Abstract
In bacteria, the translocation of tRNA and mRNA with respect to the ribosome is catalyzed by the conserved GTPase, elongation factor-G (EF-G). In order to probe the rate determining features in this process, EF-G-catalyzed translocation was imaged from two unique structural perspectives using single-molecule fluorescence resonance energy transfer. The data reveal that the rate at which the ribosome spontaneously achieves a transient, “unlocked” state is closely correlated with the rate at which the tRNA-like, domain IV/V element of EF-G engages the A site. Following these structural transitions, translocation occurs comparatively fast, suggesting that conformational processes intrinsic to the ribosome determine the rate of translocation. Experiments performed in the presence of non-hydrolyzable GTP analogues and specific antibiotics further reveal that allosterically linked conformational events in EF-G and the ribosome mediate rapid, directional substrate movement and EF-G release.
INTRODUCTION
The precise and rapid translocation of messenger RNA (mRNA) and transfer RNA (tRNA) through the ribosomal Aminoacyl (A), Peptidyl (P), and Exit (E) sites is central to translation. Fidelity in these events underpins the conversion of the mRNA codon sequence into functional proteins. Conformational changes in the ribosome are thought to modify the substrate affinities of the A, P and E sites in a manner that facilitates translocation.
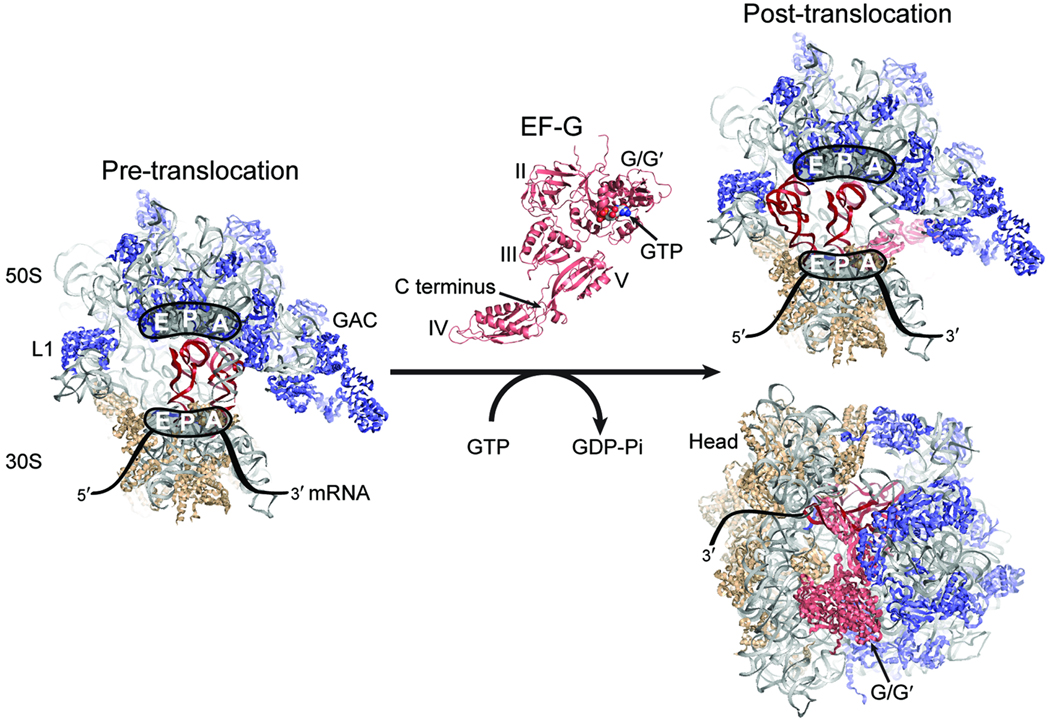
In Escherichia coli (E. coli), translocation is catalyzed by elongation factor-G (EF-G)1–3 in a manner that depends on GTP hydrolysis. EF-G is a highly conserved ribosome-dependent GTPase that operates on the pre-translocation ribosome complex containing deacylated-tRNA in the P site and peptidyl-tRNA in the A site. Through a multistep mechanism, EF-G converts the pre-translocation complex to a post-translocation complex in which P- and A-site tRNAs occupy the E and P sites, respectively1–3 (Fig. 1).
Figure 1. Structural models of the ribosome and EF-G.
(Left) The pre-translocation ribosome complex showing the large (50S) and small (30S) subunits, and the A, P, and E sites. The rRNA is shown in grey, the 50S proteins in blue, and the 30S proteins in tan. The A- and P-site tRNAs are in red. The GTPase activating center (GAC) and L1 stalk are indicated. (Center) EF-G with structural domains and GTP labeled. EF-G binds at the GAC of the pre-translocation complex, hydrolyzes GTP, and promotes formation of the post-translocation complex shown at right, in which the tRNAs have moved to the P and E sites and EF-G domains IV/V protrude into the A site41. Structural models of the ribosome and EF-G were constructed from PDB accession codes 2WRI and 2WRJ. The A-site tRNA is from PDB 1GIX.
In line with observations that slow, spontaneous translocation occurs in the absence of translation factors or nucleotide hydrolysis4–7, structural, biochemical, and single-molecule fluorescence resonance energy transfer (smFRET) studies indicate that the ribosome is intrinsically capable of achieving conformations necessary and sufficient for translocation8–16. Such observations suggest that translocation occurs by way of an unstable or rarely-achieved configuration. Consistent with this notion, the ribosome spontaneously achieves a translocation intermediate known as the unlocked state that requires the convergence of at least three loosely coupled structural transitions8–10,13,14, including P/E hybrid state formation, closure of the large subunit L1 stalk domain and a ratchet-like rotation of the subunit interface (subunit ratcheting). Formation of the P/E hybrid state requires a ~40 Å motion of the 3′ CCA terminus of P-site tRNA into the large subunit E site17. Closure of the L1 stalk domain, comprised of 23S rRNA helices 76–79 and the L1 protein, occurs via a hinge-like rearrangement at the base of the stalk18 moving it from an open position distal to the E site inward toward the subunit interface to contact the P/E tRNA elbow19. Subunit ratcheting entails a ~20° rotation of the small ribosomal subunit19. The rate of unlocked state formation is accelerated by P-site tRNA deacylation, peptidyl tRNA occupancy in the A site, and EF-G binding10,13,14.
