Abstract
Silicon photonic microring resonators have emerged as a sensitive and highly multiplexed platform for real-time biomolecule detection. Herein, we profile the evanescent decay of device sensitivity towards molecular binding as a function of distance from the microring surface. By growing multilayers of electrostatically bound polymers extending from the sensor surface, we are able to empirically determine that the evanescent field intensity is characterized by a 1/e response decay distance of 63 nm. We then applied this knowledge to study the growth of biomolecular assemblies consisting of alternating layers of biotinylated antibody and streptavidin, which follow a more complex growth pattern. Additionally, by monitoring the shift in microring resonance wavelength upon the deposition of a radioactively labeled protein, the mass sensitivity of the ring resonator platform was determined to be 14.7 ± 6.7 [pg/mm2]/Δpm. By extrapolating to the instrument noise baseline, the absolute mass limit of detection is found to be 1.5 ± 0.7 pg/mm2. Taking the small surface area of the microring sensor into consideration, this value corresponds to an absolute mass detection limit of 97 ag (i.e. 0.6 zmol of IgG), demonstrating the remarkable sensitivity of this promising label-free biomolecular sensing platform.
Keywords: Biosensor, label-free, microring resonator, evanescent field, silicon photonics
1. Introduction
Waveguides, including fiber optics, have become valuable tools for detecting chemical and biological species through a range of optical transduction mechanisms, including absorbance, fluorescence, and refractive index-induced phase or transmission intensity modulation (Fan et al. 2008; Mukundan et al. 2009; Passaro et al. 2007; Wolfbeis 2002, 2006, 2008). For all waveguide materials and geometries, light propagates through the waveguide on account of total internal reflection that occurs due to the contrast in refractive index between the core and cladding materials. At the core/cladding interface, an evanescent optical field extends from the surface and decays exponentially with distance (Tsai et al. 1990). Regardless of the particular transduction mechanism utilized for sensing, the measured signal is elicited through light-matter interactions between this evanescent field and proximal target molecules.
Most relevant to this report are label-free transduction methods whereby the presence of a particular analyte causes a fundamental change in the optical transmission properties of the waveguide, thus eliminating requirements for chromophoric or fluorescent labels (Qavi et al. 2009). In most examples, the waveguide is chemically modified to present a target-specific capture element, and localization of the analyte at the core-cladding interface leads to an attenuation in the power of the transmitted light.
Recently there have been reports of chemical and biomolecular sensors based upon waveguides fabricated from a range of different materials, many of which leverage advances in semiconductor processing for micro- or nano-scale device fabrication (Passaro et al. 2007). A particularly promising waveguide material is silicon-on-insulator (SOI), a feedstock of the microelectronics industry (Jokerst et al. 2009). SOI waveguides are patterned into the top layer of silicon, and light is effectively guided due to the high refractive index contrast between Si and the cladding layers, which include the buried oxide and the top/side cladding layers. In addition to the obvious advantages of SOI in terms of scalability and potential for mass production, silicon photonic devices also feature good modal overlap between the guided optical mode and an analyte recognition layer (Densmore et al. 2006). The high refractive index of the waveguide core layer leads to a high sensitivity towards surface-confined binding events. Several groups have demonstrated silicon photonic waveguide configurations for chemical or biological sensing applications, including Mach-Zehnder interferometers (Densmore et al. 2009; Densmore et al. 2008; Densmore et al. 2006), resonant gratings (Schmid et al. 2009), and microcavity resonators (Carlborg et al. 2010; De Vos et al. 2009; De Vos et al. 2007; Ramachandran et al. 2008; Xu et al. 2008). Using arrays of SOI microring resonators, we previously reported a bulk refractive index sensitivity of 7.6 × 10-7 refractive index units (RIU) (Iqbal et al. 2010) and have demonstrated the sensitive, label-free detection of multiple proteins and nucleic acid sequences in both single and multiparameter analytical applications (Luchansky and Bailey 2010; Qavi and Bailey 2010; Washburn et al. 2009; Washburn et al. 2010).
Another label-free optical method that is particularly well-suited to analyzing biomolecular binding, thus serving as a basis for comparison, is surface plasmon resonance (SPR). SPR measures the interaction of an evanescent field extending from a metallic film, usually gold, in contact with the analyte-containing solution. Binding events that change the refractive index at the gold-solution interface are measured either as a change in the angle or wavelength of light which is maximally coupled into the metal film. SPR has been actively investigated as a biosensing technique over the past several decades with a number of reported sensor geometries and modes of operation, several of which are available commercially (Homola 2008; Piliarik and Homola 2006; Scarano et al. 2010). For SPR sensors, the evanescent field penetration depth, which is a function of wavelength and dielectric environment, is on the order of hundreds of nanometers (Homola 2006) with bulk refractive index sensitivities usually varying between 10-5 and 10-7 RIU (Fu et al. 2008; Homola 2008). For reference, a bulk index sensitivity of 10-6 RIU corresponds to a surface coverage resolution of 0.91 pg/mm2 (Homola 2008).
In this paper, we present a simulation of the waveguide optical intensity profile as well as a direct, experimental interrogation of the evanescent intensity decay profile of a silicon photonic microring resonator. We empirically measure the distance dependence of the sensing platform utilizing layer-by-layer electrostatic polymer multilayer deposition (Ganesan et al. 2006; Mandal et al. 2009). We also directly determine the bound mass sensitivity and limit of detection (LOD) of our system by correlating the resonance wavelength shifts with the amount of bound 125I-lableled streptavidin, measured via radiometric methods. Importantly, this paper presents key, experimentally determined sensitivity metrics for an emerging silicon photonic biomolecular sensing platform. In addition to providing a benchmark for comparison, the dependence of device sensitivity on distance from the sensor surface will be important when designing coatings for optimal analyte recognition and localization within the evanescent intensity profile of the guided optical mode.
