Abstract
Frontotemporal lobar degeneration (FTLD) is a progressive neurodegenerative disease characterized by behavioral abnormalities, personality changes, language dysfunction, and can co-occur with the development of motor neuron disease. One major pathological form of FTLD is characterized by intracellular deposition of ubiquitinated and phosphorylated TAR DNA binding protein-43 (TDP-43), suggesting that dysregulation in phosphorylation events may contribute to disease progression. However, to date systematic analysis of the phosphoproteome in FTLD brains has not been reported. In this study we employed immobilized metal affinity chromatography (IMAC) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify phosphopeptides from FTLD and age-matched control postmortem human brain tissue. Using this approach we identified 786 phosphopeptides in frontal cortex (control and FTLD), in which the population of phosphopeptides represented approximately 50% of the total peptides analyzed. Label free quantification using spectral counts revealed six proteins with significant changes in the FTLD phosphoproteome. N-myc-downstream regulated gene 2 (NDRG2) and glial fibrillary acidic protein (GFAP) had an increased number of phosphospectra in FTLD, whereas microtubule associated protein 1A (MAP1A), reticulon 4 (RTN4; also referred to as neurite outgrowth inhibitor (Nogo)), protein kinase C gamma (PRKCG), and heat shock protein 90kDa alpha, class A member 1(HSP90AA1) had significantly fewer phosphospectra compared to control brain. To validate these differences, we examined NDRG2 phosphorylation in FTLD brain by immunoblot analyses, and using a phosphoserine-13 (pSer13) GFAP monoclonal antibody we show an increase in pSer13 GFAP levels by immunoblot concomitant with increased overall GFAP levels in FTLD cases. These data highlight the utility of combining proteomic and phosphoproteomic strategies to characterize postmortem human brain tissue.
Keywords: Frontotemporal dementia, phosphorylation, immobilized metal-affinity chromatography (IMAC), proteomics, neurodegeneration
Introduction
Frontotemporal lobar degeneration (FTLD), the second leading cause of dementia in the population under age 65, is a progressive neurodegenerative disease characterized by intracellular deposition of protein aggregates 1, 2. In Alzheimer’s disease (AD) and Parkinson’s disease (PD) intraneuronal inclusions are composed predominantly of pathologic forms of the microtubule-associated tau and the presynaptic protein α-synuclein, respectively. In contrast, the most common underlying pathology in FTLD is ubiquitin positive inclusions that lack tau or α-synuclein and contain the RNA binding protein TAR DNA-binding protein 43 (TDP-43) 3, 4. A similar pathology is also observed in amyotrophic lateral sclerosis (ALS), a severe form of motor neuron disease that often co-occurs with FTLD 5. Comparison of detergent-insoluble fractions from postmortem brains indicated that short TDP-43 C-terminal fragments are enriched in FTLD cases compared to control or AD brains 3. Furthermore, TDP-43 is modified by phosphorylation and ubiquitination in FTLD and ALS cases 3, 4, 6–8. These data suggest that post-translational modification of TDP-43 may play a role in disease pathogenesis; however, the underlying molecular mechanisms contributing to FTLD disease progression and neurodegeneration remain to be elucidated. Many critical events involved in cellular response, differentiation, and growth are often regulated by changes in posttranslational protein modifications. Moreover, alterations in protein modifications can influence protein – protein interactions, enzymatic activity, protein conformation, stability and degradation, and an array of human diseases, including neurodegeneration and cancer. Protein phosphorylation is the most widely studied posttranslational modification 9, and aberrant phosphorylation of several proteins including tau 10, 11, neurofilaments 12, and microtubule-associated protein 1B 13 are associated with AD pathogenesis. However to date, no large-scale analysis of the FTLD phosphoproteome has been reported.
Advances in phosphopeptide enrichment strategies and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) 14, 15 provide unique opportunities for systematic mapping of protein phosphorylation sites. Several approaches include immobilized metal-affinity chromatography (IMAC) enrichment incorporating Fe3+ ion 16, 17, or TiO2 18, 19, strong cation exchange (SCX) chromatography 20, 21, antibody capture 22, 23, chemical derivation 24, 25, and combinations of these strategies. Alternatively, calcium phosphate precipitation (CPP) can be employed as a simple, inexpensive approach to enrich phosphopeptides through formation of an insoluble calcium phosphate deposit 26. Previously, we reported the phosphoproteomic analysis of postmortem AD brain tissue using CPP enrichment directly coupled with LC-MS/MS. Although this approach identified 551 phosphopeptides on 185 proteins, the population of phosphopeptides only represented approximately 8.6% of all peptides analyzed 27.
Here we report a comparative phosphoproteome analysis of postmortem FTLD cases with age-matched unaffected control cases using IMAC enrichment followed by LC-MS/MS. Using this strategy the population of phosphopeptides represented approximately 50% of the total peptides analyzed from control and FTLD brain. Label free quantification using spectral counts revealed six proteins with significant changes in the FTLD phosphoproteome. N-myc-downstream regulated gene 2 (NDRG2), and glial fibrillary acidic protein (GFAP), had an increased number of phosphospectra in FTLD, whereas microtubule associated protein 1A (MAP1A), reticulon 4 (RTN4), protein kinase C gamma (PRKCG), and heat shock protein 90kDa alpha, class A member 1(HSP90AA1) had significantly fewer phosphospectra compared to control brain. In our best efforts to validate these differences we used alkaline phosphatase treatment followed by western blot analyses to examine NDRG2 phosphorylation in FTLD brain. Using a GFAP serine-13 (pSer13) phosphospecific monoclonal antibody, we demonstrate increased levels of pSer13 in FTLD cases by western blotting, and immunohistochemistry analyses revealed pSer13 GFAP positive astrocytes in the frontal cortex superficial layer. These data highlight the utility of IMAC coupled with LC-MS/MS for the identification of phosphopeptides in human postmortem brain tissue.
Materials and Methods
Immunohistochemistry
Paraffin-embedded sections of hippocampus and frontal cortex (8 μm thick) were deparaffinized and microwaved in citrate buffer (10 mM, pH 6) for 10 minutes. After cooling to room temperature for 1 hour, sections were rinsed and endogenous peroxidase activity was blocked with 3% hydrogen peroxide at 40°C. Sections were then incubated with normal goat serum for 15 minutes at 40°C, followed by primary antibody polyclonal rabbit TDP-43 antibody (Proteintech, Chicago, IL) or Serine 409/410 phosphorylated TDP-43 mouse monoclonal antibody (CosmoBioUSA, Carlsbad, CA) (diluted in 1% BSA) overnight at 4°C. The following day, sections were incubated with biotinylated goat secondary antibody for 30 minutes at 37°C and finally with avidin-biotin peroxidase complex (Vector Laboratories) for 60 minutes at 37°C. 3,3′-Diaminobenzoic acid (DAB) was used as the chromogen for color development and was followed with hematoxylin counterstain. For pan-GFAP and phosphoserine13 GFAP staining, human free floating brain sections (50 μm) were prepared with a freezing microtome (Microm, Heidelberg, Germany). Control and FTLD frontal cortex sections were incubated in 3% hydrogen peroxide (H2O2) to quench endogenous peroxidase activity. Sections were subsequently incubated with goat serum followed by rabbit GFAP antibody (Biocare Medical, Concord, CA), or mouse monoclonal antibody against serine 13 phosphorylated GFAP (clone KT13, Medical & Biological Laboratories, Japan) (1:20) overnight at 4°C. After extensive washes with tris-buffered saline (TBS), sections were incubated with biotinylated secondary antibody for 1 hour at 4°C and an additional hour with avidin-biotin-peroxidase complex (Vector Elite ABC Kit, Vector Laboratories, Burlingame, CA). Staining was visualized by light microscopy using DAB.