Bulk measurements demonstrate that the molecular mechanism of translocation is accelerated by EF-G’s interaction with the pre-translocation complex by as much as 10,000-fold20. Although linked in some fashion, GTP hydrolysis and translocation are distinct molecular events as they can be effectively uncoupled by the antibiotic viomycin and mutations in the ribosome that inhibit unlocked state formation21,22. On the ribosome, EF-G undergoes large-scale conformational rearrangements19,23,24. Such events may facilitate interaction of EF-G’s G-domain with the large subunit GTPase Activating Center (GAC) as well as GTP hydrolysis25. Crosslinking studies26 and structural changes between free and ribosome-bound EF-G suggest that a 20–40 Å movement of EF-G’s IV/V domain toward the A site is important to translocation1,24. In addition, conformational events in the post-translocation complex may be required for EF-G dissociation from the ribosome as an intramolecular crosslink between domains I and V of EF-G and the antibiotic fusidic acid have been shown to inhibit EF-G turnover26.
Here, in order to understand the order and timing of events underpinning EF-G’s interaction with the E. coli pre-translocation complex, the process of translocation was imaged under pre-steady state conditions from two, independent structural perspectives using smFRET. Formation of the unlocked state was monitored using complexes labeled on the L1 stalk and P-site tRNA. EF-G’s interaction with fluorescently labeled A-site tRNA was monitored by site-specifically labeling EF-G’s C-terminal domain near the center of its tRNA-like, domain IV/V element. The rate of translocation was measured by a change in quantum yield in dye-labeled A-site tRNA concomitant with post-translocation complex formation. The observed kinetics of EF-G and ribosome motions support a model in which the intrinsic rate of unlocked state formation limits the rate at which EF-G’s domain IV/V can engage the A site. Upon doing so, EF-G can then efficiently mediate translocation followed by release from the ribosome and L1 stalk opening. Unlocked state formation and conformational changes in EF-G are found to be linked processes that can each become rate limiting to translocation. These data highlight the need to understand the nature of intrinsic conformational events in the components of translation and their role in translational control.
RESULTS
The unlocked state can be achieved spontaneously
To probe the formation of the unlocked state during translocation, pre-translocation ribosome complexes containing tRNAfMet(Cy3-s4U8) in the P site, fMet-Phe-tRNAPhe in the A site, and L1(Cy5-S55C) were prepared as previously described13 and surface-immobilized in microfluidic reaction chambers linked to stopped-flow instrumentation. Complexes were imaged prior to, and during, stopped-flow delivery of EF-G using a prism-based total internal reflection fluorescence microscope15. Unless otherwise stated, all experiments were performed at 10 ms time resolution and at room temperature in Tris-Polymix buffer in the presence of 15 mM Mg2+. Such conditions provide an optimum between complex stability and function and minimize the likelihood of missed events due to fast conformational processes. Thus, while the translocation events reported under these conditions are slower than those obtained at elevated temperatures, at lower Mg2+ concentrations, and in the absence of physiological polyamines22,27,28, the mechanism of translocation is not meaningfully affected (Supplementary Fig. 1). The translocation rates observed are also consistent with those reported by other groups under similar conditions27–30. As discussed below, and elsewhere13, the rate and extent of translocation were also unaffected by the fluorescent labeling strategies employed (Supplementary Figs. 1a and 8b).
As previously described13,14, prior to the addition of EF-G(GTP), smFRET trajectories displayed stable dwells in a low-FRET (0.1) state, punctuated by transient excursions to intermediate- (0.25 and 0.4 FRET) and high-FRET (0.65) states (Supplementary Figs. 2 and 3a). The high-FRET configuration, which structural modeling has suggested minimally requires P/E hybrid state formation, L1 stalk closure and subunit ratcheting, has been termed the unlocked state. The relative occupancies in these four FRET states, as well as their rates of interconversion, were determined using established hidden Markov modeling (HMM) procedures31,32 (Supplementary Table 1). For simplicity, the kinetic parameters of motion are summarized as the rates of transition out of the low-FRET state, and into the high-FRET state (Supplementary Table 2). The transition rate into the high-FRET state (k→high) was 0.7±0.2 s−1, and transition out of high-FRET configurations was 46±2 s−1.
EF-G does not accelerate the rate of unlocked state formation
Upon stopped-flow delivery of saturating EF-G and GTP (10 µM and 1 mM, respectively)22,33, pre-translocation complexes initially displayed dynamics indistinguishable from those observed prior to its addition. In approximately 70% of complexes observed, this period was followed by the formation of a relatively stable high-FRET unlocked state with a lifetime of 370±14 ms (Fig. 2a, Supplementary Fig. 3b). The extended lifetime of such FRET events (; Supplementary Table 2) required the inclusion of a second high-FRET state during HMM analysis (Supplementary Fig. 4). The appearance of this state only after EF-G addition is consistent with conversion of the majority of molecules to the post-translocation complex during the imaging period (~7 s) (Supplementary Fig. 1), where a relatively long-lived interaction between deacylated tRNA and the L1 stalk is achieved either during or after translocation. As previously foreshadowed12–14, approximately 20% of the molecules observed to achieve the stable high-FRET state returned to a low-FRET state, likely indicating that the L1 stalk can open prior to tRNAfMet release from the E site. The evolution of FRET for the remaining 80% could not be fully determined due to loss of Cy3 or Cy5 fluorescence. Strikingly, the rate at which excursions to the stable unlocked state occurred (0.9±0.2 s−1) closely approximated the rate of unlocked state formation observed in the absence of EF-G (Supplementary Table 2) and the rate of translocation observed in bulk (Supplementary Fig. 1). In line with previous suggestions10,13,28, these data imply that formation of the unlocked state lies along the translocation reaction coordinate and that the rate of translocation may be limited by the ribosome’s intrinsic ability to achieve the unlocked state.