2. Materials and methods
2.1. Chemicals and materials
N-hydroxysuccinimidobiotin (NHS-Biotin), streptavidin, Zeba spin filter columns, and pre-coated iodination tubes were obtained from Pierce (Rockford, IL). 3-aminopropyltriethoxysilane (APTES) was purchased from Gelest (Morrisville, PA). Succinimidyl 4-formylbenzoate (S-4FB) and 3-N-((6-(N′-Isopropylidenehydrazino))nicotinamide)propyltriethoxysilane (HyNic silane), were purchased from SoluLink (San Diego, CA). Shipley 1813 photoresist, MF 319 developer, and 1165 photoresist stripper were purchased from MicroChem Corp. (Newton, MA). Poly(sodium 4-styrene-sulfonate) (PSS, MW∼70,000 Da), polyethyleneimine (PEI, 50% w/w in H2O, MW∼750,000 Da), and poly(allylamine hydrochloride) (PAH, MW∼56,000 Da) were obtained from Sigma Aldrich (St. Louis, MO). Biotinylated, monoclonal mouse anti-human IL-2 antibody (catalog# 555040, clone B33-2 was purchased from BD Biosciences (San Jose, CA). All other chemicals were used as received from Sigma Aldrich (St. Louis, MO) and.
All buffers were made with purified water (ELGA PURELAB filtration system; Lane End, UK), and the pH was adjusted using 1 M HCl or 1 M NaOH. Tris buffer consisted of 0.5 mM Tris and 100 mM NaCl adjusted to pH 7.1. Tris/EDTA buffer was made by dissolving 1.21 g Tris base, 0.558 g disodium EDTA, and 0.2 g sodium azide in 1 L water and adjusting to pH 7.4. PBS was made by dissolving 9.6 g Dulbecco's phosphate buffered saline mixture into 1 L water and adjusting to pH 7.4. Antibody immobilization buffer consisted of 100 mM PBS with 150 mM NaCl adjusted to pH 6.0. BSA-PBS buffer consisted of 0.1 mg/mL BSA in PBS. For blocking, StartingBlock blocking buffer (Pierce) was used.
2.2. Device: Microring resonator sensors and instrumentation
The design and fabrication of microring resonator array chips, the accompanying instrumentation, and the fluidics have been described previously (Iqbal et al. 2010; Washburn et al. 2009). Briefly, chips having 32 individually addressable microrings were used for the experiments. The entire substrate was coated with a fluoropolymer cladding layer that was removed from annular openings over 24 of the sensors. The remaining eight microrings were left occluded by the cladding and were utilized as thermal controls, since they were not exposed to the solution. Microring resonance frequencies were measured as described previously. Briefly, the beam of a 1560-nm center wavelength tunable, external cavity diode laser is focused onto an input grating coupler on the chip surface to couple light into the linear waveguide adjacent to a given microring. The laser output is then rapidly swept through a 12-nm spectral window, and the intensity of light projected from the output grating coupler is monitored as a function of laser wavelength. Resonance wavelengths are determined as minima in output coupler intensity. This process is repeated for each interrogated microring sensor by rastering the laser across all 32 input grating couplers, allowing resonance determination with ∼250-ms time resolution. Thermal control ring responses are used to control for ambient thermal drift.
2.3. Layer-by-layer electrostatic polymer deposition for evanescent decay profiling
For the layer-by-layer electrostatic deposition of polymers, PEI, PSS, and PAH were dissolved in Tris buffer to 5 mg/mL. After cleaning the microring surface with piranha solution (3:1 H2SO4:H2O2)1 and loading the chip into a previously described microfluidic flow cell (Washburn et al. 2009), chips are exposed to PEI for 5 min at 30 μL/min. Maintaining constant flow conditions with a P625 peristaltic pump from Instech Laboratories (Plymouth Meeting, PA), the surface is then rinsed with Tris buffer and exposed to PSS for 10 min followed by a buffer rinse. The surface is then exposed to PAH for 4 minutes with a 5 minute buffer rinse. The PSS and PAH deposition cycles (with the PSS time reduced to 6 minutes) and buffer rinses were repeated until a total of 72 bilayers had been grown. To facilitate automated solution switching, we constructed a robot using the LEGO (Billund, Denmark) MINDSTORMS NXT 2.0 system that automatically moved the inlet tubing between solutions. More details of the robot's design are given in the supplementary information.
2.4. Deposition of protein multilayers for evanescent field characterization
Prior to performing layer-by-layer deposition of proteins, hydrazine moieties were installed on the silicon oxide chip surface by flowing a solution of HyNic-silane (1 mg/mL in 95% ethanol and 5% dimethyl formamide) over the surface for 30 min at 5 μL/min followed by rinsing with ethanol. Separately aldehyde groups were introduced to biotinylated IgG antibodies by reaction with a 5-fold molar excess of 0.2 mg/mL S-4FB for 2 h at room temperature. After buffer exchanging with a Zeba spin column into 100 mM PBS pH 6.0 to remove excess S-4FB, the antibody solution was diluted to 0.1 mg/mL and flowed over the entire sensor array for covalent attachment via a hydrazone linkage. Aniline (100 mM) was added to catalyze the formation of the hydrazone bond between the antibody aldehyde and the hydrazine-functionalized surface (Byeon et al. 2010). After rinsing the chip in buffer, the surface was blocked in StartingBlock overnight at 4 °C.
For layer-by-layer protein deposition, biotinylated anti-IL-2 and streptavidin were diluted in BSA-PBS to 2 μg/mL. After loading the antibody-functionalized chip into the flow cell, BSA-PBS buffer was flowed at 23 μL/min. Maintaining the same flow conditions, the surface was exposed to streptavidin for 20 min, followed by a 2-min buffer rinse, and then to biotinylated antibody for 20 min, followed by a 2-min buffer rinse. This cycle was repeated for a total of 75 layers (37 bilayers on top of an antibody-functionalized surface). The LEGO robot was again used to facilitate automated solution switching over the ∼30-hr multilayering process.
2.5. Determination of bound mass sensitivity using radio-labeled streptavidin
Streptavidin was labeled with 125I, a γ-emitter, according to the manufacturer's instructions using pre-coated iodination tubes (Pierce). Labeled streptavidin was purified from free 125I using a G-25 Sephadex-packed glass column. The concentration of radiolabeled streptavidin (> 90% purity) was determined to be 1-2 μg/mL for each of three trials.