Protein extraction and digestion from brain tissue
The inclusion of FTLD cases in this study was based on the extensive neuropathologic characterization required for diagnosis based on consensus criteria in the Alzheimer’s Disease Research Center Neuropathology Core 28, 29. The cases characterized as FTLD exhibited small, pleomorphic, ubiquitin/TDP-43-positive, tau- and α-synuclein-negative cytoplasmic inclusions in neurons of both the superficial frontal cortex and dentate gyrus of the hippocampus. Additionally, these cases did not meet criteria for the neuropathological diagnosis of AD (CERAD 30 or NIA-Reagan 31) or Lewy Body with dementia 32, nor did they show tau pathology consistent with a tauopathy 33, 34. Frontal cortex from four FTLD and four control, matched as closely as possible by age and post-mortem interval (PMI) (Supplemental Table S1), were weighed individually (~ 1 g) and homogenized (Dounce homogenizer) in low salt lysis buffer (50 mM HEPES, 25 mM NaCl, 10 mM iodoacetamide (IAA), 250 mM Sucrose, 1mM EDTA, including phosphatase and protease inhibitor cocktails (Sigma, St. Louis MO). N-lauryl sacrosine (10% v/v) and 5 M NaCl were added to give a final concentration of 1% Sarkosyl and ~500mM NaCl. Samples were sonicated to disrupt nucleic acids and centrifuged at 200,000 g for 30 min at 4°C using a Beckman ultracentrifuge. The soluble supernatant was removed and protein concentration was determined by bicinchoninic acid (BCA) method (Pierce, Rockford, IL) and used to equally pool the 4 cases of FTLD and 4 cases of control (500 μg total protein per case). The protein samples (2 mg) were transferred to 15 ml Falcon tubes and four volumes of ice-cold acetone was added. Following overnight incubation at −20°C, samples were centrifuged at 20,000 g for 3 min and acetone was removed. Next, samples were uncapped and incubated at room temperature for 30 min in a fume hood to allow residual acetone to evaporate. Precipitated protein was resuspended in urea-buffer (8 M Urea, 50 mM Tris-HCl, pH 7.8) and protein concentrations were re-determined by BCA method. Pooled samples were normalized to 800 μg of total protein, treated with 5 mM dithiothreitol (DTT) for 30 min at 37°C followed by 20 mM IAA for 30 min at 37°C in the dark, and digested with 1:100 (w/w) endopeptidase LysC (Wako Chemicals, Japan) for 3 hours at 37°C. Samples were diluted with 100 mM NH4HCO3 to a final concentration of 1.2 M Urea and digested overnight with 1:50 (w/w) trypsin (Promega) at 37°C. The next day, samples were acidified with 5% formic acid, 0.2% trifluoroacetic acid (TFA).
Immobilized metal-affinity chromatography (IMAC)
The IMAC method was modified from a previously reported protocol as follows 35. The peptides from proteolytic digestion were desalted by a C18 column (Sep-Pak® Cartridges, Waters, Milford, MA), vacuum dried, and dissolved into 1 ml 80% acetonitrile (ACN), 0.1% TFA. One ml Ni-NTA magnetic beads (Qiagen, Valencia, CA) was washed 3 times with 1 ml deionized water, treated with 1 ml 40 mM EDTA pH 8.0 for 30 min on vortex at 80% strength, and washed 3 additional times with 1 ml deionized water. Next, beads were incubated with 1 ml 100 mM FeCl3 (Sigma Aldrich, MO) for 30 min on vortex at 80% strength, washed once with 1:1:1 (v/v/v) ACN, methanol, 0.01% acetic acid, and washed 3 times with 80% ACN, 0.1% TFA. Beads were incubated with peptides for 30 min on vortex at 80% strength, washed 3 times with 1 ml 80% ACN, 0.1% TFA, and resuspended in 100 μl elution buffer (1:1 ACN: 1:20 (v/v) ammonia water) while vortex on and off for 1 min. Finally, beads were softly centrifuged and supernatant containing eluted peptides was transferred to ACN pretreated auto-sampler tubes containing 100 μl 50% ACN, 5% formic acid. Samples were frozen, vacuum dried at low temperature, and dissolved in 10 μl of 6% acetic acid 0.01% TFA for LC-MS/MS analyses.
Peptide analysis by LC-MS/MS
The purified peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and each sample (control and FTLD) was analyzed in technical replicate 36. Briefly, peptide mixtures were loaded onto a C18 column (100 μm i.d., 10 cm long, 2.7 μm HALO resin from Michrom Bioresources, Inc., Auburn, CA) and eluted over a 5–30% gradient (Buffer A: 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 5% AcN; Buffer B: 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 95% AcN). Eluates were monitored in a MS survey scan followed by nine data-dependent MS/MS scans on an LTQ-Orbitrap ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). The LTQ was used to acquire MS/MS spectra (2 m/z isolation width, 35% collision energy, 5,000 AGC target, 150 ms maximum ion time). The Orbitrap was used to collect MS scans (300–1600 m/z, 1,000,000 AGC target, 750 ms maximum ion time, resolution 60,000).
Peptide filtering and protein identification
The acquired MS/MS spectra were searched against the human reference database of the National Center for Biotechnology Information (November 19, 2008) using the SEQUEST Sorcerer algorithm (version 2.0, SAGE-N) 37. Searching parameters included: partially tryptic restriction, parent ion mass tolerance (± 50 ppm), product ion tolerance (± 0.5 m/z), fixed modification on carboxyamidomethylated Cys (+57.0215 Da); dynamic modifications for oxidized Met (+15.9949 Da), phosphorylation (+79.9663 Da) on Ser/The/Tyr, and deamidation (+0.9840 Da) on Asn/Gln,), five maximal modification sites and three maximal missed cleavages. Only b and y ions were considered during the database match. To evaluate false discovery rate (FDR), all original protein sequences were reversed to generate a decoy database that was concatenated to the original database 38, 39. The FDR was estimated by the number of decoy matches (nd) and total number of assigned matches (nt). FDR = 2*nd/nt, assuming mismatches in the original database were the same as in the decoy database. To remove false positive matches, assigned peptides were grouped by a combination of tryptic state (fully and partial) and precursor ion-charge state (2+, 3+, and 4+). Each group was first filtered by mass accuracy (10 ppm for high-resolution MS), and then XCorr (1.0 minimal) and ΔCn (0.0 minimal) values to reduce protein FDR to less than 1%. XCorr is the cross-correlation score between experimental and theoretical spectra, and ΔCn is the normalized difference of XCorr scores between the 1st and 2nd peptide matches. All accepted proteins sharing peptides were grouped together, in which only the top protein with highest spectral counts was selected to represent the group. The MS/MS spectra of matched phosphopeptides were also manually inspected as described previously 27. Phosphorylated Ser and Thr spectra without signature phosphate neutral losses (−49 for doubly charged, −32.7 for triply charged, and −24.5 for quadruply charged) were removed. In some cases, the product ions on MS/MS spectra did not provide sufficient information to distinguish the phosphorylation site(s) within the same peptide sequence. The higher the ΔCn the more certain the phosphorylation site assignment 40. Thus, when the two top matches of a spectrum were both phosphopeptides with the same sequence and the ΔCn was ≤0.15, there was a higher frequency of ambiguity regarding the site of phosphorylation 40. The identified peptides/proteins are listed in supplemental data (Supplemental Table S2–S3) with accession number, amino acid sequence, mass shift, matching scores and potential site(s) of phosphorylation. Every peptide in the tables was linked to assigned MS/MS spectra, precursor ion mass and charge state.
Label-free quantification
To assess differences between the control (CTL) and FTLD phosphoproteome we compared identified proteins using spectral counts. Several of the phosphoproteins identified in this study were found exclusively in FTLD or CTL. For those proteins not identified in either CTL or FTLD samples, a spectral count of 1 was applied. The spectral counts were normalized to ensure that average spectral count ratio per protein was the same in the two datasets 41. G-test was used to judge statistical significance of protein abundance difference as previously described 27, 42, 43. Briefly, the G-value of each protein was calculated as shown in equation (1):
| (1) |
The p-value of each protein was subsequently calculated as the probability of observing a random variable larger than G from the Chi-square distribution (one degree of freedom). The frequency histogram of the p-values was afterward created and 0.01 was determined as the cutoff for p-values to detect significant changes. Proteins, normalized spectral count ratios, and p values can be viewed in Supplemental Table S3.
Western blotting
Immunoblotting was performed according to standard procedures as reported previously 44. Briefly, samples were resolved by SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Blots were blocked with TBS plus blocking buffer (USB Corporation, Cleveland, OH) at room temperature for 30 min and probed with primary antibodies in TBS plus 0.1 % Tween-20 plus blocking buffer overnight at 4°C. The following day, blots were rinsed and incubated with secondary antibodies conjugated to fluorophores (Molecular Probes/Invitrogen) for one hour at room temperature. Images were captured using an Odyssey Image Station (LiCor, Lincoln, NE), and band intensities were quantified using Scion Image. Statistical analysis was performed using Student’s t test for independent samples. To assess NDRG2 phosphorylation, FTLD brain homogenate was incubated with 0 or 23 ug of bovine alkaline phosphatase Type VII-S (3411 Units/mg protein; Sigma Aldrich, St. Louis, MO) in 100 mM Tris pH 8.0 for 2 hr at 37°C and immediately analyzed by western blot as described above. Antibodies used: mouse monoclonal NDRG2 (Abcam, USA), rabbit calnexin (Assay Designs, Ann Arbor, MI), rabbit pan-GFAP antibody (Biocare Medical, Concord, CA), clone KT13 Serine 13 phosphorylated GFAP antibody (Medical & Biological Laboratories, Japan).