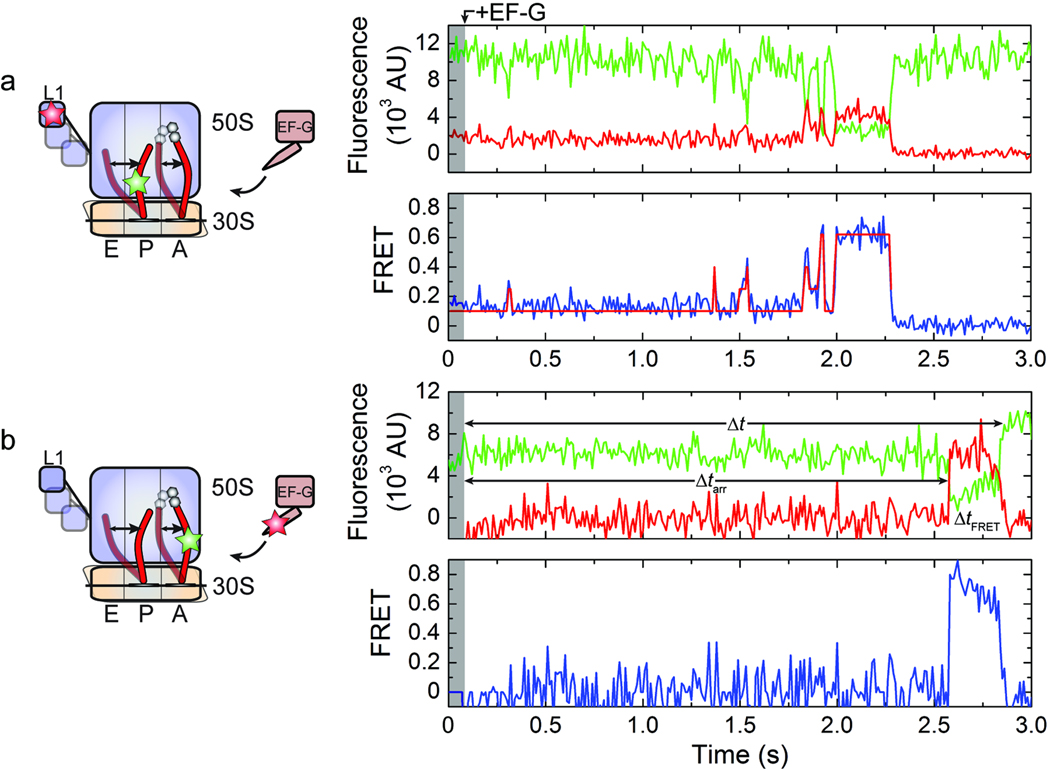
Figure 2. Observation of the translocation reaction from two unique structural perspectives.
(a) The dynamics of the ribosome complex with P-site tRNAfMet(Cy3-s4U8), A-site fMet-Phe-tRNAPhe, and L1(Cy5-S55C) following the addition of 10 µM EF-G and 1 mM GTP. (b) The apparent formation of an EF-G-ribosome interaction obtained by delivering 0.2 µM C-terminally labeled EF-G and 1 mM GTP to pre-translocation complexes with P-site tRNAfMet and A-site fMet-Phe-tRNAPhe(Cy3-acp3U47). The intervals Δt, Δtarr and ΔtFRET were used to estimate the apparent rate of translocation, the apparent on-rate , and the apparent off-rate , respectively. (Left) Cartoon diagrams indicating the sites of labeling (Cy3, green star; Cy5, red star) and the putative dynamic elements. (Right) Single-molecule fluorescence (Cy3, green; Cy5, red) and FRET (blue) trajectories. Overlaid on the FRET traces are the idealizations (red) generated during kinetic analysis (in (a) only).
Unlocked state formation and EF-G binding are correlated
In order to probe the timing of long-lived unlocked state formation and EF-G binding to the ribosome, an orthogonal labeling strategy was employed to image EF-G’s interaction with the A site. For such experiments, EF-G was enzymatically labeled with a Cy5 fluorophore at its C terminus (Online Methods)34,35, located within the domain IV/V element, which comes within close proximity to peptidyl-tRNA in the A site during translocation1. For such experiments, pre-translocation complexes were prepared containing tRNAfMet in the P site and fMet-Phe-tRNAPhe(Cy3-acp3U47) in the A site, and EF-G binding to the ribosome was monitored via FRET. To minimize background fluorescence, translocation experiments were performed at relatively low Cy5-EF-G concentration (0.2 µM with 1 mM GTP). To maximize the number of translocation events observed at this EF-G concentration, imaging was performed at 100 ms time resolution.
Consistent with the rate of unlocked state formation observed in L1- and P-site tRNA labeled complexes, a period of latency was observed subsequent to Cy5-EF-G addition followed by a FRET event that closely approximated the lifetime of the long-lived unlocked state (Figs. 2b and 3a; Supplementary Fig. 5). Direct inspection of these FRET events, as well as the distribution of all FRET values observed (Supplementary Fig. 6a), revealed somewhat higher FRET at the beginning of the events (~0.75) followed by somewhat lower FRET (~0.55) just prior to the disappearance of FRET. Here, the lower FRET value closely approximated the anticipated distance between EF-G and tRNA (~50±5 Å) in the post-translocation ribosome complex (Supplementary Fig. 6). At the present time resolution, the transition rate between FRET states could not be accurately determined. The close correspondence in rates observed for FRET appearance between EF-G and A-site tRNA and that of unlocked state formation suggests that EF-G’s interaction with the ribosome may be rate-limited by formation of the unlocked state (Fig. 3a). Moreover, the close correspondence observed between rates of EF-G dissociation and L1 stalk opening suggests that such processes are also closely correlated (Fig. 3a). As shown in Supplementary Table 2, similar correlations were observed at 5 mM Mg2+ concentration.
