Abstract
Background & Aims
CD166 (also called activated leukocyte cell adhesion molecule, ALCAM) is a marker of colorectal cancer (CRC) stem cells; it is expressed by aggressive tumors. Although the presence of CD166 at the tumor cell surface has been correlated with shortened survival, little is known about its function and expression in normal intestinal epithelia.
Methods
We characterized the expression pattern of CD166 in normal intestinal tissue samples from humans and mice using immunohistochemisty, flow cytometry and quantitative reverse transcription PCR. Human and mouse intestinal tumors were also analyzed.
Results
CD166 was expressed on the surface of epithelial cells within the stem cell niche and along the length of the intestine; expression was conserved across species. In the small intestine, CD166 was observed on crypt-based Paneth cells and intervening crypt-based columnar cells (putative stem cells). A subset of CD166-positive, crypt-based columnar cells co-expressed the stem cell markers Lgr5, Musashi-1, or Dcamkl-1. CD166 was located in the cytoplasm and at the surface of cells within human CRC tumors. CD166-positive cells were also detected in benign adenomas in mice; rare cells co-expressed CD166 and CD44 or epithelial-specific antigen.
Conclusions
CD166 is highly expressed within the endogenous intestinal stem cell niche. CD166-positive cells appear at multiple stages of intestinal carcinoma progression, including benign and metastatic tumors. Further studies should investigate the function of CD166 in stem cells and the stem cell niche, which might have implications for normal intestinal homeostasis. CD166 has potential as a therapeutic target for CRC.
Keywords: colon cancer, intestinal stem cells, cancer stem cell
Colorectal cancer (CRC) is the third most prevalent cancer in the United States, with nearly 150,000 new cases diagnosed annually. Despite efforts to improve early detection and treatment, over one-third of patients die annually from this disease1. The focus on cancer initiation and progression has dominated the effort to better understand disease pathology and guide therapeutic approaches. As such, the cancer stem cell (CSC) theory, which suggests that cancer is driven by cells harboring stem cell-like qualities, offers one explanation for why many current therapeutic approaches ultimately result in relapse of disease. In this model, some CSCs or cancer-initiating cells may be quiescent and, thus, evade eradication by standard cytotoxic therapies designed to target proliferating cells. These surviving cells can then proceed to support tumor growth and have potential to initiate recurrent or metastatic disease2-4. The reinvigoration of the CSC theory5-7 has led to identification, isolation and characterization of subsets of intestinal cancer cells that can recapitulate tumorigenesis in xenograft models8-12. While some cell surface molecules, such as CD133 and CD44, have been shown to mark CSCs in multiple organs, an additional number of markers have shown promising CSC expression in intestinal cancer including Dcamkl-1, ESA and CD16610,11,13.
CD166 or Activated Leukocyte Cell Adhesion Molecule (ALCAM) expression is pathologically correlated with aggressive disease in a variety of cancers including melanoma, prostate, breast, ovarian, esophageal, and bladder cancers14-20. In human CRC, aberrant cell surface CD166 expression is strongly correlated with a 15 month shortened survival21. Further, isolation of CD166/CD44 or CD166/ESA double-positive cells from human CRCs cells can recapitulate tumorigenesis when xenografted at low numbers into immune-deficient mice10, a hallmark of a CSC population. Although these findings suggest that CD166 may have a role in the progression of CRC, little is known about its endogenous function and cellular localization within the intestine.
In other organ systems, CD166 has a myriad of functions. This conserved cell adhesion protein participates in physiologic processes including leukocyte intravasation across the blood brain barrier, monocyte migration across endothelial junctions, angiogenesis, capillary formation, protection against apoptosis in breast cancer cells, and T-cell activation by both antigen presenting and tumor cells22-28. Further, CD166 has been described as a ligand that binds to CD6 on thymic epithelium29-31, acting in homophilic adhesion complexes between epithelial cells32, and as a cell surface marker for both a subset of hematopoietic progenitor cells33,34 and multipotent mesenchymal stem cells35,36. Based upon the intriguing CD166 expression pattern in multiple stem cell populations, this molecule has a potential role in maintaining stem cells in both normal and disease states. However, the potential overlap between CD166 normal and tumorigenic physiologic function have not been defined because the normal intestinal expression pattern has not been reported. Further, based upon its multiple roles in tumor-related processes, it is possible CD166 plays an important role in tumor pathology.
Correlation of the CD166 expression pattern with aggressive disease has led to efforts for targeting this molecule as a cancer therapeutic. Treatment of cancer cells with a CD166-internalizing antibody conjugated to chemotherapy filled lipid vesicles was shown to effectively target and kill CD166-expressing ovarian cancer cells and prostate cancer cells in vitro37,38. While early results from these types of targeted cancer therapies appear promising, it necessitates an even more careful understanding of the endogenous expression pattern and function of CD166. In the current study, we analyzed CD166 expression in normal human and mouse intestine. We identified enriched cell surface CD166 expression in the colon and small intestine (SI) crypt-base. Interestingly in the SI, CD166 is expressed on the cell surface of the differentiated Paneth cell population and the intervening crypt-based columnar cells. Notably, both normal and tumor CD166 expression patterns were conserved in mice, highlighting the value of using a mouse model for studying CD166 function within the stem cell niche and in cancer. Further, we show that a subset of CD166-expressing cells residing in the stem cell niche co-express other putative stem cell markers, including Musashi-1 (Msi-1), Dcamkl-1 and Lgr513,39-41. We propose that CD166 defines the normal intestinal stem cell niche and encompasses both differentiated Paneth cells as well as stem cell and progenitor populations. A possible function for CD166 may be to maintain the epithelial microenvironment of the stem cell niche. Therefore, targeting this cell surface antigen in cancer therapy requires careful consideration of potential effects on normal tissues.