Results and Discussion
IMAC enrichment strategy for brain samples
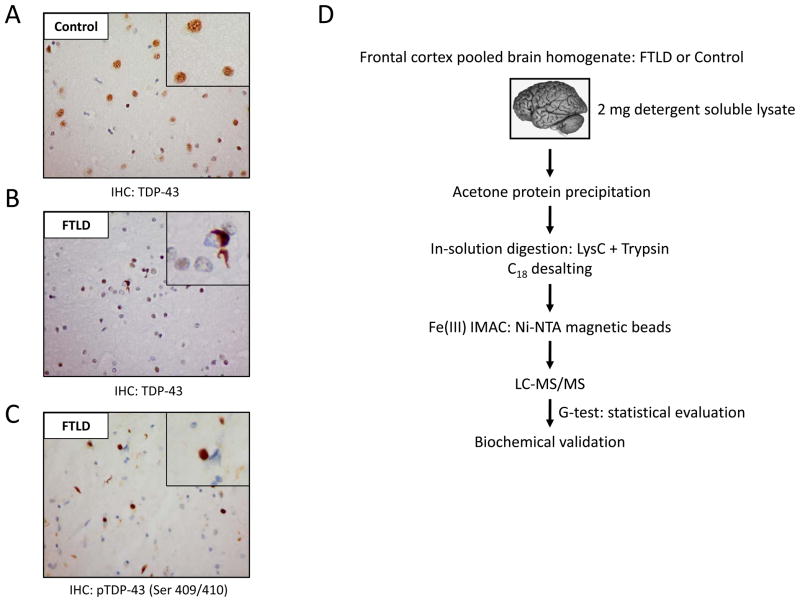
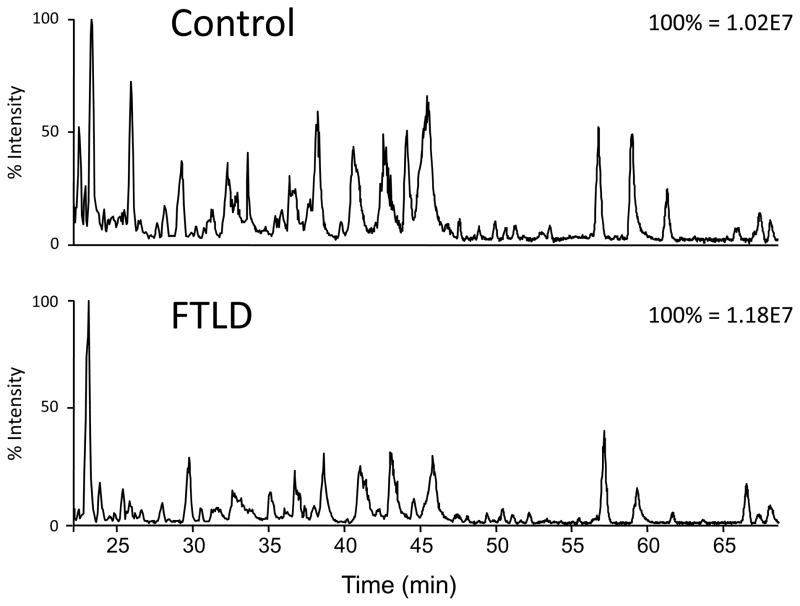
Pathological analyses of FTLD cases demonstrate cytoplasmic phosphorylated TDP-43 inclusions in contrast to normal control tissue, where TDP-43 is exclusively localized to the nucleus (Fig. 1A–C). The phosphoproteomic analyses of pooled FTLD cases in comparison to unaffected, age-matched control brain homogenate include several major steps that are illustrated in Fig. 1D. In our strategy, FTLD and control cases were individually homogenized and examined for protein quality prior to pooling by SDS-PAGE (Supplemental Fig. S1). By pooling cases in discovery-mode proteomic experiments, the between-subject variability regarding pathophysiological heterogeneity, age, sex, and race can be diluted 45, 46. Importantly, our strategy provides biochemical validation on a case-by-case basis, including a number of control and FTLD cases outside of the pooled samples. An equal amount of postmortem frontal cortex protein (500 μg) from each case was combined for pooled FTLD and control samples (2 mg per sample), and lysates were treated with acetone overnight to precipitate proteins and remove lipids (mainly from myelin). Protein loss was observed after acetone precipitation; therefore, pooled samples were re-normalized for protein amount (800 μg). Samples were sequentially in-solution digested with endopeptidase LysC (3 hours) and trypsin (overnight), and phosphopeptides were enriched by an optimized IMAC protocol using FeCl3 followed by LC-MS/MS35. Representative base peak elution profiles from control and FTLD samples are provided in Fig. 2. Previously we reported approximately 8.6% enrichment of phosphopeptides from a single postmortem AD brain sample (10 total fractions) using calcium phosphate precipitation (CPP) enrichment strategy 27. In the present study approximately 40,000 MS/MS spectra were collected and searched against a concatenated target-decoy database from two technical replicates of FTLD and control brain following IMAC enrichment. Analyses revealed 1645 total peptides of which 786 were phosphopeptides (341 protein groups) representing approximately 50% of the total peptides analyzed. Peptides and grouped proteins are provided in Supplemental Tables S2 and S3, respectively. Consistent with our previous report, in-solution protein digestion followed by CPP only allowed for approximately10% enrichment of phosphopeptides from pooled postmortem FTLD and control brain samples (data not shown). Thus, the IMAC procedure employed in this study greatly enhanced phosphopeptide enrichment when compared directly to CPP. Strong-cation exchange chromatography (SCX) at low pH prior to IMAC enrichment would have undoubtedly increased the overall number of phosphopeptides identified, however appropriate protein sample sizes for SCX is on the magnitude of 8–10 mg 20, 21. The limited quantity of control and FTLD samples prevented the use of SCX in this study and therefore likely restricted the phosphoproteome coverage to the most abundant phosphoproteins in the sample mixture.
Figure 1. TDP-43 pathology observed in FTLD and phosphopeptide enrichment strategy.
Immunohistochemical analyses of FTLD and control cases using a pan-TDP-43 (A and B) or a phosphospecific TDP-43 Ser(409/410) antibody in FTLD (C). (D) Flowchart of experimental design to enrich phosphopeptides from human brain tissue. IHC, immunohistochemistry.
Figure 2. IMAC enriched phosphopeptides from human brain samples analyzed by LC-MS/MS.
Representative base peak elution profiles of pooled control and FTLD samples.
Identification and quantification of phosphopeptides in human brain
Phosphoproteomic profiling of human postmortem brain tissue has not been extensively reported 27, 47. One concern of these studies is the length of postmortem interval (PMI) before tissue cryopreservation. During this time, acid-base imbalance, electrolyte depletion, and hypoxia may activate protein kinases and phosphatases in the cellular environment 48. Notably Li, J. et al report that phosphatases may reduce the overall level of protein phosphorylation during the PMI 49. Previously we reported phosphoproteome analysis of an AD brain specimen with a relatively long PMI of 20 hrs using the CPP approach, and 551 phosphopeptides were identified 26,27. In this report, PMIs among FTLD and control cases ranged from approximately 3 to 18 hours (Supplemental Table S1). Label free quantification of spectral counts using G-test as a measure of significance revealed six differences in the FTLD and control samples (Table 1), yet there are obvious limitations using this method of quantitative analysis. Label free quantification of spectral counts considers the total number of all spectral counts for a given phosphoprotein between samples (e.g. control and FTLD), irrespective of the site(s) of phosphorylation. Thus, the analysis is biased toward abundant phosphoproteins/peptides. Low abundant phosphopeptides that could have profound biological significance in brain will not be considered significant by G-test due to lack of experimental observations (MS/MS spectra) 50. To characterize the identified phosphoproteins in this study, we selected and classified the most frequently identified proteins (as indicated by spectral counts) according to the cellular function. The proteins were divided into five groups: neurofilaments (NFs), microtubule-associated proteins, cytoskeletal proteins, signaling/synaptic components, and chaperones. Additionally, approximately 64% of the phosphopeptides that we identified are not included in the Phospho.ELM database (Supplemental Table S2) 51. Although the FTLD cases subjected to phosphoproteomics in this study displayed signature phosphorylated TDP-43 inclusions by immunohistochemistry (Fig. 1C), TDP-43 phosphospectra was expectedly not observed. Phosphorylated TDP-43 is predominantly found in the sarkosyl-resistant (insoluble) tissue homogenate and was therefore not identified 3, 44.
Table 1.