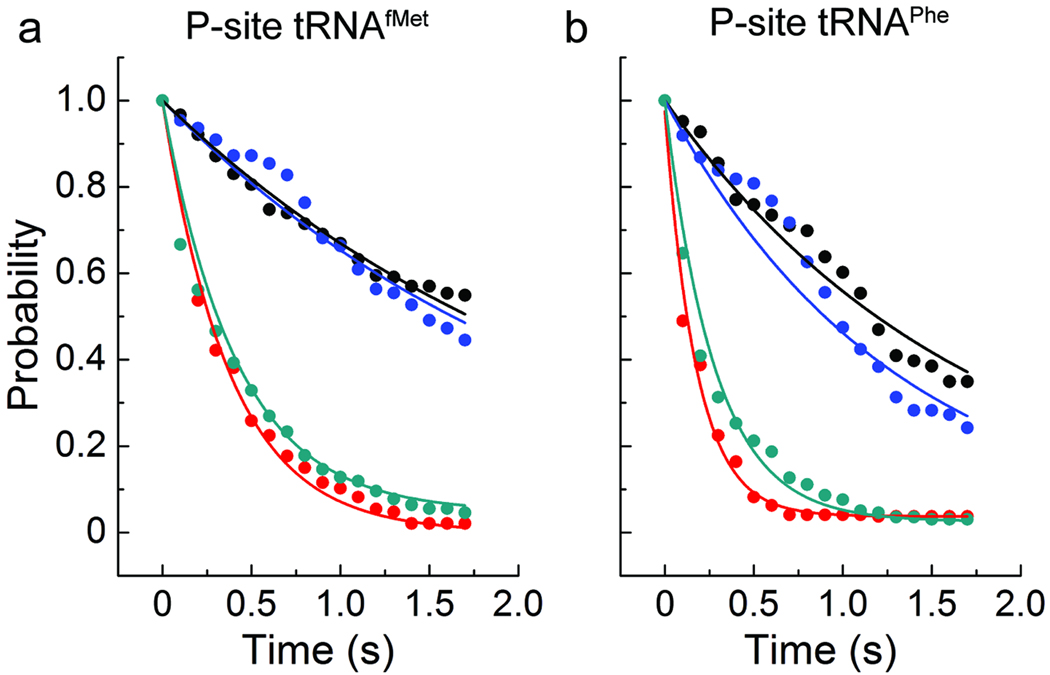
Figure 3. The kinetics of unlocked state formation and decay and EF-G-ribosome interactions are correlated.
Shown here is the distribution of dwell-times leading to the formation of the unlocked state (black), and those leading to the formation of the EF-G-ribosome interaction determined from delivery of labeled EF-G to complexes with labeled A-site tRNA (blue). Also shown is the distribution of the lifetime of the stable unlocked state (red), and that of the EF-G-ribosome interaction (cyan). Overlaid on the distributions are exponential functions with the rate constants shown in Supplementary Table 2. Data is presented for complexes with (a) P-site tRNAfMet and A-site fMet-Phe-tRNAPhe, and (b) P-site tRNAPhe and A-site NAcPhe-Lys-tRNALys.
To further examine the correlation between unlocked state formation and EF-G’s ability to engage the A site, experiments were repeated on pre-translocation complexes bearing deacylated tRNAPhe in the P site and NAc-Phe-Lys-tRNALys in the A site. Complexes of this nature have been shown to achieve the unlocked state and translocate more rapidly than those containing P-site initiator tRNAfMet 13,29,30. Here, a strong correlation was again observed between the rate of unlocked state formation and the rate at which EF-G engages the A site; an approximately similar timing was also observed between the rates of EF-G dissociation and L1 stalk opening (Fig. 3b; Supplementary Table 2). Evidence of faster single-turnover translocation is supported by the observed increase in the rate of unlocked state formation and the rate at which EF-G engaged the A site . Faster multiple turnover translocation may also be aided by the observed increase in the rate of EF-G dissociation . These data support the conclusion that unlocked state formation is rate-limiting for EF-G engaging the A site of the pre-translocation complex and that EF-G release from the post-translocation complex correlates in time with L1 stalk opening.
A-site tRNA fluorescence increases upon translocation
As observed in established bulk fluorescence assays of the translocation reaction coordinate28,29,33, termination of FRET between EF-G and tRNA was found to be accompanied by a ~20% increase in mean Cy3 fluorescence intensity (Fig. 4a). As such events were not observed in the absence of EF-G, increased Cy3 fluorescence reports directly on an EF-G dependent process and not spontaneous hybrid state fluctuations in the pre-translocation complex15,16. An increase in Cy3 fluorescence upon translocation could be explained by a change in local environment of the Cy3-labeled region of peptidyl-tRNA in the P site. Consistent with this notion, the fluorescence intensity of Cy3-labeled peptidyl-tRNAPhe directly loaded into the P site was enhanced with respect to that observed in the A site (Supplementary Figs. 7a–b). Such an enhancement could be attributed to a change in quantum yield of Cy3, reflected by an increase in fluorescence lifetime (Supplementary Fig. 7c).
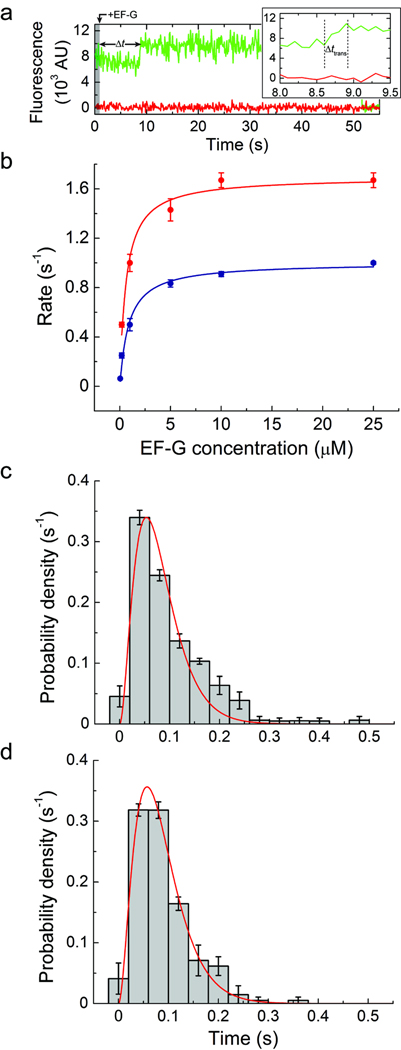
Figure 4. A step-like increase in Cy3 fluorescence accompanies peptidyl-tRNA movement to the P site.