The Storm 840 PhosphorImager (GE Healthcare) utilized to image the amount of 125I-streptavidin deposited on the substrate did not have adequate spatial resolution to allow visualization of individual sensing elements. Therefore, the fluoropolymer cladding layer was removed in its entirety from half of each chipusing photolithography and reactive ion etching (RIE), as described in the supplementary information.
Before flowing 125I-streptavidin over the sensor surface, the chip was piranha cleaned as and loaded in the microfluidic flow cell with a gasket that allowed exposure of the entire array to a single solution volume. The surface was first modified with an amine-terminated silane by flowing a 2% solution of APTES in ethanol over the surface at 30 μL/min for 10 minutes, followed by rinsing with ethanol. A 0.4-mM NHS-Biotin solution (in 100mM PBS, pH 7.4 with 4% DMF) was flowed over the surface at 30 μL/min for 30 min, followed by rinsing with PBS. Finally, the sensor surface was exposed to the 125I-streptavidin solution for 10-30 min and washed with buffer. After the microring resonance shift determination, the chip was imaged on the phosphorimager along with a set of spotted protein standard solutions that were allowed to evaporate onto a glass slide. The bound streptavidin surface density was determined by comparison to protein standards and correlated with the wavelength shifts from rings on the side of the chip from which the cladding layer had been removed. This process was performed in triplicate.
3. Calculation
3.1. Evanescent field theory: sensing modality and distance dependence
For waveguide sensors, the sensing modality is based on the interaction of the propagating evanescent field with the molecules of interest at or near the waveguide surface. In our silicon-on-insulator platform, light is coupled into linear Si waveguides that access the microring waveguides. As shown in Figure 1, each 30-μm ring is separated from the linear waveguide by 200 nm to allow for efficient light coupling between the waveguides only at wavelengths (λ) that match the resonance condition defined by:
Fig. 1.

Scanning electron micrograph of a Si microring resonator waveguide adjacent to a linear waveguide. An annular opening in the fluoropolymer cladding layer exposes the surrounding SiOx surface only in the immediate vicinity of the ring.
| Eq. 1 |
where m is an integer, r is the microring radius, and neff is the effective refractive index of the optical mode. Since organic and biological molecules have higher refractive indices (n = 1.4-1.6) than water (n = 1.33), their binding to the sensor surface increases the effective refractive index sampled by the optical mode (Vörös 2004). The monitoring of resonant wavelength shift as a function of target binding provides the label-free sensing modality.
The equations describing the evanescent field propagation are derived from the boundary conditions of the electromagnetic wave equations that require a non-zero electric field at the reflective dielectric interface. As a result, the evanescent wave decays exponentially from the surface and, ignoring light scattering, the rate at which the field intensity decays can be described by:
| Eq.2 |
where I(z) is the intensity of the evanescent as a function of perpendicular distance z from the surface, I0 is the initial intensity, and γ is the exponential decay constant. As described previously, the shift in resonance wavelength is proportional to the intensity of the evanescent field extending into the surrounding environment (Arnold et al. 2003).
3.2. Simulation of evanescent field depth
Simulations of the waveguide optical intensity profile were performed using the classic eigenfunction approach as described previously (Marcatili 1969). Briefly, Maxwell's equations are solved satisfying the boundary conditions at the dielectric interfaces of the waveguides, resulting in a set of transcendental equations that were solved for the wavenumber of the transverse optical mode. The mode is oscillatory in the core of the waveguide, but in the evanescent region external to the core, it decays exponentially as shown in Figure 2. The physical constants used in the calculation were matched to those in the experimental system, characterized by a 200-nm thick by 500-nm wide waveguide having ncore = 3.43, nsubstrate=1.46, nsurroundings=1.33, and λ=1.56 μm. The exponential decay coefficient, γ, in Eq. 2 was found to be 9.56 μm-1 for the optical field at the surface of the waveguide extending into the analyte sensing region. At a distance of z = 1/2γ= 52.3 nm from the waveguide, the intensity of the evanescent field, I(z), has decreased to (1/e) × I0 or 36.7% of its initial value.
Fig. 2.
Cross-sectional plot of the evanescent field intensity for the 0.2 × 0.5- micron optical waveguide. The dashed lines represent the top and bottom extents of the waveguide core. The top surface of the waveguide exposed to the chemical reactions is at x = 0. Refractive indices of each region utilized in the simulation of the evanescent profile are depicted.