Most enriched phosphoproteins in control and FTLD human brain tissue1
| NCBI Reference | Gene Symbol | Description | Control | FTLD | SC ratio2 | p value3 | Mass | ||
|---|---|---|---|---|---|---|---|---|---|
| TP | SC | TP | SC | ||||||
| Neurofilaments | |||||||||
| NP_005373.2 | NEFM | neurofilament, medium polypeptide 150kDa | 14 | 148 | 9 | 141 | −0.106 | 5.18E-01 | 102 |
| NP_066554.2 | NEFH | neurofilament, heavy polypeptide 200kDa | 13 | 29 | 7 | 18 | 0.512 | 2.16E-01 | 112 |
| Microtubule associated proteins | |||||||||
| NP_005900.2 | MAP1B | microtubule-associated protein 1B | 27 | 133 | 28 | 105 | 0.165 | 3.65E-01 | 270 |
| NP_002365.3 | MAP2 | microtubule-associated protein 2 | 14 | 62 | 11 | 51 | 0.105 | 6.89E-01 | 199 |
| NP_058525.1 | MAPT | microtubule-associated protein tau | 9 | 43 | 9 | 41 | −0.108 | 7.25E-01 | 37 |
| NP_002364.5 | MAP1A | microtubule-associated protein 1A | 13 | 32 | 5 | 8 | 1.824 | 2.24E-04 | 305 |
| NP_005593.2 | CLDN11 | claudin 11 | 2 | 24 | 1 | 26 | −0.292 | 4.61E-01 | 22 |
| NP_005554.1 | STMN1 | stathmin 1 | 7 | 46 | 3 | 59 | −0.535 | 5.09E-02 | 17 |
| Cytoskeletal Proteins | |||||||||
| NP_002347.5 | MARCKS | myristoylated alanine-rich protein kinase C substrate | 22 | 83 | 14 | 73 | 0.009 | 9.68E-01 | 32 |
| NP_003119.2 | SPTBN1 | spectrin, beta, non-erythrocytic 1 | 17 | 53 | 13 | 51 | −0.121 | 6.60E-01 | 274 |
| NP_054908.2 | ADD1 | adducin 1 (alpha) | 19 | 44 | 16 | 42 | −0.109 | 7.18E-01 | 84 |
| NP_001020252.1 | MBP | myelin basic protein isoform 1 | 11 | 19 | 8 | 15 | 0.165 | 7.32E-01 | 21 |
| NP_002046.1 | GFAP | glial fibrillary acidic protein | 1 | 1 | 6 | 18 | −4.346 | 3.52E-06 | 50 |
| Signaling/Synaptic Components | |||||||||
| NP_115667.2 | SGIP1 | SH3-domainGRB2-like (endophilin)interacting protein 1 | 10 | 33 | 11 | 23 | 0.345 | 3.61E-01 | 89 |
| NP_006308.3 | BASP1 | brain abundant, membrane attached signal protein 1 (GAP43) | 8 | 26 | 6 | 15 | 0.617 | 1.66E-01 | 23 |
| NP_057648.2 | CEND1 | cell cycle exit and neuronal differentiation 1 (BM88 antigen) | 10 | 29 | 9 | 41 | −0.676 | 4.49E-02 | 15 |
| NP_004296.1 | BIN1 | bridging integrator 1 (amphiphysin II) | 5 | 30 | 5 | 45 | −0.761 | 1.97E-02 | 50 |
| NP_653080.1 | AKAP12 | A-kinase anchor protein 12 (AKAP12) | 7 | 18 | 4 | 9 | 0.824 | 1.38E-01 | 182 |
| NP_055656.1 | SNAP91 | synaptosomal-associated protein, 91kDa homolog | 7 | 19 | 5 | 8 | 1.072 | 5.72E-02 | 92 |
| NP_963835.1 | NDRG2 | N-myc downstream-regulated gene 2 | 1 | 5 | 3 | 27 | −2.609 | 6.41E-06 | 39 |
| NP_065393.1 | RTN4 | reticulon 4 | 9 | 16 | 1 | 4 | 1.824 | 9.07E-03 | 130 |
| NP_002730.1 | PRKCG | protein kinase C, gamma | 1 | 17 | 1 | 4 | 1.911 | 5.49E-03 | 78 |
| Chaperones | |||||||||
| NP_079495.1 | DNAJC5 | DnaJ (Hsp40) homolog, subfamily C | 3 | 53 | 3 | 55 | −0.230 | 3.94E-01 | 22 |
| NP_031381.2 | HSP90AB1 | heat shock protein 90kDa alpha, class B member 1 | 3 | 26 | 3 | 18 | 0.354 | 4.05E-01 | 83 |
| NP_005339.3 | HSP90AA1 | heat shock protein 90kDa alpha, class A member 1 | 5 | 18 | 2 | 4 | 1.994 | 3.29E-03 | 85 |
Total phosphorylated peptides (TP) and spectral counts (SC).
Normalized SC ratio: natural log (control/FTLD).
Determined by G-test and significant if <0.01 (highlighted in grey).
NDRG2 phosphorylation in FTLD
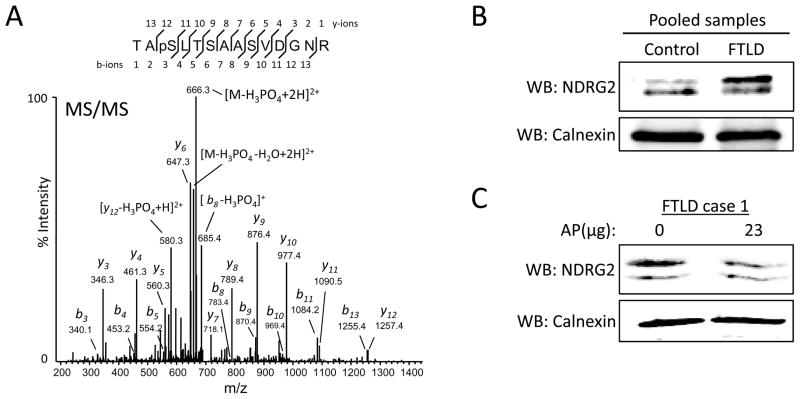
Label free quantification using spectral counts revealed six proteins with significant changes in the FTLD sample (Table 1). NDRG2 is a member of the N-myc downstream-regulated gene family and was initially cloned from a rat hippocampal cDNA library. It is suggested that NDRG2 inhibits glioblastoma cell proliferation 52 and promotes differentiation 53; however, NDRG2 is not repressed by N-myc 54. Notably, NDRG2 is up-regulated at the RNA and protein levels in AD brains 55 and is a proposed target for antidepressant and electroconvulsive therapy 56. NDRG2 possesses multiple phosphorylation sites and is a substrate for Akt and PKC 57. Our LC-MS/MS data revealed 27 NDRG2 phosphospectra in the FTLD sample and 5 in the control sample. Manual inspection of NDRG2 phosphospectra revealed that the majority of NDGR2 is phosphorylated on Ser332 in control and FTLD, while some phosphospectra were observed in FTLD that can be assigned to either Thr330 or Thr334 (Fig. 3A, Supplemental Table 2). To further investigate these findings, we pooled equivalent amounts (12.5 μg) of lysate from each FTLD or control case (Supplemental Table S1) and performed SDS-PAGE followed by immunoblot to examine NDRG2 (Fig. 3B). Western blot analysis reveals an abundant higher molecular weight form of NDRG2 in the pooled FTLD sample that suggests hyperphosphorylation. To investigate the phosphorylation status of the higher molecular weight NDRG2 isoform, lysate FTLD case 1 was treated with alkaline phosphatase and analyzed by western blot (Fig. 3C). A mobility shift, representative of phosphate group loss (80 Da), can sometimes be observed by western blot for phosphoproteins after alkaline phosphatase treatment. Although no mobility shift was observed (Fig. 3C), the ratio of the higher molecular weight isoform relative to the lower molecular weight isoform was decreased. Loss of NDRG2 immunoreactivity in the higher molecular weight isoform and no change in the lower molecular weight isoform after alkaline phosphatase treatment is difficult to interpret. Generation of an antibody against phosphorylated Ser332 NDRG2 could help resolve this issue. There are two alternative splicing isoforms of NDRG2 in humans, isoform a (NDGR2a) and isoform b (NDRG2b), that encode putative proteins that differ in length by 14 amino acids 55, 58; however, our LC-MS/MS data cannot distinguish the origin of phosphopeptides to NDRG2a or NDRG2b. Therefore, elevated levels of NDRG2a in FTLD samples may explain the abundance of the higher molecular weight isoform. Notably, NDRG2 levels were shown to increase in late-onset AD cases within brain regions vulnerable to pathology 55, however whether or not NDRG2 contributes to FTLD pathology remains unclear. The physiological role of NDRG2 and the impact of its phosphorylation in brain are unknown, but NDRG2 mRNA is highly expressed in a pattern similar to GFAP mRNA. Specifically, in situ hybridization combined with immunohistochemistry demonstrated high expression of NDRG2 in GFAP expressing astrocytes in the subgranular zone of the dentate gyrus 59. The role of reactive astrocytes in FTLD progression is a topic of great interest 60, 61; therefore, studying the role of NDRG2 with respect to astrocytic functions such as neurogenesis and regeneration is imperative.
Figure 3. Examination of NDRG2 phosphorylation in FTLD.
(A) NDRG2 phosphopeptide spectrum (pSer332) from FTLD displaying dominant precursor neutral loss peak (49 m/z). (B) Western blot analyses of pooled lysates from FTLD or control cases reveal an abundant higher molecular weight form of NDRG2 in the pooled FTLD sample, suggesting phosphorylation. (C) Lysate from FTLD case 1 was incubated with or without 23 μg of alkaline phosphatase (AP) and analyzed by western blot. A decrease in the ratio of the higher molecular weight isoform relative to the lower molecular weight isoform was observed in the AP treated sample. Blots are representative of three independent experiments. Calnexin used as a loading control for all blots. Human sample information listed in Supplemental Table S1. WB, western blot; AP, alkaline phosphatase.