(a) Single-molecule fluorescence trajectories (Cy3, green; Cy5, red) obtained during delivery of 10 µM unlabeled EF-G and 1 mM GTP to complexes with P-site tRNAfMet and A-site fMet-Phe-tRNAPhe(Cy3-acp3U47). (b) The rate of observing the increase in Cy3 fluorescence in complexes with either (blue) P-site tRNAfMet and A-site fMet-Phe-tRNAPhe(Cy3-acp3U47) or (red) P-site tRNAPhe and A-site NAcPhe-Lys-tRNALys(Cy3-acp3U47), across a range of EF-G concentrations (0.05–25 µM). The data were fit to the hyperbolic function Rate = Ratemax[EF − G]/(K1/2 + [EF − G]) with Ratemax = 1.0 ± 0.1 s−1 and K1/2 = 0.9 ± 0.1 µM for the case of P-site tRNAfMet, and Ratemax = 1.7 ± 0.1 s−1 and K1/2 = 0.6 ± 0.1 µM for P-site tRNAPhe. (c) The distribution of time over which the increase in Cy3 fluorescence occurs for complexes containing P-site tRNAfMet. In agreement with a previous report36, the distribution is well fit (R2 ≈ 0.95) by the distribution predicted for a model with three successive steps with equal rate constants (f(t) = (k3t2/2) exp(−kt), k = 37 ± 3 s−1). (d) The distribution obtained from complexes containing P-site tRNAPhe was fit to the same function with k = 35 ± 3 s−1 (R2 ≈ 0.94). Error bars represent the standard error.
This step-like Cy3 fluorescence intensity increase was further correlated with translocation by examining its rate of occurrence in the pre-translocation complex as a function of unlabeled-EF-G concentration (Fig. 4b). In the presence of 1 mM GTP, EF-G accelerated the occurrence of this event in a concentration-dependent fashion. Histograms of time intervals preceding the fluorescence increase were characterized by two time constants, where only the faster of the two components increased as a function of EF-G concentration (Supplementary Fig. 8b). At saturating EF-G concentrations, the faster population represented ~70% of ribosomes. In line with previous bulk experiments13,22,29,30, the EF-G-dependent rate was well described by a hyperbolic function saturating at a maximum rate of approximately 1.0±0.1 s−1, where K1/2 = 0.9±0.1 µM. Complexes containing tRNAPhe in the P site and NAcPhe-Lys-tRNALys in the A site showed similar behavior, except both fast and slow time components were more rapid. At saturating EF-G concentrations the EF-G-dependent rate was 1.7±0.1 s−1 and showed a slightly lower K1/2 of 0.6±0.1 µM.
Finally, the increased Cy3 fluorescence intensity was directly linked to translocation by delivering EF-G(GTP) to surface-immobilized pre-translocation complexes containing P-site tRNAfMet and A-site fMet-Phe-tRNAPhe(Cy3-acp3U47) in the presence of 20 nM ternary complex of EF-Tu(GTP)-Lys-tRNALys(Cy5-acp3U47). As anticipated, FRET between P- and A-site tRNA was only observed after the increase in Cy3 fluorescence. Prior to this event, Cy5-labeled tRNALys was unable to accommodate into the A site (Supplementary Fig. 9). These data indicate that the increased Cy3 fluorescence intensity resulted from translocation of the Cy3-labeled tRNA from the A site to the P site. In line with recent optical trapping experiments showing that the ribosome’s transit along the mRNA template occurs rapidly via three sequential steps with equal rates36, the distribution of time intervals over which the Cy3 fluorescence intensity increased (Δttrans) for both pre-translocation complexes investigated were well fit by a three-step process, each occurring at a rate of ~40 s−1 (Fig. 4c–d). Such observations provide compelling evidence that conformational events in the ribosome and EF-G are rate limiting in translocation, while substrate movements on the small ribosomal subunit are relatively rapid and largely independent of the nature of bound substrates in the pre-translocation complex.
Initial EF-G binding is not observed
EF-G’s observed on-rate, marked by the FRET event between the C terminus of EF-G and A-site tRNA (Fig. 2b, Supplementary Table 2), is ~25-fold slower than the established bimolecular rate constant of the initial EF-G-ribosome interaction20 (~150 µM−1s−1). This finding suggests that the FRET signal reports on events subsequent to initial binding. This could be explained if EF-G reversibly samples the A site at a rate substantially faster than the time resolution of the experiment. Alternatively, EF-G could occupy a conformation on the ribosome that does not yield FRET to A-site tRNA.
To differentiate between these two models, pre-steady state experiments were repeated at increased time resolution (10 ms). However, no evidence was found for reversible EF-G binding events. To directly probe whether EF-G binds the pre-translocation complex in a zero-FRET state, Cy5-labeled EF-G was delivered to pre-translocation complexes containing Cy3-labeled A-site tRNA under conditions in which both Cy3 and Cy5 were directly illuminated. Excluding the possibility that EF-G is bound stably to the ribosome prior to the FRET event, such experiments showed that the Cy5-EF-G colocalization to the ribosome occurred synchronously with the observation of FRET (Supplementary Fig. 10). Taken together, these data are consistent with established models of translocation, in which EF-G(GTP) rapidly binds and dissociates non-productively from the ribosome faster than the imaging rate (ca. ≥200 s−1)2,37,20.