Material and Methods
Mice
Mice were housed in a specific pathogen-free environment under strictly controlled light cycle conditions, fed a standard rodent Lab Chow (#5001 PMI Nutrition International), and provided water ad libitum. All procedures were performed in accordance to the OHSU Animal Care and Use Committee. The C57Bl/6 and ApcMin/+ mice42 were purchased from The Jackson Laboratory (Bar Harbor, ME).
Immunohistochemical and histochemical analyses of intestinal tissue
Adult (>6 weeks) and embryonic [(E)14.5, 15.5, 16.5, 17.5, 18.5] mouse intestines were dissected and prepared for paraffin and frozen tissue analyses as we have previously described43. Human SI and colonic tissue was fixed in 10% buffered formalin and embedded in paraffin or OCT. Five or 50μm tissue sections were stained with antibodies to CD166, Msi-1, Ki67, Serotonin, ChromograninA, ESA, CD44, Lysozyme and Laminin (Antibody information listed in Supplemental Table1). Antigen retrieval (10mM citrate buffer, pH=6 or 10mM Tris/1mM EDTA, pH=9 at 100°C for 20 minutes) was performed on paraffin embedded tissues. Visualization was performed using either fluorescent-conjugated species-specific secondary antibodies [Indocarbocyanine3 (Cy3), Indocarbocyanine5, Fluorescein isothiocyanate (FITC)] (1:500; Jackson ImmunoResearch), or brightfield diaminobenzidine (DAB) detection (Vectastain ABC kit; Vector) and Methyl green counter staining (Vector). Nuclear counterstaining with Hoechst dye (33258; Sigma; 0.1μg/ml) was performed for fluorescent analyses. For detection of CD166-positive Paneth cells, human SI tissue sections were incubated with antibodies to CD166, visualized with secondary Cy3-conjugated antibodies and images captured using a Leica DMR fluorescent microscope (Leica Microsystems). The tissue was then re-stained with Lendrum’s Phloxine Tartrazine according to standard procedures44, images recaptured and superimposed using Canvas X software (ACD). Confocal images were acquired as 0.5μm planes using an IX81 Inverted Microscope equipped with Fluoview FV1000-Spinning Disc Confocal (Olympus) scan head and FV10 ASW 1.7 software (Olympus).
To test specificity of CD166 antibodies, recombinant mouse CD166 (1 μg/mL; R&D Systems) was co-incubated with the primary CD166 antibody (15 μg/ml; R&D Systems) at room temperature for 30 minutes. Tissue sections were then incubated with the antibody peptide solution and visualized as described above.
Analyses of isolated intestinal epithelial cells
The differentiated (villus or colon cuff) and undifferentiated (crypt) epithelial cells of the mouse SI and colon were independently isolated using a modified Weiser preparation45, stained with antibodies to CD166, and sorted using a Cytopeia Influx to collect CD166+ SI villus or colonic cuff epithelia and CD166+ SI and colonic crypt epithelia, or crypt CD166− epithelia. Briefly, SI villus cells were isolated from 1mM EDTA washes and SI/colon crypt cells were isolated with subsequent 10mM EDTA washes at 4°C. Cells were resuspended in modified HBSS and gently filtered through a 45μm filter, then incubated on ice for 20 min with antibodies against CD45 conjugated to Allophycocyanin (APC) and CD166, followed by incubation with secondary antibodies conjugated to FITC for CD166 detection. Cells were resuspended in modified HBSS/5μg/ml propidium iodide/1% bovine serum albumin and sorted using a 150 μm nozzle, 4.5 psi and Spigot software. To insure sorting and analysis of single cells, a doublet discriminator was used to exclude cells based on pulse width. FACS data was analyzed using FCS Express Version 3 Research Edition (DeNovo Software). CD166+, CD45−, PI− cells (105) were collected and spun onto glass slides using a Shandon Cytospin 4 (Thermo Electron) and subsequently analyzed for expression of Paneth cell markers or expression of Lgr5 as described in the previous section.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) CD166+ populations
qRT-PCR was used to evaluate gene expression in isolated crypt and villus epithelium or in CD166+ FACS-isolated cells. Epithelial cell isolated described above. For crypt and villus epithelial expression, total RNA was isolated using RNeasy Mini kit (Qiagen) and cDNA was then generated using standard protocols45. For stem cell marker expression total RNA was isolated from CD166+ and CD166− FACS isolated cells using an RNAqueous kit (Ambion) and cDNA generated using a High Capacity cDNA Reverse Transcription kit (ABI). qRT-PCR was performed using a SYBR Green-based assay, a 7900HT Sequence Detector and analyzed according to established protocols45. Each cDNA sample was analyzed in triplicate, along with triplicate samples of the endogenous reference gene, Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and reported as an average of n=2 runs. Data was normalized to Gapdh, then for CD166 expression, calibrated against levels within the villus epithelium, or for stem cell genes, calibrated against CD166− expression. Primers listed in Table1.