4. Results and discussion
4.1. Experimental profiling of the microring resonator evanescent intensity decay using layer-by-layer electrostatic polymer deposition
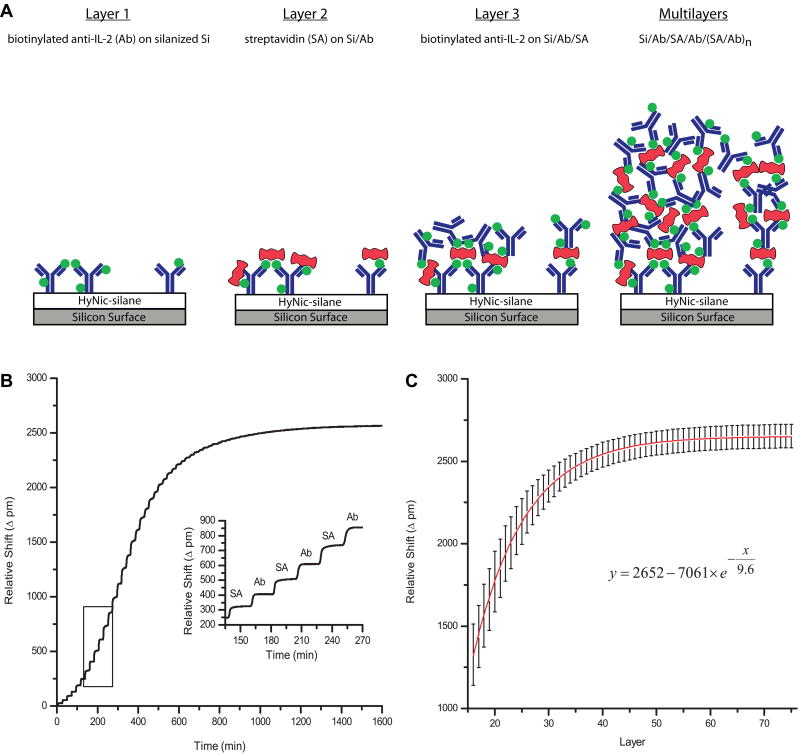
To experimentally profile the evanescent decay from our microrings, we utilized layer-by-layer assembly of alternating anionic and cationic polymers onto the surface to map the intensity of the sensor response as a function of z. Specifically, PSS and PAH were selected due to the extensive literature describing PSS/PAH multilayers with linear growth over a wide range of thicknesses (Decher and Schmitt 1992; Ladam et al. 1999; Wong et al. 2004). Following these previous reports, we prepared our silicon oxide-passivated surface with a layer of the highly branched polymer PEI to ensure sufficient coverage of the initial polyelectrolyte, followed by the addition of subsequent layers of PSS and PAH. Figure 3A illustrates the growth process, starting with the negatively charged silicon oxide surface. After addition of a PEI layer, PSS and then PAH are added in sequence. This process is repeated to form 72 PSS/PAH bilayers. Figure 3B shows the real-time shift in resonance wavelength from a representative microring corresponding to the addition of each electrostatic polymer layer. With each additional layer, the differential sensor response decreases as multilayer assembly occurs at a greater distance from the surface. The decreasing sensitivity to refractive index changes corresponds to the evanescent intensity decay profile extending off of the microring waveguide. Figure 3C shows the average net sensor response (n = 23 rings) for each successive polymer bilayer as a function of the number of PSS/PAH bilayers added.
Fig. 3.
(A) Schematic showing the layer-by-layer addition of polyelectrolytes to a negatively charged silicon oxide surface. Layer 1 involves addition of positively charged, highly branched PEI, and Layer 2 involves addition of negatively charged PSS on top of the PEI. Layer 3 consists of positively charged PAH adhering to the PSS layer. Multilayers are added by alternating deposition of PSS and PAH layers. (B) Real time data showing the relative shift in resonance frequency (Δ pm) upon addition of PAH and PSS layers. Inset shows six layer-by-layer depositions at higher resolution. (C) Plot showing the relative resonance wavelength shift for each successive PAH/PSS bilayer as a function of bilayer number. The exponential fit shown in the plot models the decay rate as the polyelectrolyte multilayers grow further from the surface and experience the decreasing evanescent field intensity, as evidenced by the reduced response for each subsequent layer. Error bars represent the standard deviation for n = 23 rings.
By fitting the points in Figure 3C to an exponential function, it becomes apparent that the response for each subsequent layer decays exponentially. From the exponential fitting function, the 1/e decay length for sensor response is 20.9 bilayers, which can be converted to a decay distance of 63 ± 4 nm using a 3.0 ± 0.2 nm thickness for each hydrated bilayer (Ladam et al. 1999; Wong et al. 2004). The literature value for PSS/PAH bilayer thickness was also experimentally confirmed through ellipsometry performed at several points during a layer-by-layer deposition process on a large area substrate (supplementary information). This experimentally determined decay distance of 63 nm is in reasonable agreement with the evanescent intensity simulation, which yielded a value of 52 nm. Possible sources of discrepancy include: uncertainty in hydrated bilayer thickness and film uniformity; the difference in surrounding index of buffer solution (experimental) versus water (simulation); dynamic changes in the mode profile during the course of multilayer growth; and other experimental deviations from idealized conditions. However, both simulation and empirical measurement provide sufficient information regarding the evanescent decay to interpret the results from the following protein multilayer study.
4.2. Monitoring the growth of protein multilayers
After verifying the exponential decay length experimentally using the electrostatic polymer system, we turned to a biological layer-by-layer system with more complex growth characteristics. As a test case, we constructed multilayers of an antibody (biotinylated mouse anti-human IL-2 IgG) functionalized with ∼10 biotin molecules, and streptavidin, which binds with high-affinity (Kd ∼ 10-15 M) to up to four biotin molecules. The multivalent nature of both biomolecules ensures that extended assemblies can be grown. As shown in Figure 4A, the surface is first functionalized with SFB-modified biotinylated antibody (Ab) as has been described previously (Luchansky and Bailey 2010), followed by addition of a streptavidin (SA) layer. Additional biotinylated Ab was attached to the bound SA. This process was repeated, resulting in multilayer growth (75 total layers). The resonance wavelength shift associated with the growth of each layer was monitored in real time under conditions empirically optimized to insure complete layer formation. As shown in Figure 4B, the resonant wavelength shift for each layer initially follows a non-linear growth trend, as will be further discussed below, before the effect of the decaying evanescent intensity dominates as the distance from the ring increases. This leads to a pseudo-inflection point in Figure 4B at ∼300 min. Following this inflection point, the average cumulative shift arising from protein layers 16 through 75 can be fit to an exponential function, as shown in Figure 4C. This fit is in the same form as the exponential function used to model the response to polymer multilayers. The shifts corresponding to layers 1-15 were not included since layer growth during this stage not only involves propagation through a decaying evanescent field, but also likely variations in the physical characteristics of each protein layer (packing density, surface coverage, protein orientation, etc.).
Fig. 4.
(A) Schematic showing the layer-by-layer addition of biotinylated antibody (Ab) and streptavidin (SA) to the ring resonator surface. Layer 1 shows covalent attachment of 4FB-modified, biotinylated antibody to the HyNic-silane functionalized silicon surface. We hypothesize that initial antibody functionalization yields incomplete surface coverage, which is followed by an annealing process that initially results in non-linear layer growth as vacancies are filled. Layers 2 and 3 show subsequent addition of streptavidin and biotinylated antibody, respectively. Extended multilayers are formed by alternating deposition of streptavidin and biotinylated antibody. Though each antibody is functionalized with ∼10 biotins, fewer are shown for clarity. (B) Real time data showing the relative shift in resonance frequency (Δ pm) upon addition of SA and Ab layers. Inset shows six layer-by-layer depositions at higher resolution. (C) Plot showing the cumulative relative shift per antibody or streptavidin layer (each individual addition of streptavidin or antibody is counted as a single layer). The exponential fit shown in the plot indicates the decay rate starting from layer 16. Error bars represent the standard deviation for n = 4 rings.