Viomycin decouples EF-G binding from translocation
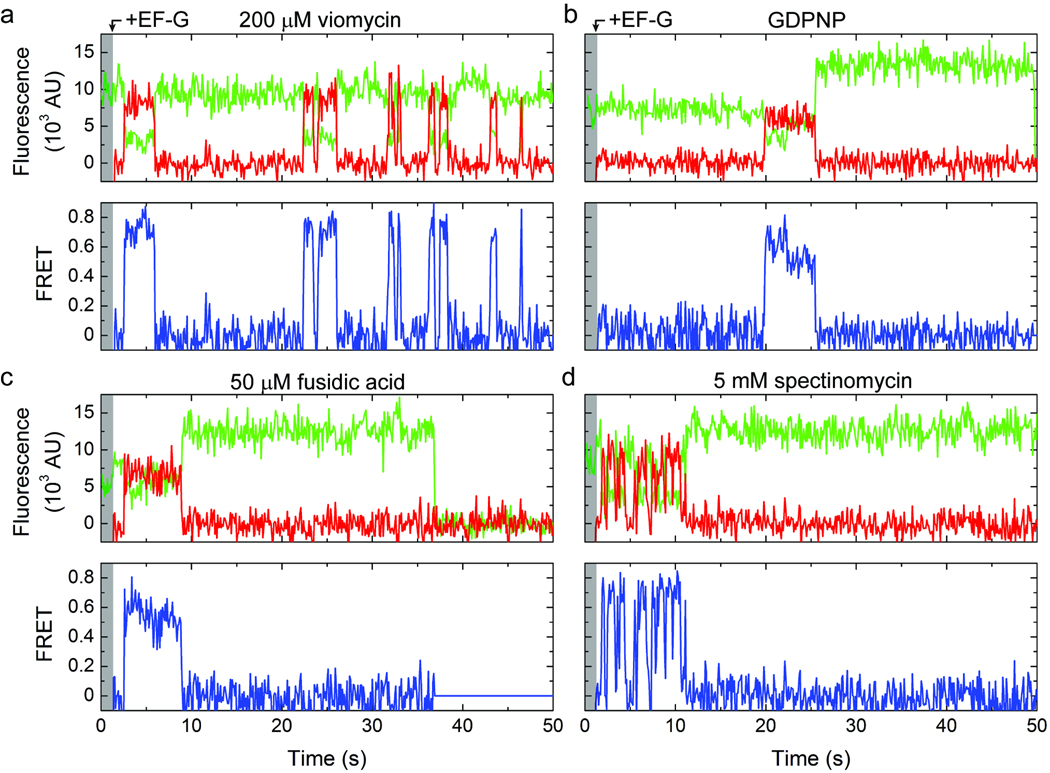
Viomycin binds the subunit interface of the ribosome and efficiently inhibits translocation subsequent to GTP hydrolysis21,38. While its molecular mechanism of action is not fully understood, viomycin increases P/E hybrid state occupancy, promotes a ratcheted ribosome conformation39,40 and stabilizes peptidyl-tRNA in the A site28. As expected for efficient translocation inhibition, in the presence of saturating concentrations of viomycin (0.2 mM) increased Cy3 fluorescence was not observed upon stopped flow delivery of EF-G(GTP) (Fig. 5a). Consistent with viomycin stabilizing the ribosome in a conformation on path to the unlocked state11,28,39,40, the rate of unlocked state formation was modestly accelerated as was the appearance of FRET between EF-G and A-site tRNA (Supplementary Tables 1–3). The distribution of FRET values observed was strongly skewed towards higher-FRET (Supplementary Fig. 6b), in line with pre-translocation complex interactions. The lifetime of the FRET event was increased ~10-fold (Supplementary Table 2, Supplementary Fig. 11a). Taken together, these findings suggest that in the presence of viomycin, EF-G is able to hydrolyze GTP and enter the A site of the unlocked ribosome, but is unable to facilitate translocation.
Figure 5. Translocation in the presence of inhibitors.
Single-molecule fluorescence (Cy3, green; Cy5, red) and FRET (blue) trajectories acquired during delivery of labeled EF-G to complexes with labeled A-site tRNAPhe in the presence of (a) 200 µM viomycin, (b) GDPNP, (c) 50 µM fusidic acid or (d) 5 mM spectinomycin.
Notably, the EF-G-ribosome interaction observed in the presence of viomycin was also characterized by intermittent fluctuations to zero FRET (Fig. 5a). Such events, although substantially diminished, were also observed at 10-fold lower EF-G concentration (20 nM), suggesting that fluctuations to zero FRET represent a mixture of dissociation events and dynamic conformational processes in the EF-G-bound complex (Supplementary Fig. 12). Although spurious photophysical events cannot presently be excluded, such observations suggest that in the presence of viomycin, EF-G can be bound to the ribosome in a configuration that does not yield detectable FRET to the A-site tRNA. These data support a model in which EF-G’s domain IV/V element may move with respect to classically-bound, peptidyl-tRNA in the A site in the EF-G-bound, viomycin-stalled complex24.
Thiostrepton prevents EF-G from engaging the A site
An EF-G-bound configuration with zero-FRET requires the distance between fluorophores to be on the order of 85 Å or greater. While the structural fold of EF-G, as well as previous kinetic26 and structural studies19,41, suggest that EF-G may bind the ribosome such that domain IV/V initially resides outside the decoding region, a transition from zero FRET to high-FRET requires a conformational event moving domain IV/V towards the A site by ~40 Å. To test this model, translocation experiments were performed in the presence of GTP and the antibiotic thiostrepton (0.1 mM), which has been shown to inhibit translocation through interactions with the large subunit GAC at a step after GTP hydrolysis and prior to inorganic phosphate (Pi) release42. A low-resolution structure of the thiostrepton-inhibited pre-translocation complex revealed EF-G bound with domain IV/V located distal to the small subunit A-site decoding region24.
In the presence of thiostrepton (0.1 mM), where the rate of unlocked state formation was modestly increased (Supplementary Tables 1 and 3), no indication of FRET was evidenced between EF-G and A-site tRNA. These data are consistent with the notion that events downstream of GTP hydrolysis are required for EF-G to fully engage the A-site of the ribosome37. Results of this nature may either suggest that thiostrepton inhibits EF-G’s interaction with the GAC, thereby preventing downstream interactions required for domain IV/V to engage the A site, or that thiostrepton prevents EF-G’s domain IV/V element from interacting with the A site while having little effect on EF-G’s overall affinity for the ribosome. The available structural data for the thiostrepton-inhibited pre-translocation complex argue in favor of the latter model in which EF-G maintains some level of interaction with the GAC, while domain IV/V remains distal to the A site and proximal to the small subunit shoulder region24. Regardless, the present observations are consistent with thiostrepton inhibiting conformational events required for EF-G to effectively engage the A site at a step either concomitant with, or immediately subsequent to GTP hydrolysis and Pi release.
GTP hydrolysis allows EF-G to engage and release the A site
While it is well established that GTP hydrolysis precedes translocation, the role of this event in the mechanism of substrate movement remains poorly understood. Contemporary translocation models suggest that initial EF-G-ribosome binding kinetics are insensitive to nucleotide and that GTP hydrolysis occurs at a subsequent step, prior to translocation22,28,37. Thus, a plausible role of GTP hydrolysis may be to facilitate conformational changes in EF-G and/or the ribosome that are necessary for translocation25,37.