Results
CD166 protein expression is enhanced in the base of the human and mouse small intestinal and colonic crypt epithelium
CD166 expression has been documented in human CRC21, but extensive evaluation of its expression pattern in normal tissue has not been performed. A previous study localizes CD166 to the cytoplasm of normal colonic epithelial cells within the crypt base21. When we stained normal human SI and colonic tissue sections with antibodies to CD166, we detected a different intestinal staining pattern. Consistent with its function in immune cells, CD166-expressing cells were detected within the intestinal mesenchyme (Fig.1A, arrows). In the epithelial compartment, detection with fluorescent and brightfield immunohistochemistry revealed enriched cell surface expression of CD166 protein in epithelial cells at the base of the crypts in both the SI (Fig.1A-B,E; arrowheads) and colon (Fig.1C-D,F; arrowheads). Peptide competition with the CD166 antibody supported the specificity of the CD166 antibody recognition pattern (Supplemental Fig.1). Importantly, in contrast to previous report21, CD166 expression appeared strongest on the cell surface, and was more robustly expressed on cells that resided in the base of the crypt.
Figure 1. CD166 expression pattern in the human small intestine and colon.
(A-C) Human small intestine stained with antibodies to CD166 (red) and counterstained with Hoechst (blue). (A) CD166-positive cells are located within the mesenchymal (arrows) and epithelial compartments (arrowheads). (Ba-Bb) Enlarged view of small intestinal crypts. (C-Da,Db) Human colon stained with antibodies to CD166, visualized with DAB (brown) in (C) and with fluorescence (red) and Hoechst counterstain (blue) in (Da-Db). Arrowheads point to cell surface epithelial expression. (E,F) larger magnification views of the crypt base boxed in yellow from B and D, respectively. Solid lines demark epithelial-mesenchymal boundary and dashed lines mark the apical epithelial surface. Bar=25μm.
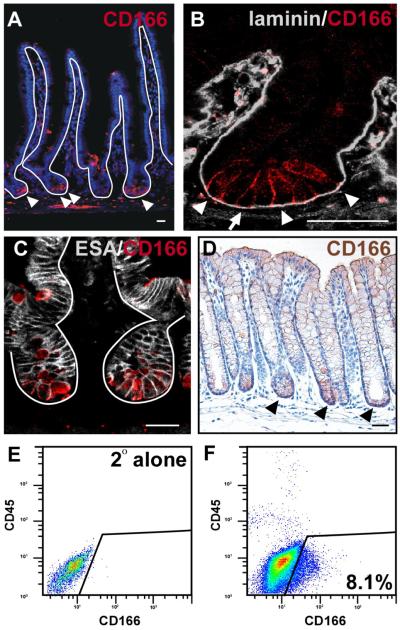
Optimal immunohistochemical expression patterns and in vivo molecular manipulations required to define functional properties are not readily obtainable within human tissues. Therefore, to validate the mouse as a viable model organism for future studies examining the role of CD166 in normal intestinal physiology and in tumorigenesis, we extended our analysis of CD166 to the mouse intestine. Because the mouse intestine is easily dissected, oriented and manipulated, it allowed for a more in-depth analysis of CD166 intestinal epithelial expression. As expected, the mouse expression pattern of CD166 recapitulated that of the human expression pattern (Fig.2). Importantly, the cell surface localization and increased expression levels on cells within the crypt-base were more readily appreciated (Fig.2A-D). Further, high expression levels on cells of the stem cell niche was confirmed on the RNA level using a differential isolation of villus and crypt epithelial cells followed by qRT-PCR and primers for CD166 (Supplemental Fig.2B). Interestingly, crypt-based expression did not vary down the length of the SI, which has not been the case for other putative stem cell markers such as Bmi146. Because the protein expression was more readily detectable in the mouse, increased resolution of the distinct expression domain in the small intestinal crypt was apparent. CD166 expression appeared to be predominantly on the cell surface of the lower crypt-base cells (Fig.2B). Low levels of cell surface CD166 were also detected on the small intestinal villus when sectioned on a tangential plane and by qRT-PCR (Supplemental Fig.2A,B). Although it is apparent that CD166-expressing cells reside in the mesenchyme, a subpopulation of CD166-expressing epithelial cells were also present within the intestine; CD166/ESA double-stained cells are apparent within the intestinal epithelial crypt base (Fig.2C).
Figure 2. CD166 expression pattern in the mouse small intestine and colon.
(A-B) Mouse small intestine (SI) stained with antibodies to CD166 (red) and (A) counterstained with Hoechst (blue) or (B) co-stained with antibodies to laminin (gray) in higher magnification of the crypt. Arrowheads mark CD166-expressing cells. Arrows mark columnar crypt-based cell in B. (C) Mouse small intestinal crypt co-stained with CD166 (red) and the pan-epithelial ESA (white). (D) Mouse colon stained with antibodies to CD166 (brown), demonstrating enhanced expression in the crypt base (black arrowheads). Solid lines denote the epithelial-mesenchymal boundary. Bar=25μm. (E) Flow cytometry isotype control on isolated intestinal epithelial crypt cells. (F) Flow cytometry analysis of isolated crypt epithelial cells stained with CD166 antibodies. Box denotes CD166-positive, CD45-negative cells.
