, in embryos from dams on a control diet, prenatal alcohol significantly decreased DNA methylation compared to maltose controls (p < .03). For dams that received ethanol, the 3ZM diet significantly increased methylation compared to dams on a control diet (p = .05).In placental tissue we observed no significant changes in DNA methylation following in utero ethanol exposure at any of the CpG sites (Table 2). However, the placental data was quite variable. In placentae from dams on a control diet that received maltose (8 litters), the methylation levels at the 4 CpG sites did not vary much. At site 1, levels ranged from 63–72%, site 2 from 48–56%, site 3 from 62–71% and site 4 from 74–81%. In contrast, in placentae from dams on a control diet that received ethanol (5 litters), methylation levels varied considerably at sites 1 and 2: site 1 from 59–80% and site 2 from 37–64%. The reason for this increased variability in control, ethanol exposed litters is unclear.
Table 2.
Methylation at CpG sites in the Igf2 DMR1.
| Site | Embryo | Placenta | ||||
|---|---|---|---|---|---|---|
| C, MD | C, E | 3SZM, E | C, MD | C, E | 3SZM, E | |
| 1. AACCCGCCTG | .7481 (.006) | .7232 (.030) | .7739 (.032) | .6769 (.013) | .6640 (.041) | .7449 (.027) |
| 2. TCAGCGTTTT | .5855 (.019) | .5136 (.027) | .6174 (.032) | .5254 (.009) | .4790 (.052) | .6016 (.044) |
| 3. TCTCCGGGGT | .7156 (.008) | .6372 (.021) | .7257 (.037) | .6564 (.011) | .6536 (.028) | .7289 (.032) |
| 4. TCAGCGGCTT | .8063 (.015) | .7892 (.021) | .8123 (.046) | .7795 (.008) | .7386 (.033) | .7876 (.030) |
| 5. Average | .7139 (.008) | .6658 (.022) | .7323 (.033) | .6595 (.009) | .6338 (.036) | .7158 (.025) |
Values represent mean (± SEM) methylation levels at each CpG site, and an average of the four sites, using the Mquant algorithm. C = control diet; 3SZM = methyl-supplemented diet; MD = maltose-dextrin; E = ethanol.
q-RT-PCR
The Igf2 gene has at least 4 transcripts. Three of these transcripts originate from the alternative use of first exons, exons 1–3, coupled to exon 4 and the remaining exons (Moore et al., 1997). There is also a placental-specific transcript found in mouse but not human (Monk et al., 2006). We used q-RT-PCR to examine expression of all 4 transcripts, using primers designed to amplify exon 1, exon 2, exon 3 and the placental specific exon (Moore et al., 1997). Relative quantification was determined using the method of Livak and Schmittgen, with Gapdh mRNA as the endogenous reference. Maltose-dextrin treated tissue was used as the calibrator for relative expression of ethanol-treated tissue. Therefore, for each transcript, ΔΔCt was calculated as:
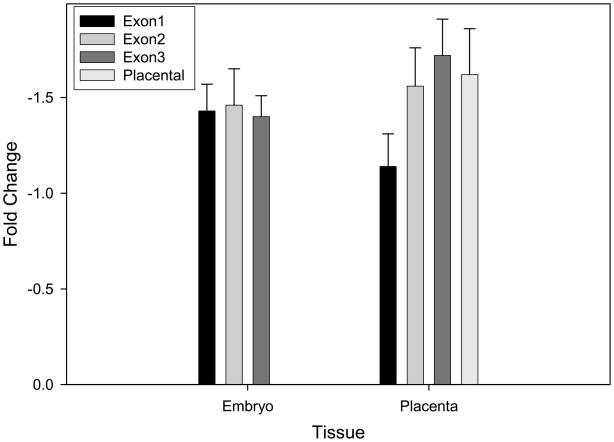
Fold changes were calculated as −(2 ΔΔCt). In embryonic tissue, the placental-specific transcript was not detected. Following prenatal alcohol exposure we observed fold decreases that ranged from 1.40–1.46 (Figure 1). In placental tissue, we observed larger decreases in expression; exon1-exon3 transcript levels were reduced 1.41–1.72 fold (Figure 1). In addition, we saw a 1.62 fold decrease in expression of the placental-specific transcript following in utero ethanol exposure. Thus, all transcripts showed approximately 1.5 fold decreases, with larger decreases observed in placenta compared to embryo.
Figure 1.