From the protein multilayer exponential fitting equation, we can determine that the 1/e decay length is 9.6 layers. In other words, after 9.6 protein layers, the evanescent intensity has decreased to 1/e (or ∼37%) of its initial value at the ring surface. Using the 1/e distance of 63 nm determined experimentally with electrostatic polymer layers, these 9.6 monolayers each represent a 6.6-nm thickness on average (or ∼13 nm per Ab/SA bilayer). IgG antibodies have a molecular weight (MW) of ∼150 kDa with approximate molecular dimensions of 15 × 7 × 3.5 nm, and their four-polypeptide-chain structure is highly flexible (Jung et al. 2008; Saphire et al. 2002). Streptavidin (MW ∼ 53 kDa) is roughly one-third the size of an antibody with dimensions of 5.8 × 5.4 × 4.8 nm (Cassier et al. 1998; Hendrickson et al. 1989), and a densely packed streptavidin monolayer has a thickness of 4.5-5 nm from studies using ellipsometry and electron microscopy (Darst et al. 1991; Herron et al. 1992). By simple addition of thicknesses, an Ab/SA bilayer is expected to be between 8 and 21 nm thick. Considering that the Abs are randomly oriented on the surface and that this disorder propagates with the addition of SA, the average thickness of 13 nm per bilayer seems quite reasonable.
Comparing the cumulative sensor response of the electrostatic polymer and the protein multilayers provides additional insights into the nature of the assemblies. In Figure 3C, the signal arising from 72 polymer bilayers effectively levels off at a maximum cumulative wavelength shift of ∼10,000 pm. These 72 bilayers extend ∼210 nm from the ring surface before the additional signal arising from subsequent layers becomes increasingly negligible due to the evanescent intensity decay. However, in the case of the protein multilayers, as depicted in Figure 4C, the cumulative wavelength shift arising from 37 protein bilayers levels off at ∼2,700 pm, approximately four times less than the maximum signal from polymer multilayers. This implies that, given the similar refractive indices of the proteins and polymers (n ≈ 1.5), the polymers form a higher density assembly that yields a larger total response within the bounds of the same evanescent field profile.
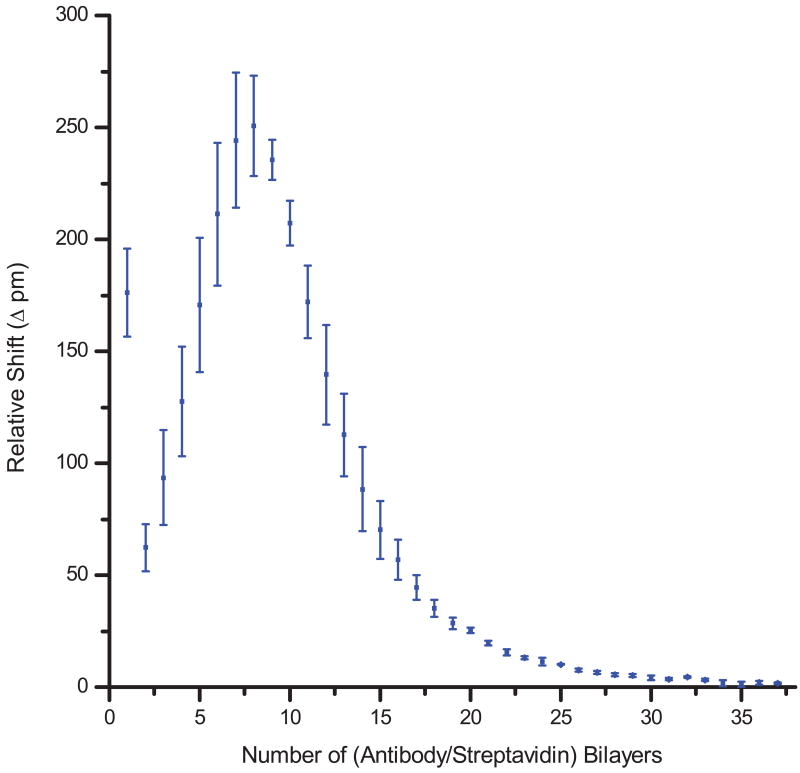
In contrast to the linear trend of the initial electrostatic polymer multilayers, the first 15 protein layers display a non-linear growth profile in which each layer yields a larger resonance wavelength shift than the previous layer. This effect can beobserved in Figure 5, where the differential resonance shift between each layer is plotted as a function of layer number. Prior to bilayer 8 (the first 15 protein layers), each successive layer corresponds to an increasing differential shift despite the decreasing evanescent field intensity, with a maximum shift of 250 pm/protein bilayer at bilayer 8. Following this maximum shift, the observed resonance shift behavior is dominated by the decay of the evanescent intensity. We attribute this interesting multilayer assembly behavior to an increasing protein binding density throughout the early phase of growth (layers 1-15) where later layers are more complete than those initially deposited. We imagine initial antibody surface loading yields an incomplete monolayer, and subsequent biomolecular layers effectively anneal the initial holes in the film. After the addition of 15 protein layers (8 bilayers), we suspect that most holes have been filled as further layer growth appears linear. This means that subsequent resonance wavelength shifts for the remaining layers are completely dictated by the decay in the evanescent intensity. This model of linear layer growth following complete annealing by layer 15 is consistent with the measured shift in resonance wavelength for layers 16-75. A simple exponential decay function effectively fits the behavior of layers 16-75 (Figure 4C), where we assume linear growth of the protein multilayers and calculate an average protein layer thickness of 6.6 nm. This analysis reveals that our understanding of the evanescent intensity decay at increasing distances from the sensor surface allows for the isolation of contributions from non-linear layer growth and estimation of the protein layer thickness. Most importantly, however, we can directly observe the diminishing signal measured as capture agents are localized farther away from the sensor surface.