To probe the timing and role of GTP hydrolysis in the translocation mechanism, experiments were performed with Cy5-EF-G and pre-translocation ribosome complexes containing Cy3-labeled A-site tRNA in the presence of the non-hydrolyzable GTP analogue, GDPNP (Fig. 5b). As anticipated from previous investigations20,33, the apparent rate of translocation, marked by the increase in Cy3 fluorescence intensity, was reduced by ~50-fold. This apparent reduction in translocation rate was principally attributed to an increased delay in the appearance of FRET between EF-G and peptidyl-tRNA (~13-fold) (; Supplementary Table 2, Supplementary Fig. 11b). Consistent with GDPNP stabilizing EF-G on the post-translocation complex28,37, the lifetime of EF-G’s interaction with the ribosome was greatly extended with respect to that observed in the presence of GTP (~25 s vs. ~300 ms) (Supplementary Table 2). Correspondingly, the distribution of FRET values observed was found to be strongly skewed toward lower FRET (Supplementary Fig. 6c) in line with modeled distance for the post-translocation complex (Supplementary Fig. 6). Similar findings were obtained in the presence of GDPCP, except the delay prior to the FRET event was increased more dramatically . This finding suggests that the apparent binding defect caused by the GTP analogues may relate to a reduction in the rate at which EF-G hydrolyzes these two compounds. Consistent with the strong influence of nucleotide on the observed EF-G-ribosome interaction, no FRET was observed between EF-G and the A-site tRNA in the presence of saturating concentrations of purified GDP (1 mM).
The role of conformational events required for EF-G release from the ribosome was further probed through experiments in the presence of fusidic acid (50 µM), an antibiotic that binds directly to EF-G at the interface of domains II and V to stabilize the post-translocation state but does not inhibit GTP hydrolysis41,43–46. In line with expectations43,44, in the presence of fusidic acid, the distribution of dwell times leading to the FRET event was unchanged and the lifetime of the EF-G-bound complex was greatly prolonged (~7 s) while no appreciable defect in translocation was observed (Fig. 5c; Supplementary Fig. 11c; Supplementary Table 2). As expected from the inhibition of conformational events required for EF-G release after translocation, the dominant FRET value in the fusidic-acid stalled complex (~0.55) was consistent with the post-translocation state (Supplementary Fig. 6d).
Spectinomycin inhibits a late step of translocation
The spontaneous rate of translocation is reported to be approximately three orders of magnitude slower than the spontaneous rate of unlocked state formation4 (Supplementary Table 2). These data suggest that conformational transitions in the ribosome in addition to those required for unlocked state formation may contribute to the process of translocation. These transitions may include small subunit motions, such as head rotation or tilt, which occur in the direction of translocation and are implicated in A-site tRNA positioning24,47,48. To examine the role of conformational events in the small subunit head domain, experiments were performed in the presence of saturating concentrations of spectinomycin (5 mM), which binds helix 34 at the base of the small subunit head domain in a manner that inhibits translocation at a step after GTP hydrolysis and Pi release27,48,49.
Consistent with these results, in the presence of spectinomycin, the nature of the EF-G-ribosome interaction subsequent to EF-G engaging the A site was found to be dramatically altered in ~25% of the FRET events observed (Fig. 5d). In this sub-population, fast conformational dynamics were observed that were similar to those seen in the presence of viomycin. As for viomycin, the rate at which EF-G engaged the A site was increased by spectinomycin (Supplementary Table 2), in line with its effect on the rate of unlocked state formation (Supplementary Tables 1 and 3). However, unlike viomycin, the high-FRET value observed in the presence of spectinomycin was somewhat lower (~0.73 as compared to ~0.81; Supplementary Fig. 6e). Clear evidence was also found for translocation, both in the increase in Cy3 fluorescence intensity observed subsequent to FRET events (Fig. 5d) and in A-site tRNA binding assays. These data suggest that spectinomycin alters conformational events required for EF-G to effectively engage the A site.
DISCUSSION
By monitoring the process of translocation from two, distinct structural perspectives new light has been shed on the role and timing of large-scale conformational changes in the ribosome and EF-G in the mechanism of translocation. These investigations included a robust approach for monitoring translocation that is based on changes in single-molecule fluorescence. In line with structural studies1,19,23,24,50, including a recent high-resolution crystal structure of the post-translocation ribosome complex41, and rapid kinetic measurements26,37, the present observations provide further evidence that increased attention must be given to understanding the allosteric linkage between conformational processes in the ribosome and EF-G that drive the basic mechanism of translocation.
Under the conditions examined, pre-steady state smFRET imaging revealed that the rate of translocation closely approximates the intrinsic rate at which the ribosome spontaneously achieves the unlocked state. These data suggest that EF-G does not measurably accelerate the rate at which this key intermediate in translocation is achieved. While our investigations support the notion that translocation is accelerated by GTP hydrolysis20,33, the data presented suggest that the stored energy in GTP does not directly influence formation of the unlocked state. Instead, the data argue that the GTP nucleotide likely plays a role in enabling EF-G to achieve a high-affinity interaction with the ribosome25, restricting intrinsic degrees of freedom in EF-G that must be liberated with proper timing of GTP hydrolysis in order for EF-G’s domain IV/V element to effectively engage the A site. Collectively, these data suggest that conformational dynamics intrinsic to the ribosome and EF-G are intimately linked and critical determinants of translocation. Antibiotics, through direct interactions with the ribosome or EF-G, uncouple ribosome and EF-G conformational events to inhibit translocation. Further experiments aimed at understanding whether domain IV/V motions impart force on A-site tRNA to catalyze translocation, or whether such motions act passively to allow it to enter the A site to prevent backward steps, will be required to delineate whether translocation follows Brownian ratchet or power stroke mechanisms, or a mixture of the two3,37.