GFAP Ser13 phosphorylation in FTLD
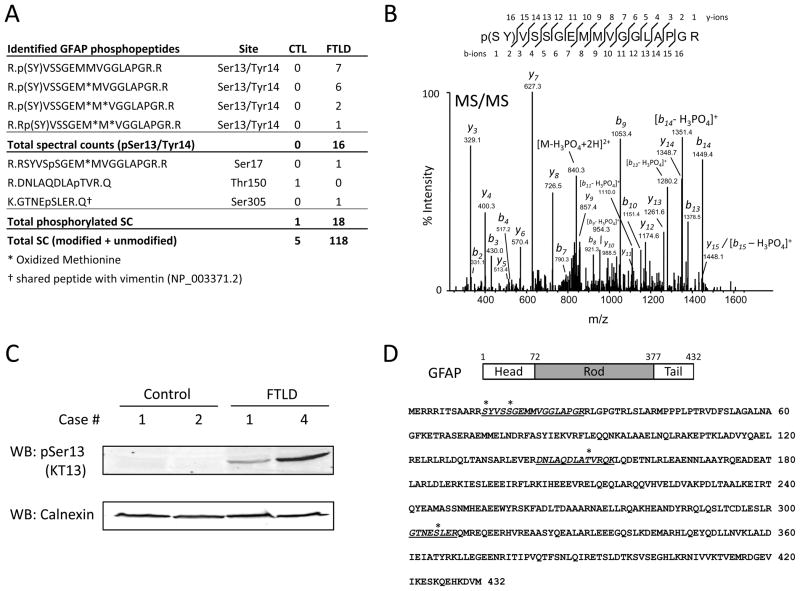
GFAP is a major glial intermediate filament in the cytoskeleton of mature astrocytes 62, and phosphorylation and dephosphorylation of GFAP regulate its self-assembly and degradation 63, 64. GFAP levels dynamically respond to aging and neurodegeneration 65–68, and previous reports have shown that GFAP levels are increased in FTLD and AD 61, 69–72. Consistent with these findings, total spectral counts (including non-phosphorylated peptides) for GFAP in our LC-MS/MS analyses were 118 and 5 for FTLD and control, respectively (Fig. 4A). Among those, 18 GFAP phosphospectra in the FTLD sample and 1 phosphospectra in the control sample were identified. Manual inspection of the observed GFAP phosphopeptides revealed that 16 GFAP phosphospectra mapped to Ser13 or Tyr14 in FTLD, while the remaining two peptides indicated phosphorylation sites at Ser17 and Ser305 (Fig. 4B). Lack of biochemical evidence of GFAP tyrosine phosphorylation as well as the presence of a strong neutral loss of a phosphate group (49 m/z) supports that the most plausible phosphorylation site is Ser13 (Fig. 4B and Supplemental table S2) 73. To assess Ser13 phosphorylation, 200 μg of lysate from two control and FTLD cases were subjected to SDS-PAGE followed by western blot. Using a phosphoserine-13 (pSer13) GFAP monoclonal antibody (clone KT13) 64, 73–76, immunoblot analysis reveals the absence of bands in control samples and immunoreactive pSer13 GFAP bands in FTLD samples (Fig. 4C). GFAP protein is comprised of three major domains, including the N-terminal head, central rod containing four α-helical regions, and C-terminal tail (Fig. 4D). Phosphorylation of residues in the head domain inhibits polymerization of GFAP filaments and promotes depolymerization 77. Specifically, GFAP Ser13 is phosphorylated at the cleavage furrow during cytokinesis in primary cultured astrocytes, possibly by Rho kinase, and phosphorylation of Ser13 also plays a role in structural plasticity of astrocytic processes 73–76, 78.
Figure 4. Examination of GFAP Ser13 phosphorylation in FTLD.
(A) Table of identified GFAP phosphopeptides in FTLD and control. (B) Representative Ser13/Tyr14 GFAP phosphopeptide spectrum from FTLD displaying precursor neutral loss peak. (C) To assess Ser13 phosphorylation, lysate from two control and FTLD cases were analyzed by western blot for pSer13 GFAP using clone KT13 antibody. The presence of immunoreactive bands in FTLD samples and absence in controls is consistent with LC-MS/MS analyses. Calnexin used as a loading control. Human sample information listed in Supplemental Table S1. (D) Protein structure and amino acid sequence of GFAP. Identified phosphorylation sites indicated by *. WB, western blot.
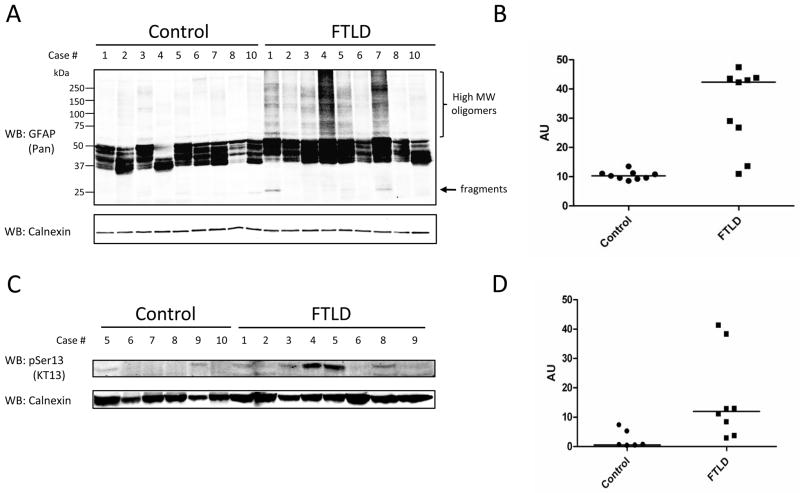
Proteomic studies are typically biased to the most abundant proteins in the sample mixture; therefore, an obvious consideration when assessing quantitative changes in a protein’s phosphorylation status is evaluating the overall levels of protein in the sample. As described above, GFAP levels are increased in FTLD, and not surprisingly, the phosphoproteome enrichment strategy used in the current report revealed higher levels of GFAP in the pooled FTLD sample as compared to the pooled control sample (Fig. 4A). To validate levels of GFAP protein in our samples, 50 μg of lysate from nine control and FTLD cases was examined by SDS-PAGE followed by western blot. Probing with a pan-GFAP antibody revealed that global levels were significantly higher in FTLD; however there was more heterogeneity among FTLD samples (Fig. 5A and B). Notably, several FTLD cases displayed high molecular weight bands that may indicate oligomeric or polyubiquitinated GFAP 79, while immunoreactivity between ~25–50 kDa represent degradation products that may be dephosphorylated, N- or O-deglycosylated, and/or oxidized 80, 81. To investigate the presence of pSer13 GFAP in additional human samples not included in the phosphoproteomic analyses, 150 μg of lysate from six control and eight FTLD cases were examined by SDS-PAGE followed by western blot using clone KT13 pSer13 mono-clonal antibody. Immunoreactivity was observed in five FTLD cases, but only two controls (Fig. 5C). Furthermore, we performed immunohistochemical analyses for pan-GFAP and pSer13 GFAP in one control and two FTLD cases (Fig. 6). In FTLD cases, intense pan-GFAP staining was observed in cortical layers I–II, with the strongest immunoreactivity in the cortical surface. In comparison pSer13 GFAP was largely associated with cortical surface astrocytes, but not observed in cortical layers I–II. FTLD case 5 displayed more intense pSer13 GFAP staining compared to case 6, which is consistent with our biochemical analyses (Fig. 5B).
Figure 5. Analyses of pan-GFAP and GFAP Ser13 phosphorylation in human brain.
(A) Western blot analysis for pan-GFAP in control and FTLD cases. High molecular weight bands in FTLD cases may indicate oligomeric/modified GFAP; immunoreactivity between ~25–50 kd may represent degradation isoforms 80, 81. (B) Quantitative analyses of western blot indicates mean GFAP levels are ~3-fold higher in FTLD (p value = 0.0001). (C) Western blot analyses for pSer13 GFAP using clone KT13 antibody in control and FTLD cases. (D) Quantitative analyses of western blot indicates mean pSer13 GFAP levels are ~8-fold higher in FTLD (p value = 0.045). Calnexin used as loading control for all blots. Human sample information listed in Supplemental Table S1. WB, western blot.
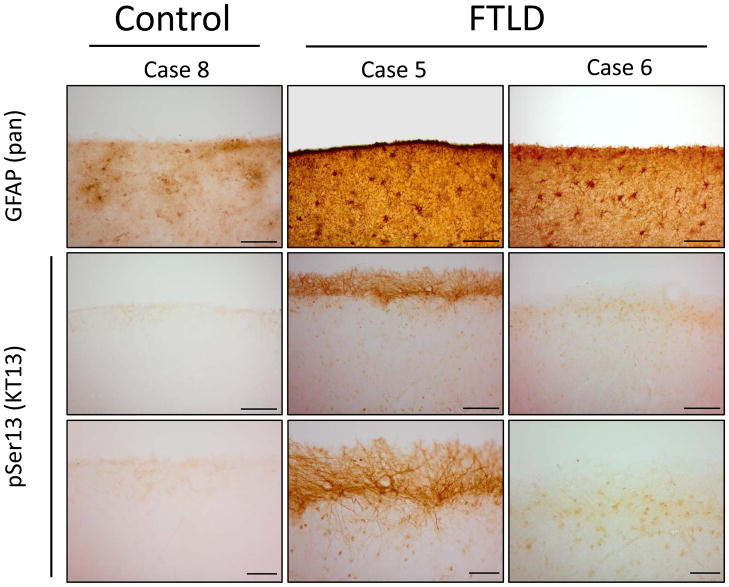
Figure 6. Immunohistochemical analyses of pSer13 GFAP in control and FTLD.