To confirm cell surface expression on epithelial cells, we isolated the intestinal epithelium using a method that disrupts epithelial cell adhesion complexes45, then performed FACS to isolate CD166-positive epithelium. Enriched populations of differentiated, villus epithelium and undifferentiated crypt-based epithelium were isolated (Fig.2E-F). The crypt epithelium contained a sizable CD166-expressing population (8.1%). Reanalysis of the CD166-expressing crypt cell population revealed two distinct populations based upon forward scatter (FSC, cell volume) and side scatter (SSC, inner complexity including type of cytoplasmic granules) (Fig.3A). Isolation of these intact crypt-based CD166-expressing cells allowed for a closer examination of cell identity.
Figure 3. FACS-isolated CD166 epithelial cells express Paneth cell granules or Lgr5.
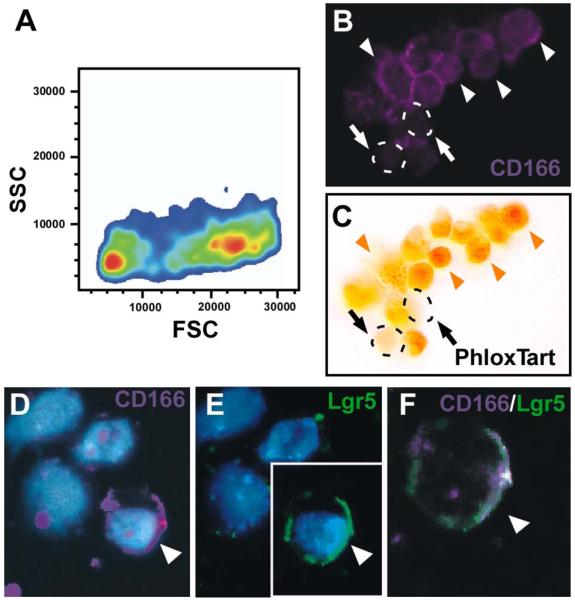
(A) Forward (FSC) and side (SSC) scatter analysis of CD166-expressing crypt epithelial cells display two distinct populations of cells (orange centers). (B-C) FACS-isolated, cytospun CD166-expressing mouse crypt cells (B; purple) stained with (C) Phloxine Tartrazine. White arrowheads designate CD166-positive cells; orange arrowheads designate CD166-positive, Phloxine Tartrazine-positive cells. Arrows and dashed circles designate CD166-positive, Phloxine Tartrazine-negative cells. (D-F) Cytospun, isolated CD166-positive cells (purple) co-stained with antibodies to Lgr5, a putative stem cell marker (green). Arrowhead designates a double-labeled cell.
Both differentiated Paneth cells and intestinal stem cells reside in the region marked by CD166-expressing cells. Isolated CD166-expressing epithelia were cytospun (Fig.3B) and subsequently analyzed for marker expression to determine if they were Paneth cells or stem cells (Fig.3C-F). A subset of FACS-isolated CD166-expressing cells stained with Phloxine Tartrazine, an established Paneth cell histochemical stain (Fig.3B-C; arrowheads). This approach bypasses non-specific cross-reactivity of antibodies with Paneth cell granules on cut tissue surface because intact cells are analyzed. For completeness, in vivo co-localization of CD166 expression and Paneth cells, co-staining of mouse small intestinal tissue with antibodies to CD166 and lysozyme was performed. As predicted, these two markers co-localized within the crypt base (Supplement Fig.3A-C). The human expression pattern was also consistent with the mouse pattern in the SI as determined by sequential staining of human tissue with antibodies to CD166 and the histochemical stain, Phloxine Tartrazine (Supplemental Fig.3D-F, arrowheads). Further, a subset of CD166-expressing crypt-based cells also co-expressed markers for differentiated or differentiating enteroendocrine cells (5-HT and Chromogranin A; Supplemental Fig.4).
Interestingly, there was a population of FACS-isolated CD166-positive cells that did not co-stain with Phloxine Tartrazine (Fig.3B,C; arrows) or enteroendocrine markers (Supplemental Fig.4). To determine if this subset of cells co-expressed putative stem cell markers, FACS-isolated CD166-expressing cells were cytospun then analyzed for Lgr5 expression using antibodies. Lgr5-positive cells represented a small fraction of CD166-positive crypt-based cells (Fig.3D-F; arrowheads). Additional putative stem cell markers, Msi-139 and Dcamkl-1, also shared overlapping expression with a subset of CD166-expressing cells (Fig.4A,C), but extended into the adjacent CD166-negative zone (Fig.4B,D). Further expression analysis of putative stem cell markers reported in the intestine, other organ systems, or in cancer stem cells showed high relative expression levels in CD166+ versus CD166− isolated crypt-based cell populations by qRT-PCR (Fig.4E); this includes Ascl247, Lgr541, CD2448,49, Klf550, CD4410, 51, Lrig152. As expected, expression of an intestinal epithelial differentiation gene, Fabpl53, is decreased in the CD166+ cell population.
Figure 4. CD166 is expressed in a subset of cells expressing stem cell markers Musashi-1 and Dcamkl-1.