Relative expression of Igf2 transcripts in embryonic and placental tissue following in utero ethanol exposure. Data were analyzed using the 2−ΔΔ Ct method as described in Livak and Schmittgen (2001). Data were normalized to Gapdh and fold-changes for alcohol-exposed tissues were determined in relation to maltose-dextrin controls.
Methyl-Supplemented Diet and Ethanol Teratogenesis
Studies have shown that placing dams on methyl-supplemented diets before pregnancy and maintaining them on these diets throughout gestation can not only alter DNA methylation at epigenetically mediated loci, but can also alter epigenetic behavioral phenotypes (Cooney et al., 2002; Cropley et al., 2006; Waterland et al., 2006a; Wolff et al., 1998). We therefore examined whether placing dams on the high dose 3SZM diet, a diet known to have strong epigenetic effects (Cooney et al., 2002; Wolff et al., 1998), would ameliorate ethanol teratogenesis. Results showed no statistically significant effects of diet (C or 3SZM) or treatment (MD or E) on maternal weight gain or prenatal mortality (PNM). However, inspection of Table 3 shows that ethanol-treated dams on the control diet had 14% PNM, while ethanol-treated dams on the 3SZM diet had only 5% PNM, which suggests that methyl-supplementation can somewhat ameliorate PNM. For fetal weight at c-section, independent t-tests showed that for ethanol-treated pups, those on the 3SZM diet weighed more than those on the control diet (1.022 g vs. 0.957 g, p < .05; Table 3).
Table 3.
Mean (± SEM) percent maternal weight gain (PMWG), prenatal mortality (PNM), fetal weight at c-section (CSWT) and morphological malformations: dilated brain ventricles (BV), hydronephrosis (KID), digit malformations (DIG), rib malformations (RIB) and vertebral malformations (VERT).
| D Control, MD | Control, E | 3SZM, MD | 3SZM, E | |
|---|---|---|---|---|
| PMWGA | 58 (11) | 49 (10) | 61 (6) | 53 (11) |
| PNMB | 8 (2) | 14 (3) | 8 (1) | 5 (2) |
| CSWT | 1.027 (.045) | 0.957 (.073) | 1.058 (.040) | 1.022 (.056) |
| BVC | 12 (2) | 46 (6) | 9 (1) | 53 (8) |
| KID | 5 (1) | 37 (4) | 3 (1) | 25 (3) |
| DIG | 3 (1) | 19 (3) | 0 | 10 (2) |
| RIB | 7 (1) | 16 (3) | 5 (1) | 11 (2) |
| VERT | 0 | 18 (2) | 0 | 2 (1) |
PMWG = [(weight day 18 − weight day 9)/weight day 9] × 100.
PNM = [(resorptions + dead)/implantation sites] × 100.
For morphological malformations, values represent percent litter malformed (± SEM).
Control = control diet; 3SZM = methyl-supplemented diet; MD = maltose-dextrin; E = ethanol. Sample sizes (litters): C, MD 10; C, E 12; 3SZM, MD 10; 3SZM, E 13.
For morphological malformations, the only significant effects we saw were on the developing skeletal system, where methyl supplementation almost completely ameliorated vertebral malformations following in utero ethanol exposure (p < .02; Table 3). Ethanol-exposed fetuses whose mothers were on a control diet had an 18% vertebral malformation rate (percent litter malformed), while ethanol-exposed fetuses whose mothers were on the 3SZM diet had a 2% malformation rate. In terms of number of fetuses, only 2 of 94 fetuses (both in the same litter) in the 3SZM group that received ethanol had vertebral malformations, whereas 14 of 79 fetuses in the control diet group that received ethanol had vertebral malformations. The 18% vertebral malformation rate in B6 fetuses is unusually low: we typically observe vertebral malformation rates of 30% or greater in B6 fetuses (Boehm et al. 1997; Downing and Gilliam 1999; Downing et al. 2009) Methyl supplementation also reduced hydronephrosis and digit malformations following in utero ethanol exposure, but these effects were not statistically significant (Table 3).