Control case 8 and FTLD cases 5 and 6 were stained with pan-GFAP and clone KT13 antibody. (Top, 20x magnification) Pan-GFAP immunoreactivity strongest in cortical layers I–II, whereas (Bottom, 40x magnification) pSer13 GFAP immunoreactivity was strongly observed in the cortical surface of case 5 (consistent with western blot analyses, Fig. 5C). 20x scale bars 100 μM, and 40x scale bars 40 μM. pSer13, phosphoserine-13. Human sample information listed in Supplemental Table S1.
Proteins exhibiting reduced phosphospectra in FTLD
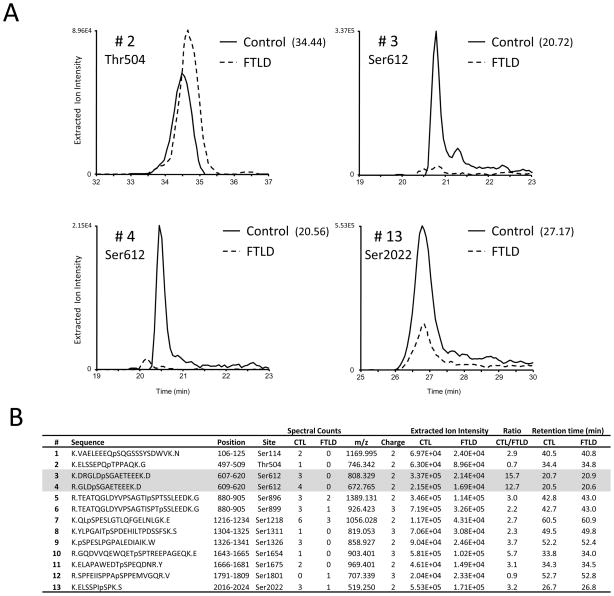
MAP1A, and its family member MAP1B, are microtubule-associated proteins that are predominantly expressed in neurons and are hypothesized to facilitate the formation of axons and dendrites by their ability to bind and stabilize microtubules. The total phosphopeptide spectral counts for MAP1A (non-phosphorylated peptides were not observed) were 32 and 8 for control and FTLD, respectively (Table 1 and Fig. 7). Analyses of extracted ion intensities for identified phosphopeptides in control and FTLD indicate sufficient signal loss for the majority of phosphopeptides identified in FTLD (Fig. 7B). The most significant change was observed for Ser612. The ratio of intensity was ~12-fold greater in the control sample and consistent between the two peptides identified. Moreover, 7 spectral counts from Ser612 containing phosphopeptides were observed in control as compared to 0 in FTLD. All other peptides, with the exception of Thr504 and Ser1801, displayed ~3 to 5-fold greater intensity in the control sample (Fig. 7B). We did not verify the accuracy of this LC-MS/MS data using orthogonal biochemical approaches. Therefore, the reduction in MAP1A phosphospectra in FTLD may be attributable to lower levels of MAP1A in FTLD sarkosyl-soluble fraction. In AD, it is suggested that abnormal hyperphosphorylated tau sequesters MAP1A/MAP1B and causes disassembly of microtubules 82. However, FTLD cases in this study lack tau pathology and no significant difference in number of phosphospectra of MAP1B and MAPT (tau) were observed (Table 1). Like tau, MAP1B is phosphorylated by glycogen synthase kinase-3 β in growing axons 83, whereas regulators of MAP1A phosphorylation remain to be elucidated.
Figure 7. MAP1A phosphopeptide analyses in control and FTLD using extracted ion intensity.
(A) Representative extracted ion chromatogram (XIC) from control and FTLD for MAP1A phosphopeptides 2, 3, 4 and 13 found in B. For XIC a user defined precursor mass tolerance of ±5 ppm was employed. Indicated in parentheses, point in time (minutes) when the MS/MS spectrum was obtained in control samples. In (B) the site of phosphorylation, spectral count(s), m/z, charge state, extracted ion intensities and retention time from control and FTLD for identified MAP1A phosphopeptides are listed.
In addition to MAP1A, a reduced number of phosphospectra (p<0.01) for reticulon 4 (RTN4), also referred to as neurite outgrowth inhibitor (Nogo), protein kinase C gamma (PKCγ, PRKCG), and heat shock protein 90kDa alpha, class A member 1 (HSP90AA1) was observed in FTLD. Reticulons are ubiquitously expressed transmembrane proteins that are involved in nerve growth inhibition in the central nervous system 84. Nogo is arguably the most well studied reticulon family member and exists in three isoforms: Nogo-A, -B, and –C 85. The total phosphospectra from 9 peptides, corresponding to 5 sites (Ser181, Ser182, Ser184, Thr188, Ser991) for Nogo-A, were 16 and 4 for control and FTLD, respectively (Table 1). Interestingly, the only Nogo-A phosphopeptide sequenced in FTLD was Thr188. PKCγ is a neuron specific member of the classical PKC family and plays a role in synaptic plasticity modulation, including long term potentiation and long term depression 86. The total phosphospectra for PKCγ were 17 for control and 4 for FTLD (Table 1). The only phosphopeptide identified was Thr655, and the ratio of extracted ion intensity was ~4-fold greater in the control sample. Thr655 is one of two autophosphorylation sites in the PKCγ kinase domain that is necessary for full activation of PKCγ in response to various stimuli 87, 88. Notably, PKCγ deficient mice display deficits in spatial and contextual learning 89. In total 22 phosphorylated kinases, according to the gene ontology (GO) classification, were identified in both control and FTLD samples (Supplemental Table S4). HSP90AA1 is a stress-induced cytosolic protein that suppresses protein aggregation and facilitates protein folding 90–92. The total phosphospectra from 5 peptides, corresponding to 2 sites (Ser231 and Ser263), were 18 and 4 in control and FTLD, respectively. For Ser263; 8 spectra were sequenced in control and 0 in FTLD. For Ser231, 10 phosphospectra were sequenced in control and 4 in FTLD. Phosphorylation activates heat shock protein 90 and likely regulates its chaperoning function 93. The accuracy of LC-MS/MS data for Nogo, PKCγ, and HSP90AA1 was not verified using orthogonal biochemical approaches; therefore, the reduction in observed phosphopeptides in FTLD may be attributable to lower levels of these proteins in disease tissue.
Conclusion
Our findings represent the first phosphoproteomic analyses of FTLD postmortem brain tissue and demonstrate the feasibility of attaining 50% enrichment of phosphopeptides from autopsy brain specimens coupling IMAC with LC-MS/MS. This strategy revealed six proteins with significant changes in the FTLD sample. NDRG2 and GFAP had an increased number of phosphospectra, whereas MAP1A, Nogo, PKCγ, and HSP90AA1 had significantly fewer compared to control brain. Furthermore, biochemical and immunohistochemical analyses validated an increase in GFAP Ser13 phosphorylation in FTLD. NDRG2 and GFAP share similar astrocytic mRNA expression patterns and support an evolving connection between astroglia and FTLD.
Supplementary Material
Protein quality for Control cases 1–3, 5–10 and FTLD cases 1–6, 8–10 (Supplemental Table S1) was evaluated by SDS-PAGE using 20 μg per case. Gel was stained with Coomassie Blue, destained, and scanned for image analysis. Control case 4 and FTLD case 7 were not included due to limited sample resource.
Acknowledgments
We thank members of the Peng and Levey/Lah labs for constructive discussion regarding this manuscript. This work was also supported by the National Institutes of Health through the Emory Alzheimer’s Disease Center grant (P50AG025688), the Emory Neuroscience NINDS Core Facilities (P30NS055077), and NIH training grant (F32AG032848-02 to N.T.S.). Parts of this research were conducted while J.H.H was an Ellison Medical Foundation/AFAR Postdoctoral Fellow. Acknowledgement is made to the donors of ADR, a program of the American Health Assistance Foundation, for support of this research.
Footnotes
Supporting Information Available
Supplemental Figure S1 and supplementary tables S1–S4 are available free of charge via the Internet at http:/pubs.acs.org.