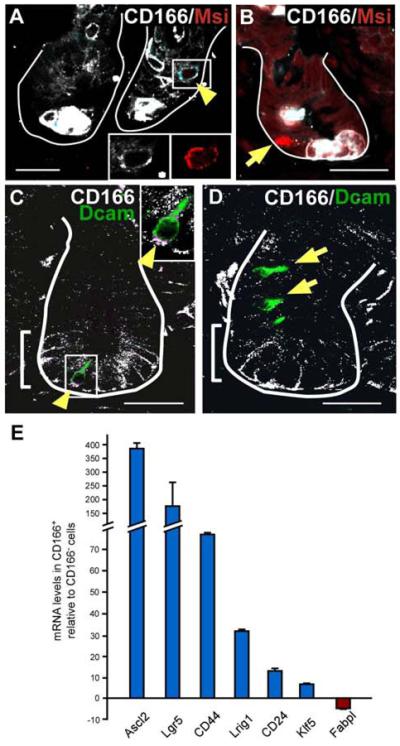
(A,B) Mouse small intestinal tissue co-labeled with antibodies to CD166 (white) and the putative stem cell marker, Musashi-1 (Msi, red). (A) White box and yellow arrowhead designates a CD166/Msi co-expressing cell with cell surface co-expressing in light blue. Bottom right insets depict higher magnification of singly stained cell, CD166 (white) and Msi (red). (B) A Msi-positive, CD166-negative cells indicated with arrow. (C,D) Mouse intestinal tissue co-labeled with antibodies to CD166 (white) and the putative stem cell marker, Dcamkl-1 (green). (C) Yellow arrowhead and box designates a CD166/Dcamkl co-expressing cell, with the co-expressing cell surface in purple. A higher magnification is provided in the upper right inset. (D) CD166-negative, Dcamkl-1-positive cells are designated by arrowhead. Brackets mark CD166-expressing region. White lines mark epithelial-mesenchymal boundary. Bar=25μm. (E) Quantitative RT-PCR analysis of stem cell markers within the CD166-positive cell population relative to the adjacent CD166-negative population. Triplicate samples of n=2 runs, and S.E.M.
Interestingly, in contrast to the adult intestine, analysis of the developing mouse intestine revealed that CD166 expression was ubiquitously expressed in the epithelium at embryonic day (E)14.5 (Supplemental Fig.5A). However, at the onset of villus formation, E16.5, CD166 expression became localized to both the villus (arrows) and intervillus region (arrowheads; Supplemental Fig.5B), and by post-natal (P) development, expression was localized in the intervillus region (P4; arrowhead; Supplemental Fig. 5C).
CD166 is highly expressed in human colon adenocarcinoma and liver metastases
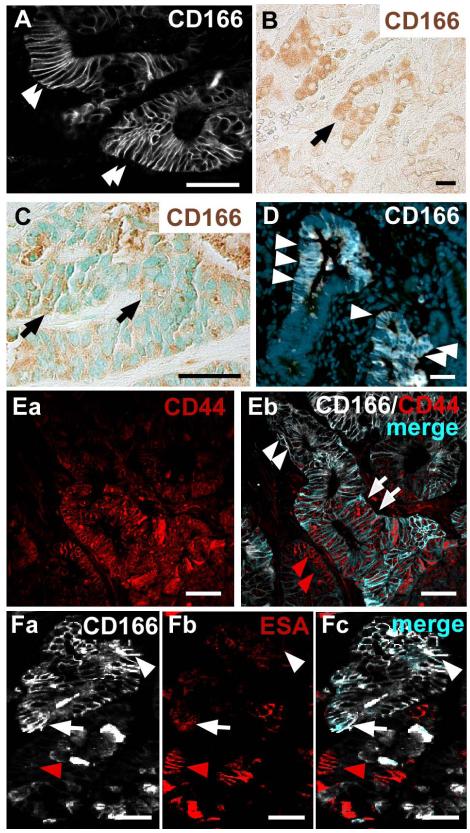
To further characterize the expression patterns of CD166 during intestinal tumorigenesis, we stained human adenocarcinoma and liver metastases with antibodies to CD166. We identified both cell surface and cytoplasmic expression in primary tumors and metastases (Fig.5A-C). Human tumors were decidedly heterogeneous in their CD166 expression. While some tumor samples exhibited only cell surface expression (Fig.5A), others exhibited cytoplasmic expression (Fig.5B). CD166-positive cells generally appeared within clustered regions of epithelium. Interestingly, CD166 expression within liver metastasis was also heterogeneous, shown here as cytoplasmic expression (Fig.5C).
Figure 5. CD166 expression of human colorectal cancer is recapitulated in mouse colorectal adenomas.
(A-B) Human primary colorectal adenomas labeled with antibodies to CD166 show (A) cell surface expression (white, arrowheads) or (B) cytoplasmic expression (brown, arrow). (C) Human colorectal liver metastasis that has both cell surface and cytoplasmic expression of CD166 (brown, arrow). Methyl Green nuclear counterstain (green). (D) Benign mouse intestinal tumor labeled with antibodies to CD166 (white, arrowheads) and Hoechst nuclear counterstain (blue). (Ea-Eb) Human primary colorectal adenoma co-stained with antibodies to CD44 (red) and CD166 (white). (Eb) Subpopulations of tumor cells express both CD44 and CD166 (merged, light blue, white arrows), or only CD44 (red staining, red arrowheads). Some tumor cells do not express CD44 but express CD166 (white staining, white arrowheads). (Fa-Fc) Mouse adenoma stained with antibodies to CD166 (white) and ESA (red). Subpopulations of cells express CD166 and ESA (merged, light blue, white arrow), ESA alone (red arrowhead) or CD166 alone (white arrowhead). Bar=25μm.