Fig. 5.
Plot displaying the relative differential shift per antibody/streptavidin bilayer. Notably, the signal increases initially to a maximum shift of ∼250 pm at bilayer 8. Beyond this point the signal from subsequent layers decreases in an exponential fashion as they are deposited further away from the surface where there is a lower evanescent field intensity. Error bars represent the standard deviation for n = 4 rings.
4.3. Determining the bound mass sensitivity for the microring resonator biosensing platform
Another metric that is valuable for comparing the relative performance of different sensing methodologies is the number of target molecules required to elicit a measureable sensor response. Clearly, the mode of signal transduction, the sensor configuration and geometry, and the size or mass of the target molecule play a critical role in determining how best to define this metric. Therefore, we decided to determine the absolute LOD of the microring resonator biosensing platform in terms of the minimum mass that can be detected. Since most biological molecules have similar refractive indices, we assert that this metric can be converted to the LOD in terms of the number of molecules for any bound analyte of known molecular weight.
In order to probe the mass sensitivity of the microring resonator platform, the amount of material bound at the surface must be carefully measured and correlated with the resonance wavelength shift upon binding. Our approach involves (1) measuring the wavelength shift of radioactive streptavidin binding to a biotinylated microring surface and then (2) determining the mass of bound streptavidin using standard radiometric techniques. Specifically, we used 125I-labeled streptavidin and phosphorimaging to determine the amount of mass bound to the surface. The relationship between 125I-streptavidin surface density and net resonance wavelength shift response was measured for three different sensor chips and used to determine the mass sensitivity and LOD for the biosensing platform.
The phosphorimager utilized for streptavidin mass quantitation has a spatial resolution of 50 μm, which is not sufficient to resolve individual microring sensing elements, especially considering that the fluoropolymer cladding material is only removed from an 8-μm wide annular opening over each microring (see Figure 1). Furthermore, we observed a non-zero radioactive response from the cladding material, meaning not all signal in a single phosphorimager pixel can be attributed to a microring. Therefore, we utilized RIE to remove the cladding layer from one half of the chip before the surface was functionalized. Etching only half of the chip allowed half of the rings to remain unaltered to ensure that no binding artifacts or inconsistencies resulted from the etching process and to preserve unexposed thermal control rings for drift correction.
Given that the silicon microrings possess a native layer of silicon oxide, we feel it is a reasonable assumption that the binding density of 125I-streptavidin on a microring will be comparable to that of the freshly-exposed bulk silicon oxide surface. To ensure that replicate measurements were independent of chip-to-chip variations, we tested the refractive index sensitivity of each chip by measuring the relative wavelength shift elicited by switching the surrounding solution from water to 1 M NaCl. All rings on all chips responded to 1 M NaCl with a 475 ± 25 pm shift. This variation falls well within the precision tolerance required to assess the absolute mass bound to the microring surface.
Each chip was functionalized with APTES and NHS-biotin, and then exposed to 125I-streptavidin, creating a protein layer as shown in Figure 6A. To obtain a range of surface densities for calculating mass sensitivity, variable streptavidin loadings were produced by monitoring the real-time binding of streptavidin and initiating a buffer rinse after different exposure times (from 10 to 30 minutes). Figure 6B shows a representative binding curve where 1.2 μg/mL 125I-streptavidin is introduced at t = 12. With varied streptavidin exposure times, we were able to obtain net sensor responses of 46 pm (low coverage), 165 pm (medium coverage), and 216 pm (high/saturated coverage). Following the buffer rinse and net binding shift determination, the chips were imaged, and the amount of bound mass was determined by comparison with spotted standards.
Fig. 6.
(A) Schematic showing the APTES-modified silicon surface coated with a layer of NHS-biotin followed by a layer of 125I-labeled streptavidin. (B) Real time data showing binding of 125I-labeled streptavidin to a biotin-functionalized surface. After rinsing the surface with buffer, the average net shift (relative shift after streptavidin minus relative shift before streptavidin) is measured for multiple rings. (C) Phosphorimage showing the relative intensity of 125I-streptavidin standards compared with that bound on the U-shaped area of the chip to which flow was directed by microfluidics. The red rectangle highlights the region of interest directly over the rings on the chip where the radioactivity intensity was measured. In this selected area, the cladding layer was etched off exposing silicon oxide over the entire surface. (D) Calibration curve relating the mass of spotted protein standards (black dots) from (C) to the phosphorimager intensity obtained. The blue square represents the signal arising from the selected area (box in C).
As shown in Figure 6C, 125I-streptavidin bound to all areas over which flow was directed, including the half of the chip where the cladding layer was intact (although to a lesser extent). A rectangular region directly over the rings on the etched half of the substrate was selected on each chip, and the integrated total image intensity from the selected area was converted to a mass of bound streptavidin based on calibration with the 125I-streptavidin standards (see Figure 6D; additional data presented in the supplementary information). Based on the pixel count of the sampled channel area and the pixel size of the phosphorimager, the bound mass was converted to a surface density. By dividing the surface density by the average net shift of the rings from within the sampled channel area, the average mass sensitivity of the three chips was determined to be 14.7 ± 6.7 [pg/mm2]/Δpm (see Table S-1 in the Supplementary Information). Thus, a resonance wavelength shift of 1 pm represents 15 ± 7 pg/mm2 of bound protein, or inversely, each 1 pg/mm2 of protein density generates a 0.07 ± 0.03 pm shift in resonant wavelength. Based on a signal noise floor of 0.1 pm (see details in the supplementary information), the platform's LOD was determined to be 1.5 ± 0.7 pg/mm2. Using the surface area of 66 μm2/ring and the surface density LOD described above, we determine the absolute mass LOD to be 97 ag of bound protein. This mass sensitivity signifies the ability to detect just 0.6 zmol (∼400 molecules) of an antibody bound to the microring in a direct and label-free assay. The use of secondary antibodies in sandwich assay formats can further augment the sensitivity of the platform to a protein target, especially for lower molecular weight protein targets (Luchansky and Bailey 2010).