Discussion
C57Bl/6J (B6) mice are susceptible to growth retardation and morphological malformations following prenatal ethanol exposure. One mechanism that may play a role in B6 susceptibility to ethanol teratogenesis is genomic imprinting, an epigenetic phenomenon whereby only one of two parental copies of an allele is expressed. In this study we examined DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure. The Igf2/H19 transcriptional unit is quite complex. Igf2 is paternally expressed and produces the insulin like growth factor 2 protein, while H19 is maternally expressed and produces a noncoding RNA. This locus is characterized by four DMRs, DMR0-DMR2 in the Igf2 region and an H19 DMR (Lopes et al., 2003). Both the H19 DMR and the Igf2 DMR1 contain methylation-sensitive insulator/silencer elements. The Igf2 DMR1 contains binding sites for GCF2, a transcriptional repressor (Eden et al., 2001; Reed et al., 1998). Methylation of this repressor element keeps the GCF2 protein from binding and facilitates transcription.
We observed only small decreases in methylation at the 4 CpG sites in DMR1 following prenatal alcohol exposure. One interpretation of our data is that prenatal alcohol exposure simply does not have large effects on DNA methylation in embryos or placentae at the Igf2 DMR1, at least at the timepoint we examined. However, embryonic tissue is quite heterogeneous; it consists of many different cell and tissue types. An alternative hypothesis is that prenatal alcohol exposure may have larger effects on DNA methylation in specific tissues of the developing embryo (i.e. the developing digits and skeletal system, where we see malformations), but these effects are “washed out” when looking in a more heterogeneous tissue, whole embryo. Future studies in our laboratory will analyze methylation in more homogeneous tissues. We observed ~ 1.5 fold decreases in expression of three different Igf2 transcripts in embryos, and similar decreases in the four transcripts in placentae following prenatal alcohol exposure. This is in line with the methylation data at DMR1; decreased methylation should allow more repressor protein to bind and decrease gene expression. However, it is unlikely that such small decreases in methylation observed at this Igf2 DMR can completely account for the 1.5 fold decreases in transcript level.
Alcohol and DNA methylation can interact in several ways. Alcohol can disrupt methyl group metabolism (Schalinske and Nieman, 2005) and can alter expression of DNA methyltransferase genes in sperm (Bielawski et al., 2002), fetuses exposed to alcohol in utero (Garro et al., 1991) and chronic alcoholics (Bonsch et al., 2006). Furthermore, alcohol can alter DNA methylation at particular genes, including HERP, SNCA and Nr2b (Bleich et al., 2006; Bonsch et al., 2005; Ravindran and Ticku, 2004). It has also been shown that genes involved in alcohol metabolism (ADH1B and ADH1C) can be regulated by DNA methylation (Dannenberg et al., 2006). Recently, the Ramsay laboratory has shown that prenatal alcohol exposure decreased DNA methylation at the H19 DMR in placental tissue but not embryonic tissue (Haycock and Ramsay 2009). Our results at the linked Igf2 gene showed just the opposite: small decreases in methylation in embryos but no changes in placentae. Unfortunately, the Ramsay studies did not look at expression of H19, so the functional significance of their methylation changes is not known. These studies demonstrate that alcohol can alter DNA methylation at specific loci in both developing tissues and adults.
We found that placing dams on a methyl-supplemented diet before mating and keeping them on the diet throughout pregnancy reversed the decrease in methylation we observed at the Igf2 DMR1 following prenatal alcohol exposure. Furthermore, methyl-supplementation ameliorated prenatal growth retardation, prenatal mortality and both digit and vertebral malformations following in utero ethanol exposure. The most dramatic effects were on vertebral malformations, which methyl-supplementation almost completely abolished. These results are consistent with other studies showing that a methyl-supplemented diet can alter DNA methylation and phenotype at epigenetically modified loci, including both the mouse agouti viable yellow and axin-fused mutations/loci (Cooney et al., 2002; Waterland et al., 2006a). In addition, in an elegant series of studies, the Thomas laboratory has shown that administering choline to either pregnant rat dams or neonatal rat pups can ameliorate some of the growth retardation and behavioral deficits observed following prenatal or neonatal ethanol exposure (Ryan et al. 2008; Thomas et al. 2000, 2004, 2007, 2009). Our methyl-supplemented diet consists primarily of cofactors and methyl donors for methyl metabolism, including methionine, betaine, choline, folic acid and zinc. It will be interesting to see the effects of each of these supplements individually (i.e., choline, betaine, folic acid) on the different morphological malformations we observe following in utero ethanol exposure.