References
- 1.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10 (10):1055–63. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296 (5575):1991–5. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 3.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314 (5796):130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, Foong C, White CL, 3rd, Schneider JA, Kretzschmar HA, Carter D, Taylor-Reinwald L, Paulsmeyer K, Strider J, Gitcho M, Goate AM, Morris JC, Mishra M, Kwong LK, Stieber A, Xu Y, Forman MS, Trojanowski JQ, Lee VM, Mackenzie IR. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171 (1):227–40. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, Chatterjee A, Hurtig HI, Karlawish JH, Rosen HJ, Van Deerlin V, Lee VM, Miller BL, Trojanowski JQ, Grossman M. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59 (6):952–62. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, Trojanowski JQ. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61 (5):427–34. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 7.Neumann M, Kwong LK, Truax AC, Vanmassenhove B, Kretzschmar HA, Van Deerlin VM, Clark CM, Grossman M, Miller BL, Trojanowski JQ, Lee VM. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J Neuropathol Exp Neurol. 2007;66 (3):177–83. doi: 10.1097/01.jnen.0000248554.45456.58. [DOI] [PubMed] [Google Scholar]
- 8.Mackenzie IR, Rademakers R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenetics. 2007 doi: 10.1007/s10048-007-0102-4. [DOI] [PubMed] [Google Scholar]
- 9.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268 (19):5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 10.Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci U S A. 1986;83 (11):4044–8. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iqbal K, Alonso Adel C, Chen S, Chohan MO, El-Akkad E, Gong CX, Khatoon S, Li B, Liu F, Rahman A, Tanimukai H, Grundke-Iqbal I. Tau pathology in Alzheimer disease and other tauopathies. Biochim Biophys Acta. 2005;1739 (2–3):198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Sternberger NH, Sternberger LA, Ulrich J. Aberrant neurofilament phosphorylation in Alzheimer disease. Proc Natl Acad Sci U S A. 1985;82 (12):4274–6. doi: 10.1073/pnas.82.12.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulloa L, Montejo de Garcini E, Gomez-Ramos P, Moran MA, Avila J. Microtubule-associated protein MAP1B showing a fetal phosphorylation pattern is present in sites of neurofibrillary degeneration in brains of Alzheimer’s disease patients. Brain Res Mol Brain Res. 1994;26 (1–2):113–22. doi: 10.1016/0169-328x(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 14.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422 (6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 15.Sadygov RG, Cociorva D, Yates JR. 3rd, Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat Methods. 2004;1 (3):195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 16.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20 (3):301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 17.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2 (11):1234–43. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4 (7):873–86. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell. 2006;127 (3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3 (11):1093–101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101 (33):12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A. 2000;97 (1):179–84. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23 (1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19 (4):375–8. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 25.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5 (5):591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Ye J, Jensen ON, Roepstorff P. Highly efficient phosphopeptide enrichment by calcium phosphate precipitation combined with subsequent serial immobilized metal ion affinity chromatography (IMAC) enrichment. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M700278-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7 (7):2845–51. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trojanowski JQ, Dickson D. Update on the neuropathological diagnosis of frontotemporal dementias. J Neuropathol Exp Neurol. 2001;60 (12):1123–6. doi: 10.1093/jnen/60.12.1123. [DOI] [PubMed] [Google Scholar]
- 29.McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol. 2001;58 (11):1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 30.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41 (4):479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 31.The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. Neurobiol Aging. 1997;18 (4 Suppl):S1–2. [PubMed] [Google Scholar]
- 32.McKeith IG, Dickson DW, Lowe J, Emre M, O’Brien JT, Feldman H, Cummings J, Duda JE, Lippa C, Perry EK, Aarsland D, Arai H, Ballard CG, Boeve B, Burn DJ, Costa D, Del Ser T, Dubois B, Galasko D, Gauthier S, Goetz CG, Gomez-Tortosa E, Halliday G, Hansen LA, Hardy J, Iwatsubo T, Kalaria RN, Kaufer D, Kenny RA, Korczyn A, Kosaka K, Lee VM, Lees A, Litvan I, Londos E, Lopez OL, Minoshima S, Mizuno Y, Molina JA, Mukaetova-Ladinska EB, Pasquier F, Perry RH, Schulz JB, Trojanowski JQ, Yamada M. Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology. 2005;65 (12):1863–72. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 33.Litvan I, Hauw JJ, Bartko JJ, Lantos PL, Daniel SE, Horoupian DS, McKee A, Dickson D, Bancher C, Tabaton M, Jellinger K, Anderson DW. Validity and reliability of the preliminary NINDS neuropathologic criteria for progressive supranuclear palsy and related disorders. J Neuropathol Exp Neurol. 1996;55 (1):97–105. doi: 10.1097/00005072-199601000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Dickson DW. Pick’s disease: a modern approach. Brain Pathol. 1998;8 (2):339–54. doi: 10.1111/j.1750-3639.1998.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81 (11):4566–75. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8 (8):3944–50. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 38.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 39.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4 (3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 40.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for highthroughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24 (10):1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 41.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125 (1):173–86. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 42.Xia Q, Liao L, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–56. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seyfried NT, Huysentruyt LC, Atwood JA, Iii, Xia Q, Seyfried TN, Orlando R. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: A membrane proteomics approach. Cancer Letters. 2008;263 (2):243–252. doi: 10.1016/j.canlet.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyUb chains. Mol Cell Proteomics. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gozal YM, Duong DM, Gearing M, Cheng D, Hanfelt J, Funderburk C, Peng J, Lah J, Levey A. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimers disease. Journal of Proteome Research. 2009 doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SI, Voshol H, van Oostrum J, Hastings TG, Cascio M, Glucksman MJ. Neuroproteomics: expression profiling of the brain’s proteomes in health and disease. Neurochem Res. 2004;29 (6):1317–31. doi: 10.1023/b:nere.0000023618.35579.7c. [DOI] [PubMed] [Google Scholar]
- 47.DeGiorgis JA, Jaffe H, Moreira JE, Carlotti CG, Jr, Leite JP, Pant HC, Dosemeci A. Phosphoproteomic analysis of synaptosomes from human cerebral cortex. J Proteome Res. 2005;4 (2):306–15. doi: 10.1021/pr0498436. [DOI] [PubMed] [Google Scholar]
- 48.Zhou JY, Hanfelt J, Peng J. Clinical proteomics in neurodegenerative diseases. Proteomics Clin Appl. 2007:1. doi: 10.1002/prca.200700378. in press. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Gould TD, Yuan P, Manji HK, Chen G. Post-mortem interval effects on the phosphorylation of signaling proteins. Neuropsychopharmacology. 2003;28 (6):1017–25. doi: 10.1038/sj.npp.1300112. [DOI] [PubMed] [Google Scholar]
- 50.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4 (10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, Au WS, Wang J, Li F, Ji S, Han H, Nie X, Li Q, Kung HF, Leung SY, Lin MC. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106 (3):342–7. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- 53.Choi SC, Kim KD, Kim JT, Kim JW, Yoon DY, Choe YK, Chang YS, Paik SG, Lim JS. Expression and regulation of NDRG2 (N-myc downstream regulated gene 2) during the differentiation of dendritic cells. FEBS Lett. 2003;553 (3):413–8. doi: 10.1016/s0014-5793(03)01030-5. [DOI] [PubMed] [Google Scholar]
- 54.Okuda T, Kondoh H. Identification of new genes ndr2 and ndr3 which are related to Ndr1/RTP/Drg1 but show distinct tissue specificity and response to N-myc. Biochem Biophys Res Commun. 1999;266 (1):208–15. doi: 10.1006/bbrc.1999.1780. [DOI] [PubMed] [Google Scholar]