5. Conclusions
Through a combination of two multilayer studies on a silicon photonic microring resonator biosensing platform, the experimentally determined evanescent intensity decay profile was found to be in reasonable agreement with simulation results. The 1/e decay distance of binding sensitivity was measured to be 63 nm, highlighting the outstanding overlap of the waveguide mode with the biomolecular sensing region. Since most biological binding interactions with a capture probe (i.e. antibody, nucleic acid strand) and target analyte have dimensions on the order of tens of nanometers, this platform is well-suited for sensing specific binding eventsnear the ring without large contributions from the surrounding environment (Kunz and Cottier 2006). Furthermore, with an empirical measure of the decay length characteristics of the microrings, it may be possible to design and optimize capture strategies to maximally utilize this property. For example, it is clear that assay designs involving non-fouling surface modifications (i.e. PEG chains, polymer brushes, etc) and secondary/tertiary signal amplification steps should take under consideration the strong evanescent decay distance dependence of sensor response. These considerations become especially clear in the protein multilayer data where the signal is reduced by 63% after only 5 SA/Ab bilayers. Understanding the decay length also enables elucidation of non-linear growth processes such as those observed in the protein multilayer system.
Beyond characterizing the sensing modality and the extent to which the evanescent intensity propagates through the sample, the inherent mass sensitivity of the platform was also empirically determined. Through the use of radio-labeled protein binding to the sensor surface, it was possible to directly relate a given resonance wavelength shift to an empirically determined protein surface density. This allowed for determination of the platform's inherent sensitivity, which is 15 [pg/mm2]/Δpm. The high sensitivity of ring resonators to biological binding events at this early stage of technology development, with a ∼1.5 pg/mm2 LOD, is comparable to that of SPR (Homola 2008) and other label-free techniques (Carlborg et al. 2010; Fan et al. 2008; Qavi et al. 2009). By improving peak resolution and reducing system noise, this sensitivity may be improved further. In both the multilayering and surface density studies, the ability to monitor each ring's response in real time was vital to the characterization ofplatform performance metrics.
This paper presents important empirical determinations of key sensitivity metrics for a promising silicon photonic microring resonator chemical and biomolecular sensing platform. This information will be useful not only in comparing this platform to others, both optical and non-optical, but also provides key insights into the current limitations of sensor response which will be used to further optimize the technology for detection in complex, real-world environments.
Supplementary Material
Acknowledgments
This work is funded by the NIH Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01, and by the Camille and Henry Dreyfus Foundation. Eastman Chemical Company provided funds used to construct the LEGO robot used for multilayer growth automation. M.S.L. is supported via a National Science Foundation Graduate Research Fellowship and a Robert C. and Carolyn J. Springborn Fellowship from the Department of Chemistry at the University of Illinois at Urbana-Champaign. A.L.W. is supported via a National Science Foundation Graduate Research Fellowship. We acknowledge F. T. Limpoco for assistance with polyelectrolyte multilayer thickness determination via ellipsometry, Ji-Yeon Byeon for acquiring the SEM image of the microring, and Abraham Qavi for assistance in creating the robot automation. This research was carried out in part in the Frederick Seitz Materials Research Laboratory Central Facilities, University of Illinois, which are partially supported by the U.S. Department of Energy under Grants DE-FG02-07ER46453 and DE-FG02-07ER46471. No funding sources had any involvement beyond financial support.
Footnotes
Caution! Piranha solutions are extraordinarily dangerous, reacting explosively with traces quantities of organics.
Appendix A. Supplementary information: Supplementary information associated with this article, including a description of the automation of multilayer formation, polyelectrolyte multilayer thickness verification via ellipsometry, fluoropolymer cladding etching, surface density calibration data, and a peak shift resolution study, can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold S, Khoshsima M, Teraoka I, Holler S, Vollmer F. Opt Lett. 2003;28(4):272–274. doi: 10.1364/ol.28.000272. [DOI] [PubMed] [Google Scholar]
- Byeon, J.Y., Limpoco, F.T., Bailey, R.C., 2010. Under review.
- Carlborg CF, Gylfason KB, Kazmierczak A, Dortu F, Polo MJB, Catala AM, Kresbach GM, Sohlstrom H, Moh T, Vivien L, Popplewell J, Ronan G, Barrios CA, Stemme G, van der Wijngaart W. Lab Chip. 2010;10(3):281–290. doi: 10.1039/b914183a. [DOI] [PubMed] [Google Scholar]
- Cassier T, Lowack K, Decher G. Supramol Sci. 1998;5(3-4):309–315. [Google Scholar]
- Darst SA, Ahlers M, Meller PH, Kubalek EW, Blankenburg R, Ribi HO, Ringsdorf H, Kornberg RD. Biophys J. 1991;59(2):387–396. doi: 10.1016/S0006-3495(91)82232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K, Girones J, Popelka S, Schacht E, Baets R, Bienstman P. Biosens Bioelectron. 2009;24(8):2528–2533. doi: 10.1016/j.bios.2009.01.009. [DOI] [PubMed] [Google Scholar]