Tumors in a mouse model for intestinal tumorigenesis, the ApcMin/+ mouse42, displayed a strikingly similar CD166 expression pattern compared to human colorectal tumors (Fig.5D). Both predominant cell surface staining and diffuse cytoplasmic expression was detected. Interestingly, only a subset of the CD166-expressing tumor cells was in the cell cycle, as determined by co-expression of the proliferative marker Ki67 (Supplemental Fig.6). This might reflect the possibility that at any one time, only a subset of CSCs were actively cycling. Supporting this notion, in crypt-like regions of the ApcMin/+ mouse intestine, Ki67 generally marked the transient-amplifying cell population (Supplemental Fig.6B, bracket), but also marked a rare subset of CD166 crypt-base columnar epithelial cells.
To further characterize the expression domain of CD166 in intestinal tumors, we performed double staining with CD166 and either CD44 or ESA on human or ApcMin/+ mouse intestinal tumor sections. CD44 and ESA were previously used in combination with CD166 to identify and isolate a CSC population in human CRC10. We found that CD44 was detectable on most human tumor epithelial cells (Fig.5Ea), but that the overlapping expression region with CD166 was somewhat more restricted (Fig.5Eb, arrows). Interestingly, CD166 expression was generally lost in the aberrant crypt structures and, therefore, CD166 and CD44 were primarily expressed in mutually exclusive cell populations. However, there was a small subset of dual-expressing cells (Fig.5Eb, arrows). In contrast, ESA was lost on large clusters of tumor cells, but a discrete double-labeled population of cells existed within both human and mouse tumors, shown here on ApcMin/+ tumors (Fig.5Fa-Fc, arrow).
Discussion
The elucidation of CD166 expression within the stem cell niche of the small intestine and colon suggests an intriguing and potentially important role for this molecule in the intestinal stem cell niche. Although we show that CD166 was expressed at low levels in differentiated intestinal cells, it is robustly expressed at high levels on the cell surface of cells within the stem cell niche at the base of the crypt. In the SI, CD166 was distinctly present on both putative stem cell populations comprised of the crypt-based columnar epithelial cells (Lgr5+, Msi-1+, Dcamkl-1+), as well as the crypt-based differentiated Paneth cell population. In light of its previously described role in cell adhesion and its capacity to form homodimers across adjacent cell membranes, it is intriguing to postulate that CD166 may have an important function in anchoring the stem cell within the intestinal stem cell niche, or in instructing stem cell behavior. In support of this, a precedent exists for the participation of adhesion molecules in establishing cell polarity or directing asymmetric stem cell division54.
A number of intestinal stem cell markers (Fig.4) were co-expressed in CD166-expressing crypt cells. In contrast, the putative stem cell marker Msi-1 was often expressed in a single crypt cell within a zone of CD166-positive cells. Interestingly, Msi-1 was also expressed in CD166-negative regions. The current number of putative stem cell markers clearly highlights the complexity of stem and progenitor cell dynamics within the intestinal crypt. While CD166 may be shown, in the future, to mark stem cells, it is more likely, based upon its broader expression pattern, that it is actually a stem cell niche marker, expressed on both the stem cell and the surrounding supporting epithelial cells. In this context, the understanding of how niche cells influence stem cell behavior during homeostasis, tissue regeneration and disease can be examined in this newly defined cluster of crypt-based cells.
CD166 may possess multiple functions within the intestinal epithelium. This is suggested by its multi-faceted expression pattern in subsets of fully differentiated Paneth and enteroendocrine cells juxtaposed to its expression in a putative stem population. Future exploration of CD166 differential function and regulation in intestinal epithelium will contribute to a better understanding of whether its dysregulation contributes to disease progression in intestinal cancer, and may provide insight into whether CD166 has active function on CRC stem cells, or whether it is merely a “coincidental” marker.
Consistent CD166 expression in both human and mouse tumors demonstrates that the mouse provides a viable model for studying the function and expression of CD166 in tumorigenesis. Interestingly, CD166 was highly expressed in early adenoma formation in the ApcMin/+ mouse. Further, we confirmed that CD166 expression was retained within human CRC and metastatic disease, and that both a cell surface and cytoplasmic expression pattern was apparent. These findings, in particular an alteration in cellular localization of CD166, support a potential functional role for this molecule in tumorigenesis in either a cell adhesion or signaling capacity. Our analyses extend these initial findings and show that the observed expression patterns are also retained in metastatic lesions.