- 55.Mitchelmore C, Buchmann-Moller S, Rask L, West MJ, Troncoso JC, Jensen NA. NDRG2: a novel Alzheimer’s disease associated protein. Neurobiol Dis. 2004;16 (1):48–58. doi: 10.1016/j.nbd.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi K, Yamada M, Ohata H, Momose K, Higuchi T, Honda K. Expression of Ndrg2 in the rat frontal cortex after antidepressant and electroconvulsive treatment. Int J Neuropsychopharmacol. 2005;8 (3):381–9. doi: 10.1017/S1461145705005134. [DOI] [PubMed] [Google Scholar]
- 57.Burchfield JG, Lennard AJ, Narasimhan S, Hughes WE, Wasinger VC, Corthals GL, Okuda T, Kondoh H, Biden TJ, Schmitz-Peiffer C. Akt mediates insulin-stimulated phosphorylation of Ndrg2: evidence for cross-talk with protein kinase C theta. J Biol Chem. 2004;279 (18):18623–32. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- 58.Boulkroun S, Fay M, Zennaro MC, Escoubet B, Jaisser F, Blot-Chabaud M, Farman N, Courtois-Coutry N. Characterization of rat NDRG2 (N-Myc downstream regulated gene 2): a novel early mineralocorticoid-specific induced gene. J Biol Chem. 2002;277 (35):31506–15. doi: 10.1074/jbc.M200272200. [DOI] [PubMed] [Google Scholar]
- 59.Nichols NR. Ndrg2, a novel gene regulated by adrenal steroids and antidepressants, is highly expressed in astrocytes. Ann N Y Acad Sci. 2003;1007:349–56. doi: 10.1196/annals.1286.034. [DOI] [PubMed] [Google Scholar]
- 60.Broe M, Kril J, Halliday GM. Astrocytic degeneration relates to the severity of disease in frontotemporal dementia. Brain. 2004;127 (Pt 10):2214–20. doi: 10.1093/brain/awh250. [DOI] [PubMed] [Google Scholar]
- 61.Martin JA, Craft DK, Su JH, Kim RC, Cotman CW. Astrocytes degenerate in frontotemporal dementia: possible relation to hypoperfusion. Neurobiol Aging. 2001;22 (2):195–207. doi: 10.1016/s0197-4580(00)00231-1. [DOI] [PubMed] [Google Scholar]
- 62.Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000) Neurochem Res. 2000;25 (9–10):1439–51. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- 63.Inagaki M, Nakamura Y, Takeda M, Nishimura T, Inagaki N. Glial fibrillary acidic protein: dynamic property and regulation by phosphorylation. Brain Pathol. 1994;4 (3):239–43. doi: 10.1111/j.1750-3639.1994.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 64.Takemura M, Gomi H, Colucci-Guyon E, Itohara S. Protective role of phosphorylation in turnover of glial fibrillary acidic protein in mice. J Neurosci. 2002;22 (16):6972–9. doi: 10.1523/JNEUROSCI.22-16-06972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eddleston M, Mucke L. Molecular profile of reactive astrocytes--implications for their role in neurologic disease. Neuroscience. 1993;54 (1):15–36. doi: 10.1016/0306-4522(93)90380-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res Bull. 1999;49 (6):377–91. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 67.Finch CE. Neurons, glia, and plasticity in normal brain aging. Neurobiol Aging. 2003;24(Suppl 1):S123–7. doi: 10.1016/s0197-4580(03)00051-4. discussion S131. [DOI] [PubMed] [Google Scholar]
- 68.Gomes FC, Paulin D, Moura Neto V. Glial fibrillary acidic protein (GFAP): modulation by growth factors and its implication in astrocyte differentiation. Braz J Med Biol Res. 1999;32 (5):619–31. doi: 10.1590/s0100-879x1999000500016. [DOI] [PubMed] [Google Scholar]
- 69.Beach TG, Walker R, McGeer EG. Patterns of gliosis in Alzheimer’s disease and aging cerebrum. Glia. 1989;2 (6):420–36. doi: 10.1002/glia.440020605. [DOI] [PubMed] [Google Scholar]
- 70.Arnold SE, Han LY, Clark CM, Grossman M, Trojanowski JQ. Quantitative neurohistological features of frontotemporal degeneration. Neurobiol Aging. 2000;21 (6):913–9. doi: 10.1016/s0197-4580(00)00173-1. [DOI] [PubMed] [Google Scholar]
- 71.Hanzel DK, Trojanowski JQ, Johnston RF, Loring JF. High-throughput quantitative histological analysis of Alzheimer’s disease pathology using a confocal digital microscanner. Nat Biotechnol. 1999;17 (1):53–7. doi: 10.1038/5225. [DOI] [PubMed] [Google Scholar]
- 72.Muramori F, Kobayashi K, Nakamura I. A quantitative study of neurofibrillary tangles, senile plaques and astrocytes in the hippocampal subdivisions and entorhinal cortex in Alzheimer’s disease, normal controls and non-Alzheimer neuropsychiatric diseases. Psychiatry Clin Neurosci. 1998;52 (6):593–9. doi: 10.1111/j.1440-1819.1998.tb02706.x. [DOI] [PubMed] [Google Scholar]
- 73.Kosako H, Amano M, Yanagida M, Tanabe K, Nishi Y, Kaibuchi K, Inagaki M. Phosphorylation of glial fibrillary acidic protein at the same sites by cleavage furrow kinase and Rho-associated kinase. J Biol Chem. 1997;272 (16):10333–6. doi: 10.1074/jbc.272.16.10333. [DOI] [PubMed] [Google Scholar]
- 74.Matsuoka Y, Nishizawa K, Yano T, Shibata M, Ando S, Takahashi T, Inagaki M. Two different protein kinases act on a different time schedule as glial filament kinases during mitosis. EMBO J. 1992;11 (8):2895–902. doi: 10.1002/j.1460-2075.1992.tb05358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishizawa K, Yano T, Shibata M, Ando S, Saga S, Takahashi T, Inagaki M. Specific localization of phosphointermediate filament protein in the constricted area of dividing cells. J Biol Chem. 1991;266 (5):3074–9. [PubMed] [Google Scholar]
- 76.Sekimata M, Tsujimura K, Tanaka J, Takeuchi Y, Inagaki N, Inagaki M. Detection of protein kinase activity specifically activated at metaphase-anaphase transition. J Cell Biol. 1996;132 (4):635–41. doi: 10.1083/jcb.132.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inagaki M, Gonda Y, Nishizawa K, Kitamura S, Sato C, Ando S, Tanabe K, Kikuchi K, Tsuiki S, Nishi Y. Phosphorylation sites linked to glial filament disassembly in vitro locate in a non-alpha-helical head domain. J Biol Chem. 1990;265 (8):4722–9. [PubMed] [Google Scholar]
- 78.Takemura M, Nishiyama H, Itohara S. Distribution of phosphorylated glial fibrillary acidic protein in the mouse central nervous system. Genes Cells. 2002;7 (3):295–307. doi: 10.1046/j.1365-2443.2002.00513.x. [DOI] [PubMed] [Google Scholar]
- 79.Tang G, Perng MD, Wilk S, Quinlan R, Goldman JE. Oligomers of mutant glial fibrillary acidic protein (GFAP) Inhibit the proteasome system in alexander disease astrocytes, and the small heat shock protein alphaB-crystallin reverses the inhibition. J Biol Chem. 285(14):10527–37. doi: 10.1074/jbc.M109.067975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korolainen MA, Auriola S, Nyman TA, Alafuzoff I, Pirttila T. Proteomic analysis of glial fibrillary acidic protein in Alzheimer’s disease and aging brain. Neurobiol Dis. 2005;20 (3):858–70. doi: 10.1016/j.nbd.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 81.Martinez A, Carmona M, Portero-Otin M, Naudi A, Pamplona R, Ferrer I. Type-dependent oxidative damage in frontotemporal lobar degeneration: cortical astrocytes are targets of oxidative damage. J Neuropathol Exp Neurol. 2008;67 (12):1122–36. doi: 10.1097/NEN.0b013e31818e06f3. [DOI] [PubMed] [Google Scholar]
- 82.Alonso AD, Grundke-Iqbal I, Barra HS, Iqbal K. Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc Natl Acad Sci U S A. 1997;94 (1):298–303. doi: 10.1073/pnas.94.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trivedi N, Marsh P, Goold RG, Wood-Kaczmar A, Gordon-Weeks PR. Glycogen synthase kinase-3beta phosphorylation of MAP1B at Ser1260 and Thr1265 is spatially restricted to growing axons. J Cell Sci. 2005;118 (Pt 5):993–1005. doi: 10.1242/jcs.01697. [DOI] [PubMed] [Google Scholar]
- 84.Schweigreiter R, Bandtlow CE. Nogo in the injured spinal cord. J Neurotrauma. 2006;23 (3–4):384–96. doi: 10.1089/neu.2006.23.384. [DOI] [PubMed] [Google Scholar]
- 85.Oertle T, Klinger M, Stuermer CA, Schwab ME. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003;17 (10):1238–47. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- 86.Saito N, Shirai Y. Protein kinase C gamma (PKC gamma): function of neuron specific isotype. J Biochem. 2002;132 (5):683–7. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- 87.Parekh DB, Ziegler W, Parker PJ. Multiple pathways control protein kinase C phosphorylation. EMBO J. 2000;19 (4):496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hofmann J. The potential for isoenzyme-selective modulation of protein kinase C. FASEB J. 1997;11 (8):649–69. doi: 10.1096/fasebj.11.8.9240967. [DOI] [PubMed] [Google Scholar]
- 89.Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell. 1993;75 (7):1253–62. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- 90.Buchner J. Hsp90 & Co. - a holding for folding. Trends Biochem Sci. 1999;24 (4):136–41. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 91.Miyata Y, Yahara I. The 90-kDa heat shock protein, HSP90, binds and protects casein kinase II from self-aggregation and enhances its kinase activity. J Biol Chem. 1992;267 (10):7042–7. [PubMed] [Google Scholar]
- 92.Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperones protein folding in vitro. Nature. 1992;358 (6382):169–70. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- 93.Miyata Y. Protein kinase CK2 in health and disease: CK2: the kinase controlling the Hsp90 chaperone machinery. Cell Mol Life Sci. 2009;66 (11–12):1840–9. doi: 10.1007/s00018-009-9152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein quality for Control cases 1–3, 5–10 and FTLD cases 1–6, 8–10 (Supplemental Table S1) was evaluated by SDS-PAGE using 20 μg per case. Gel was stained with Coomassie Blue, destained, and scanned for image analysis. Control case 4 and FTLD case 7 were not included due to limited sample resource.