The role that DNA methylation plays in differential susceptibility to ethanol teratogenesis in B6 and D2 fetuses remains to be determined. We have shown that prenatal alcohol has little effect on DNA methylation at the Igf2 DMR1 in B6 embryos and placentae four hours after alcohol exposure. We are examining the other 2 DMRs at the Igf2 locus (DMR0 and DMR2) and the H19 DMR. In order to investigate the role that genomic imprinting may play in differential B6/D2 sensitivity, we are examining changes in DNA methylation in reciprocal B6D2 and D2B6 embryos and placentae. The Igf2/H19 transcriptional unit is one of the most studied imprinted loci, but additional imprinted loci need to be examined as well..Future studies will also need to examine changes in DNA methylation at additional timepoints following prenatal alcohol exposure. These studies will further elucidate the role of DNA methylation in ethanol teratogenesis.
Acknowledgments
These studies were funded by grants R01AA016676 and R24AA01466
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002;26:347–351. [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bonsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30:587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Boehm SL, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J and A/J inbred mouse strains. Alcohol. 1997;14:389–395. doi: 10.1016/s0741-8329(97)87950-5. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Mol Neurosci. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Chasnoff IJ. Fetal Alcohol Syndrome in twin pregnancy. Acta Genet Med Gemellol (Roma) 1985;34:229–232. doi: 10.1017/s0001566000004797. [DOI] [PubMed] [Google Scholar]
- Christoffel KK, Salafsky I. Fetal alcohol syndrome in dizygotic twins. J Pediatr. 1975;87:963–967. doi: 10.1016/s0022-3476(75)80919-x. [DOI] [PubMed] [Google Scholar]
- Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- Cropley JE, Suter CM, Beckman KB, Martin DIK. Germ-line epigenetic modification of the murine AVY allele by nutritional supplementation. Proc Nat Acad Sci. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg LO, Chen HJ, Tian H, Edenberg HJ. Differential regulation of the alcohol dehydrogenase 1B (ADH1B) and ADH1C genes by DNA methylation and histone deacetylation. Alcohol Clin Exp Res. 2006;30:928–937. doi: 10.1111/j.1530-0277.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- Downing C, Gilliam DM. Cytoplasmic factors do not contribute to a maternal effect on ethanol teratogenesis. Behav Genet. 1999;29:31–39. doi: 10.1023/a:1021485821842. [DOI] [PubMed] [Google Scholar]
- Downing C, Balderrama-Durbin C, Broncucia H, Gilliam D, Johnson TE. Ethanol teratogenesis in five inbred strains of mice. Alcohol Clin Exp Res. 2009;33:1238–1245. doi: 10.1111/j.1530-0277.2009.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Constancia M, Hashimshony T, Dean W, Goldstein B, Johnson AC, Keshet I, Reik W, Cedar H. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 2001;20:3518–3525. doi: 10.1093/emboj/20.13.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the Fetal Alcohol Syndrome. Alcohol Clin Exp Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Giknis MLA, Damjanov I, Rubin E. The differential transplacental effects of ethanol in four mouse strains. Neurobehav Toxicol. 1980;2:235–237. [Google Scholar]
- Gilliam DM, Kotch LE, Dudek BC, Riley EP. Ethanol teratogenesis in selectively bred Long-Sleep and Short-Sleep mice: A comparison to inbred C57BL/6J mice. Alcohol Clin Exp Res. 1989;13:667–672. doi: 10.1111/j.1530-0277.1989.tb00402.x. [DOI] [PubMed] [Google Scholar]
- Gilliam DM, Mantle MA, Barkhausen DA, Tweden DR. Effects of acute prenatal ethanol administration in a reciprocal cross of C57BL/6J and Short-Sleep mice: Maternal effects and nonmaternal factors. Alcohol Clin Exp Res. 1997;21:28–34. [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: Effect on DNA methylation in the H19 imprinting control region. Biol Reprod. 2009;81:616–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;2(7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. The fetal alcohol syndrome. Teratology. 1975;12:1–10. doi: 10.1002/tera.1420120102. [DOI] [PubMed] [Google Scholar]
- Koren G, Nulman I, Chudley AE, Loocke C. Fetal Alcohol Spectrum Disorder. CMAJ. 2003;169:1181–1185. [PMC free article] [PubMed] [Google Scholar]
- Leakey TI, Zielinski J, Siegfried RN, Siegel ER, Fan CY, Cooney CA. A simple algorithm for quantifying DNA methylation levels on multiple independent CpG sites in bisulfite genomic sequencing electropherograms. Nuc Acid Res. 2008;36:e64. doi: 10.1093/nar/gkn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TS. Analysis of relative gene expression using real-time quantitative PCR and the 2−ΔΔ CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei M, Walter J, et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Human Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- May PA, Gossage JP. Estimating the prevalence of Fetal Alcohol Syndrome. Alcohol Res Health. 2001;25:159–167. [PMC free article] [PubMed] [Google Scholar]
- Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Human Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. PNAS. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RH, Ouellette EM, Warner L, Leichtman SR. Congenital malformations in the offspring of a chronic alcoholic mother. Pediatrics. 1974;53:490–494. [PubMed] [Google Scholar]
- Paoloni-Giacobino A, D’Aiuto L, Cirio MC, Reinhart B, Chaillet JR. Conserved features of imprinted differentially methylated domains. Gene. 2007;399:33–45. doi: 10.1016/j.gene.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran CRM, Ticku MK. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Mol Brain Res. 2004;121:19–27. doi: 10.1016/j.molbrainres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Reed AL, Yamazaki H, Kaufman JD, Rubinstein Y, Murphy B, Johnson AC. Molecular cloning and characterization of a transcription regulator with homology to GC-binding factor. J Biol Chem. 1998;273:21594–21602. doi: 10.1074/jbc.273.34.21594. [DOI] [PubMed] [Google Scholar]
- Riikonen RS. Difference in susceptibility to teratogenic effects of alcohol in discordant twins exposed to alcohol during the second half of gestation. Pediatr Neurol. 1994;11:332–336. doi: 10.1016/0887-8994(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Ryan SH, Williams JK, Thomas JD. Choline supplementation attenuates learning deficits associated with neonatal alcohol exposure in the rat: effects of varying the timing of choline administration. Brain Res. 2008;1237:91–100. doi: 10.1016/j.brainres.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson PD, Streissguth AP, Bookstein FL, Little RE, Clarren SK, Dehaene P, Hanson JW, Graham JM., Jr Incidence of fetal alcohol syndrome and prevalence of alcohol-related neurodevelopmental disorder. Teratology. 1997;56:317–326. doi: 10.1002/(SICI)1096-9926(199711)56:5<317::AID-TERA5>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Schalinske KL, Nieman KM. Disruption of methyl group metabolism by ethanol. Nutr Rev. 2005;63:387–391. doi: 10.1111/j.1753-4887.2005.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KD. Analyzing real-time PCR data by the comparative CT method. Nature Prot. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sha K. A mechanistic view of genomic imprinting. Ann Rev Genomics Hum Genet. 2008;9:197–216. doi: 10.1146/annurev.genom.122007.110031. [DOI] [PubMed] [Google Scholar]
- Sokol RJ, Delaney-Black V, Nordstrom B. Fetal alcohol spectrum disorder. JAMA. 2003;290:2996–2999. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet. 1993;47:857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- Thomas JD, La Fiette MH, Quinn VRE, Riley EP. Neonatal choline supplementation ameliorates the effects of prenatal alcohol exposure on a discrimination learning task in rats. Neurotoxicol Teratol. 2000;22:703–711. doi: 10.1016/s0892-0362(00)00097-0. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Garrison M, O’Neill TM. Perinatal choline supplementation attenuates behavioral alterations associated with neonatal alcohol exposure in rats. Neurotoxicol Teratol. 2004;26:35–45. doi: 10.1016/j.ntt.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Biane JS, O’Bryan KA, O’Neill TM, Dominguez HD. Choline supplementation following third-trimester equivalent alcohol exposure attenuates behavioral alterations in rats. Behav Neurosci. 2007;121:120–130. doi: 10.1037/0735-7044.121.1.120. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Abou EJ, Dominguez HD. Prenatal choline supplementation mitigates the adverse effects of prenatal alcohol exposure on development in rats. Neurotoxicol Teratol. 2009;31:303–311. doi: 10.1016/j.ntt.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren K, Li TK. Genetic polymorphisms: impact on the risk of fetal alcohol spectrum disorders. Birth Defects Res A Clin Mol Teratol. 2005;73:195–203. doi: 10.1002/bdra.20125. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation Axin Fused. Genesis. 2006a;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Human Mol Genet. 2006b;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- Webster WS, Walsh DA, Lipson AH, McEwen SE. Teratogenesis after acute alcohol exposure in inbred and outbred mice. Neurobehav Toxicol. 1980;2:227–234. [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in AVY/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]