Implications for the mechanism of translocation
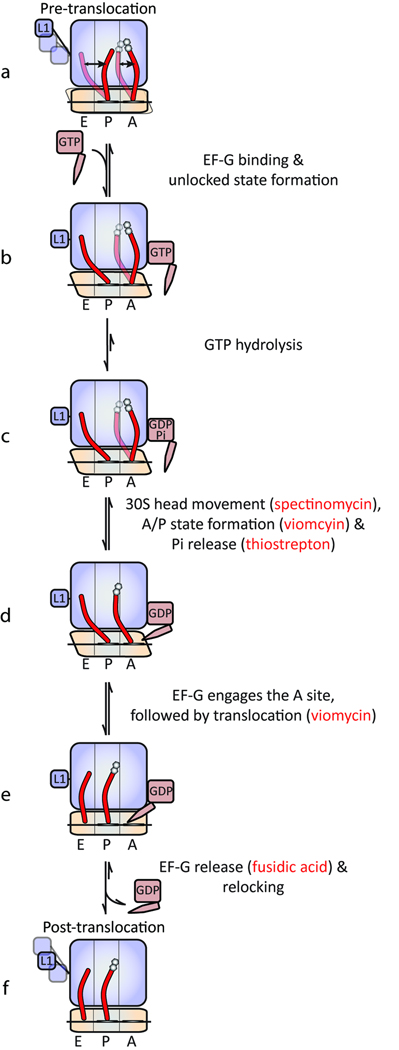
While the present observations generally support existing models of translocation1–3, they provide the added insight that correlated conformational changes in EF-G and the ribosome regulate translocation. Figure 6 provides a summary of these findings in the context of contemporary knowledge of the translocation mechanism. At sub-saturating EF-G concentrations, translocation is rate limited by initial binding kinetics. However, at saturating EF-G concentrations, EF-G’s ability to engage the A site appears to be rate-limited by intrinsic dynamic events in the ribosome, in particular, the rate at which the unlocked state is achieved. Direct interactions between the L1 stalk and P-site tRNA in this state facilitate translocation by stabilizing and/or acting on P/E hybrid tRNA in a manner that lowers the activation barrier for subsequent conformational events in the ribosome and/or substrate movement. Concomitant with, or immediately subsequent to unlocked state formation, EF-G undergoes conformational changes enabling it to productively interact with the large subunit GAC domain rapidly followed by GTP hydrolysis. Temporally linked with Pi release, EF-G’s domain IV/V element productively engages the A site. Non-hydrolyzable GTP analogues and thiostrepton uncouple the conformational linkage between EF-G and the ribosome immediately prior to GTP hydrolysis and Pi release, respectively. Consistent with the uncoupled nature of A- and P-site tRNA motions15,28, viomycin stabilizes peptidyl-tRNA in the A site and and/or alters ribosome conformation in a manner that inhibits translocation after GTP hydrolysis and Pi release and just prior to tRNA movement.
Figure 6. Schematic of the translocation mechanism highlighting points of antibiotic inhibition.
Closely timed with EF-G entering the A site, conformational events in the small ribosomal subunit head domain enable the tip of domain IV/V to enter the small subunit decoding region. Such events may include, but are not limited to, tilt or rotation of the small subunit head domain that may accompany peptidyl-tRNA adopting a puromycin-reactive A/P hybrid state28. Such subunit head motions, altered by the binding of spectinomycin48, may be directly or indirectly related to the unlocking step described previously27,37. Upon engaging the small subunit decoding region, EF-G(GDP) efficiently captures the unlocked, head-swiveled state of the ribosome to promote the rapid translocation of mRNA and tRNA substrates. Upon reaching the post-translocation state, conformational events in EF-G and the ribosome facilitate relocking events, including subunit unratcheting. Consistent with previous bulk studies20,37 the relative ratios of FRET states observed (0.75 vs. 0.55) assigned to EF-G-bound pre- and post-translocation complexes suggests that EF-G release after translocation may occur more rapidly than, or on a similar time scale to, the translocation reaction itself. Relocking secures mRNA and tRNAs in their respective post-translocation positions, restoring the ribosome to an energetically favorable configuration. Through related structural events, EF-G undergoes conformational changes that enable it to release from the particle. Subsequent to EF-G release the ribosome may freely sample an ensemble of post-translocation complex native state structures. L1 stalk dynamics in the post-translocation state, in particular the formation of open stalk configurations, may contribute to relocking as well as EF-G and tRNA release from the A and E sites, respectively.
Implications for the regulation of translation elongation
The order and timing of translocation events described here suggest that the rate of translation elongation in the cell, and thus the rate of protein synthesis, may be controlled by conformational processes intrinsic to the ribosome. Future experiments designed to further explore this possibility will demand additional structural perspectives and increased imaging resolution. While the observation that intrinsic conformational events in the ribosome may control the rate of translation is entirely consistent with the energy landscape view of protein synthesis control40,51, it will be important to understand how this dynamic landscape changes with experimental conditions. At elevated temperatures, where conformational events occur more rapidly, the events rate-limiting to translocation may be distinct. Systematic studies exploring how ribosome and EF-G dynamics change over condition space are therefore warranted.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank C. Walsh (Harvard University) and members of his laboratory for providing the Sfp expression vector, and for their assistance with the fluorescent-labeling of EF-G. The authors would also like to thank K.Y. Sanbonmatsu and P. Whitford for structural models of the E. coli ribosome. This work was supported by National Institutes of Health Grant GM079238.
Footnotes
Author contributions
J.B.M. and S.C.B. designed the experiments. J.B.M. and M.R.W. performed the experiments and analyzed the data. R.B.A. and L.W. prepared reagents. J.B.M., M.R.W., R.B.A., and S.C.B. wrote the paper.
REFERENCES
- 1.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoji S, Walker SE, Fredrick K. Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wintermeyer W, et al. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- 4.Fredrick K, Noller H. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 5.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. Factor-free ("non-enzymic") and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 7.Bergemann K, Nierhaus KH. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J. Biol. Chem. 1983;258:15105–15113. [PubMed] [Google Scholar]
- 8.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010 doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 18.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 20.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 21.Modolell J, Vazquez D. The inhibition of ribosomal translocation by viomycin. Euro J Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 22.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 25.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053–2065. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 27.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 28.Pan D, Kirillov S, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 30.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, et al. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 35.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 36.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 38.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2'-O-methylations in 16S and 23S rRNAs. Mol Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 39.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 40.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2009;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodnina MV, et al. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc Natl Acad Sci U S A. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodley JW, Zieve FJ, Lin L, Zieve ST. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun. 1969;37:437–443. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 44.Inoue-Yokosawa N, Ishikawa C, Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J Biol Chem. 1974;249:4321–4323. [PubMed] [Google Scholar]
- 45.Wang Y, et al. Single-molecule structural dynamics of EF-G--ribosome interaction during translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- 46.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 47.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 48.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JHD. A Steric Block in Translation Caused by the Antibiotic Spectinomycin. ACS Chem. Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 50.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 51.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating Unwanted Photophysical Processes for Improved Single-Molecule Fluorescence Imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.