- De Vos KM, Bartolozzi I, Bienstman P, Baets R, Schacht E. Optical biosensor based on silicon-on-insulator microring cavities for specific protein binding detection. Nanoscale Imaging, Spectroscopy, Sensing, and Actuation for Biomedical Applications IV 2007 [Google Scholar]
- Decher G, Schmitt J. Fine-Tuning of the film thickness of ultrathin multilayer films composed of consecutively alternating layers of anionic and cationic polyelectrolytes. Trends in Colloid and Interface Science VI. 1992:160–164. [Google Scholar]
- Densmore A, Vachon M, Xu DX, Janz S, Ma R, Li YH, Lopinski G, Delage A, Lapointe J, Luebbert CC, Liu QY, Cheben P, Schmid JH. Opt Lett. 2009;34(23):3598–3600. doi: 10.1364/OL.34.003598. [DOI] [PubMed] [Google Scholar]
- Densmore A, Xu DX, Janz S, Waldron P, Mischki T, Lopinski G, Delage A, Lapointe J, Cheben P, Lamontagne B, Schmid JH. Opt Lett. 2008;33(6):596–598. doi: 10.1364/ol.33.000596. [DOI] [PubMed] [Google Scholar]
- Densmore A, Xu DX, Waldron P, Janz S, Cheben P, Lapointe J, Delage A, Lamontagne B, Schmid JH, Post E. IEEE Photon Technol Lett. 2006;18(21-24):2520–2522. [Google Scholar]
- Fan X, White IM, Shopoua SI, Zhu H, Suter JD, Sun Y. Anal Chim Acta. 2008;620(1-2):8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu E, Chinowsky T, Nelson K, Yager P. SPR Imaging for Clinical Diagnostics. In: Schasfoort RBM, Tudos AJ, editors. Handbook of Surface Plasmon Resonance. Royal Society of Chemistry; 2008. pp. 313–332. [Google Scholar]
- Ganesan PG, Wang X, Nalamasu O. Appl Phys Lett. 2006;89(21):213107–213103. [Google Scholar]
- Hendrickson WA, Pähler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Proc Natl Acad Sci U S A. 1989;86(7):2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron JN, Mueller W, Paudler M, Riegler H, Ringsdorf H, Suci PA. Langmuir. 1992;8(5):1413–1416. [Google Scholar]
- Homola J. Electromagnetic Theory of Surface Plasmons. Surface Plasmon Resonance Based Sensors. 2006:3–44. [Google Scholar]
- Homola J. Chem Rev. 2008;108(2):462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J Sel Top Quantum Electron. 2010;16(3):654–661. [Google Scholar]
- Jokerst NM, Luan L, Palit S, Royal M, Dhar S, Brooke MA, Tyler T. IEEE Trans Biomed Circuits Syst. 2009;3(4):202–211. doi: 10.1109/TBCAS.2009.2020693. [DOI] [PubMed] [Google Scholar]
- Jung Y, Jeong JY, Chung BH. Analyst. 2008;133:697–701. doi: 10.1039/b800014j. [DOI] [PubMed] [Google Scholar]
- Kunz R, Cottier K. Anal Bioanal Chem. 2006;384(1):180–190. doi: 10.1007/s00216-005-0196-1. [DOI] [PubMed] [Google Scholar]
- Ladam G, Schaad P, Voegel JC, Schaaf P, Decher G, Cuisinier F. Langmuir. 1999;16(3):1249–1255. [Google Scholar]
- Luchansky MS, Bailey RC. Anal Chem. 2010;82(5):1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Goddard JM, Erickson D. Lab Chip. 2009;9(20):2924–2932. doi: 10.1039/b907826f. [DOI] [PubMed] [Google Scholar]
- Marcatili EAJ. Bell Labs Technical Journal. 1969;48(21):2071–2102. [Google Scholar]
- Mukundan H, Anderson AS, Grace WK, Grace KM, Hartman N, Martinez JS, Swanson BI. Sensors. 2009;9(7):5783–5809. doi: 10.3390/s90705783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro VMN, Dell'Olio F, Casamassima B, De Leonardis F. Sensors. 2007;7(4):508–536. doi: 10.3390/s7112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piliarik M, Homola J. SPR Sensor Instrumentation. Surface Plasmon Resonance Based Sensors. 2006:95–116. [Google Scholar]
- Qavi AJ, Bailey RC. Angew Chem Int Ed. 2010;49(27):4608–4611. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qavi AJ, Washburn AL, Byeon JY, Bailey RC. Anal Bioanal Chem. 2009;394(1):121–135. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Wang S, Clarke J, Ja SJ, Goad D, Wald L, Flood EM, Knobbe E, Hryniewicz JV, Chu ST, Gill D, Chen W, King O, Little BE. Biosens Bioelectron. 2008;23(7):939–944. doi: 10.1016/j.bios.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Saphire EO, Stanfield RL, Crispin MDM, Parren PWHI, Rudd PM, Dwek RA, Burton DR, Wilson IA. J Mol Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- Scarano S, Mascini M, Turner APF, Minunni M. Biosensors Bioelectron. 2010;25(5):957–966. doi: 10.1016/j.bios.2009.08.039. [DOI] [PubMed] [Google Scholar]
- Schmid JH, Sinclair W, García J, Janz S, Lapointe J, Poitras D, Li Y, Mischki T, Lopinski G, Cheben P, Delâge A, Densmore A, Waldron P, Xu DX. Opt Express. 2009;17(20):18371–18380. doi: 10.1364/OE.17.018371. [DOI] [PubMed] [Google Scholar]
- Tsai DP, Jackson HE, Reddick RC, Sharp SH, Warmack RJ. Appl Phys Lett. 1990;56(16):1515. [Google Scholar]
- Vörös J. Biophys J. 2004;87(1):553–561. doi: 10.1529/biophysj.103.030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn AL, Gunn LC, Bailey RC. Anal Chem. 2009;81(22):9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal Chem. 2010;82(1):69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfbeis OS. Anal Chem. 2002;74(12):2663–2677. doi: 10.1021/ac020176e. [DOI] [PubMed] [Google Scholar]
- Wolfbeis OS. Anal Chem. 2006;78(12):3859–3873. doi: 10.1021/ac060490z. [DOI] [PubMed] [Google Scholar]
- Wolfbeis OS. Anal Chem. 2008;80(12):4269–4283. doi: 10.1021/ac800473b. [DOI] [PubMed] [Google Scholar]
- Wong JE, Rehfeldt F, Hanni P, Tanaka M, Klitzing RV. Macromolecules. 2004;37(19):7285–7289. [Google Scholar]
- Xu DX, Densmore A, Delâge A, Waldron P, McKinnon R, Janz S, Lapointe J, Lopinski G, Mischki T, Post E, Cheben P, Schmid JH. Opt Express. 2008;16(19):15137–15148. doi: 10.1364/oe.16.015137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.