CD166 expression relative to other CSC and proliferative markers in the ApcMin/+ mouse recapitulated previous findings in humans10 that cells positive for both CD166, CD44 and ESA constitute a small subpopulation of total tumor mass. By analyzing the expression pattern of these markers in the ApcMin/+ mouse, a model of pre-neoplastic intestinal cancer, our findings suggest that mere co-expression of these markers may not be sufficient to promote invasive tumorigenesis. Alternatively, the presence of cells harboring these markers might suggest that these benign tumors have the potential for metastatic advance. Analysis of CD166 expression in ApcMin/+ polyps found that crypt-like structures near the muscularis tend to be low or lack expression of CD166 although they are high in CD44 and Ki67 expression. While the significance of this observation is not known, it is possible that loss of CD166 cell surface expression is a precursor for tumor progression. While our study does not evaluate in depth the percentage of CD166/ESA/CD44 triple positive cells within a benign tumor, we do not rule out the possibility that early or benign tumor states may harbor few numbers or lack cells capable of acting as CSCs. Importantly, our data reveals robust cell surface expression of CD166 within the endogenous intestinal stem cell niche, suggesting that as a cell adhesion molecule, it may play an important role in maintaining the integrity of the stem cell niche, or in directing cells to the crypt base. Inarguably, future studies examining the epithelial function of CD166 within the stem cell niche remain a critical focus for understanding the importance of this molecule in homeostasis and in disease; studies in cell lines and in enterospheres will facilitate this understanding. Ultimately, however, based upon the robust expression of CD166 within the stem cell niche, functional inhibition of this molecule has the potential to disrupt the maintenance of the stem cell niche and thereby compromise the epithelial barrier. Our data provides caution for therapeutically targeting CD166 for cancer.
Conclusions
Cell surface antigen expression of CD166 was recently identified as an important marker on human intestinal CSCs10. Along with this observation and its history in cancer progression as a marker for aggressive disease21, CD166 has been proposed as an intriguing molecule for therapeutic targeting in the treatment of cancer. For effective targeting of any cell surface antigen, its endogenous expression pattern must first be elucidated. Here, we report a broad range of CD166 expression patterns in the human and mouse intestine. We show that CD166 is expressed on a number of intestinal cells, including putative stem cells and differentiated crypt-based cells. This discovery provides important implications for future targeting of CD166 in disease therapy and, significantly, provides insight into the potential functional role of this critical molecule.
Supplementary Material
Acknowledgements
We are grateful to the Wong laboratory for insightful discussions and to Pam Canaday and Mandy Boyd for excellent FACS assistance.
Grant acknowledgment: TGL: CA106195; AEP: HD049309; ECA: HL007781; MHW: DK068326; CA118235; DK085525.
Abbreviations
- 5-HT
5-hydroxytryptamine
- ALCAM
Activated leukocyte cell adhesion molecule
- APC
Allophycocyanin
- APC/Min mouse
Adenomatous polyposis coli, multiple intestinal neoplasm mouse
- CRC
Colorectal cancer
- CSC
Cancer stem cell
- Cy3
Indocarbocyanine-3
- DAB
Di-aminobenzidine
- Dcamkl-1
Doublecortin and calmodulin kinase-like 1
- E
Embryonic day
- EDTA
Ethylenediaminetetraacetic acid
- ESA
Epithelial specific antigen
- FACS
Fluorescent-activated cell sorting
- FITC
Fluorescein isothiocyanate
- HBSS
Hanks buffered saline solution
- Msi-1
Musashi-1
- SI
Small intestine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors state they have no disclosures.
References
- 1.Cancer Facts & Figures 2008. American Cancer Society; Atlanta: 2008. [Google Scholar]
- 2.Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev. 2008;18:48–53. doi: 10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Dylla SJ, Beviglia L, Park IK, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 6.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990s. Cancer Metastasis Rev. 2000;19:I–XI. 193–383. [PubMed] [Google Scholar]
- 7.Vermeulen L, Sprick MR, Kemper K, et al. Cancer stem cells--old concepts, new insights. Cell Death Differ. 2008;15:947–58. doi: 10.1038/cdd.2008.20. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 10.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 12.Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–9. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 13.May R, Riehl TE, Hunt C, et al. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–7. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 14.van Kempen LC, van den Oord JJ, van Muijen GN, et al. Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am J Pathol. 2000;156:769–74. doi: 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein WM, Wu BP, Zhao S, et al. Increased expression of stem cell markers in malignant melanoma. Mod Pathol. 2007;20:102–7. doi: 10.1038/modpathol.3800720. [DOI] [PubMed] [Google Scholar]
- 16.Kristiansen G, Pilarsky C, Wissmann C, et al. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate. 2003;54:34–43. doi: 10.1002/pros.10161. [DOI] [PubMed] [Google Scholar]
- 17.Burkhardt M, Mayordomo E, Winzer KJ, et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J Clin Pathol. 2006;59:403–9. doi: 10.1136/jcp.2005.028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mezzanzanica D, Fabbi M, Bagnoli M, et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin Cancer Res. 2008;14:1726–33. doi: 10.1158/1078-0432.CCR-07-0428. [DOI] [PubMed] [Google Scholar]
- 19.Verma A, Shukla NK, Deo SV, et al. MEMD/ALCAM: a potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology. 2005;68:462–70. doi: 10.1159/000086989. [DOI] [PubMed] [Google Scholar]
- 20.Tomita K, van Bokhoven A, Jansen CFJ, et al. Activated Leukocyte Cell Adhesion Molecule (ALCAM) Expression is Associated with a Poor Prognosis for Bladder Cancer Patients. Urooncology. 2003;3:121–129. [Google Scholar]
- 21.Weichert W, Knosel T, Bellach J, et al. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57:1160–4. doi: 10.1136/jcp.2004.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cayrol R, Wosik K, Berard JL, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat Immunol. 2008;9:137–45. doi: 10.1038/ni1551. [DOI] [PubMed] [Google Scholar]
- 23.Masedunskas A, King JA, Tan F, et al. Activated leukocyte cell adhesion molecule is a component of the endothelial junction involved in transendothelial monocyte migration. FEBS Lett. 2006;580:2637–45. doi: 10.1016/j.febslet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, Quertermous T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J Biol Chem. 2004;279:55315–23. doi: 10.1074/jbc.M407776200. [DOI] [PubMed] [Google Scholar]
- 25.Ohneda O, Ohneda K, Arai F, et al. ALCAM (CD166): its role in hematopoietic and endothelial development. Blood. 2001;98:2134–42. doi: 10.1182/blood.v98.7.2134. [DOI] [PubMed] [Google Scholar]
- 26.Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med Sci Monit. 2006;12:BR263–73. [PubMed] [Google Scholar]
- 27.Hassan NJ, Barclay AN, Brown MH. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur J Immunol. 2004;34:930–40. doi: 10.1002/eji.200424856. [DOI] [PubMed] [Google Scholar]
- 28.Kato Y, Tanaka Y, Hayashi M, et al. Involvement of CD166 in the activation of human gamma delta T cells by tumor cells sensitized with nonpeptide antigens. J Immunol. 2006;177:877–84. doi: 10.4049/jimmunol.177.2.877. [DOI] [PubMed] [Google Scholar]
- 29.Bowen MA, Patel DD, Li X, et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med. 1995;181:2213–20. doi: 10.1084/jem.181.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanki JP, Chang S, Kuwada JY. The molecular cloning and characterization of potential chick DM-GRASP homologs in zebrafish and mouse. J Neurobiol. 1994;25:831–45. doi: 10.1002/neu.480250708. [DOI] [PubMed] [Google Scholar]
- 31.Patel DD, Wee SF, Whichard LP, et al. Identification and characterization of a 100-kD ligand for CD6 on human thymic epithelial cells. J Exp Med. 1995;181:1563–8. doi: 10.1084/jem.181.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Degen WG, van Kempen LC, Gijzen EG, et al. MEMD, a new cell adhesion molecule in metastasizing human melanoma cell lines, is identical to ALCAM (activated leukocyte cell adhesion molecule) Am J Pathol. 1998;152:805–13. [PMC free article] [PubMed] [Google Scholar]
- 33.Corbel C, Cormier F, Pourquie O, et al. BEN, a novel surface molecule of the immunoglobulin superfamily on avian hemopoietic progenitor cells shared with neural cells. Exp Cell Res. 1992;203:91–9. doi: 10.1016/0014-4827(92)90043-8. [DOI] [PubMed] [Google Scholar]
- 34.Uchida N, Yang Z, Combs J, et al. The characterization, molecular cloning, and expression of a novel hematopoietic cell antigen from CD34+ human bone marrow cells. Blood. 1997;89:2706–16. [PubMed] [Google Scholar]
- 35.Bruder SP, Ricalton NS, Boynton RE, et al. Mesenchymal stem cell surface antigen SB-10 corresponds to activated leukocyte cell adhesion molecule and is involved in osteogenic differentiation. J Bone Miner Res. 1998;13:655–63. doi: 10.1359/jbmr.1998.13.4.655. [DOI] [PubMed] [Google Scholar]
- 36.Arai F, Ohneda O, Miyamoto T, et al. Mesenchymal stem cells in perichondrium express activated leukocyte cell adhesion molecule and participate in bone marrow formation. J Exp Med. 2002;195:1549–63. doi: 10.1084/jem.20011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piazza T, Cha E, Bongarzone I, et al. Internalization and recycling of ALCAM/CD166 detected by a fully human single-chain recombinant antibody. J Cell Sci. 2005;118:1515–25. doi: 10.1242/jcs.02280. [DOI] [PubMed] [Google Scholar]
- 38.Roth A, Drummond DC, Conrad F, et al. Anti-CD166 single chain antibody-mediated intracellular delivery of liposomal drugs to prostate cancer cells. Mol Cancer Ther. 2007;6:2737–46. doi: 10.1158/1535-7163.MCT-07-0140. [DOI] [PubMed] [Google Scholar]
- 39.Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 40.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009 doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 42.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 43.Wong MH, Rubinfeld B, Gordon JI. Effects of forced expression of an NH2-terminal truncated beta-Catenin on mouse intestinal epithelial homeostasis. J Cell Biol. 1998;141:765–77. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luna LG. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. McGraw-Hill Book Co.; 1968. [Google Scholar]
- 45.Davies PS, Dismuke AD, Powell AE, et al. Wnt-reporter expression pattern in the mouse intestine during homeostasis. BMC Gastroenterol. 2008;8:57. doi: 10.1186/1471-230X-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Flier LG, van Gijn ME, Hatzis P, et al. Transcription Factor Achaete Scute-Like 2 Controls Intestinal Stem Cell Fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 48.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2010;298 doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 50.Jiang J, Chan YS, Loh YH, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nature Cell Biology. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 51.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Research. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen KB, Collins CA, Nascimento E, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon TC, Roth KA, Gordon JI. Use of transgenic mice to map cis-acting elements in the liver fatty acid-binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. J Biol Chem. 1993;268:18345–58. [PubMed] [Google Scholar]
- 54.Picco V, Hudson C, Yasuo H. Ephrin-Eph signalling drives the asymmetric division of notochord/neural precursors in Ciona embryos. Development. 2007;134:1491–7. doi: 10.1242/dev.003939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.