Abstract
HIV-1 envelope glycoprotein gp41 undergoes large conformational changes to drive fusion of viral and target cell membranes, thereby exhibiting at least three distinct conformations during the viral entry process. Neutralizing antibodies against gp41 block HIV-1 infection by targeting its membrane proximal external region in a fusion-intermediate state. Here we report biochemical and structural evidence that non-neutralizing antibodies, capable of binding with high affinity to an immunodominant segment adjacent to the neutralizing epitopes in the membrane-proximal region, only recognize a gp41 conformation when membrane fusion is complete. We propose that these non-neutralizing antibodies are induced in HIV-1 infected patients by gp41 antigens in a triggered, postfusion form and contribute to production of ineffective humoral responses. These results have important implications for gp41-based vaccine design by rational strategies.
Introduction
The first critical step of HIV-1 infection is fusion of viral and target cell membranes. Viral attachment and membrane fusion are mediated by viral envelope glycoprotein upon engagement with cellular receptors1,2. The envelope protein is synthesized as a precursor, gp160, which trimerizes and undergoes cleavage into two, noncovalently-associated fragments, the receptor-binding fragment gp120 and the fusion fragment gp413,4. Three copies of each fragment make up the mature viral spike, which constitutes the sole antigen on the virion surface. Sequential binding of gp120 to the primary receptor CD4 and coreceptor (e.g. CCR5 and CXCR4) induces large conformational changes, which then trigger dissociation of gp120 and a cascade of refolding events in gp411,5. Gp41, with its C-terminal transmembrane segment inserted in the viral membrane, is folded into a prefusion conformation within the precursor, gp160. Cleavage between gp120 and gp41 makes this pre-fusion conformation metastable with respect to a rearranged, postfusion conformation. When triggered by the binding of gp120 to the coreceptor, the N-terminal fusion peptide of gp41 translocates and inserts into the target cell membrane. The extended conformation of the protein, with the fusion peptide inserted into cell membrane and the transmembrane anchor in the viral membrane, is referred to as the “prehairpin intermediate”6. It can be targeted by T-20/Enfuvirtide, the first approved fusion-inhibiting antiviral drug, as well as by certain broadly neutralizing antibodies7–9. Subsequent rearrangements involve folding back of the C-terminal heptad repeat 2 (HR2) region of gp41 into a hairpin conformation, creating a six-helix bundle, which places the fusion peptide and the transmembrane segment at the same end of the molecule 10,11. This irreversible refolding of gp41 effectively brings the two membranes together. During the fusion process, gp41 exhibits at least three distinct conformational states: the prefusion conformation, an extended, prehairpin intermediate, and the postfusion conformation. The conformational differences among these states are so great that each of them likely presents distinct antigenic surfaces to the immune system.
HIV-1 infected patients typically generate strong antibody responses to the envelope glycoprotein, but most of these antibodies are either non-neutralizing or strain-specific, and many recognize epitopes occluded on mature trimeric spikes or epitopes located in the highly variable loops. Extensive glycosylation, sequence diversity, and receptor-triggered conformational changes and epitope masking pose great challenges to generation of broadly reactive neutralizing antibodies (NAbs)12–14. Some patient sera show broadly neutralizing activity, but immunogens that can induce such antibody responses have remained elusive15. Nevertheless, a number of broadly reactive neutralizing monoclonal antibodies (mAb) have been isolated that recognize regions of the HIV-1 envelope glycoprotein. Some are located on gp120: the CD4 binding site (CD4bs), the V2 and V3 loops and the carbohydrates on the outer domain of gp12016–22. Additional neutralizing antibodies target regions on gp41 adjacent to the viral membrane and called the membrane-proximal external region (MPER; residues 662–683 (HXB2 numbering))23–25. Our previous studies on the molecular mechanism of neutralization by two of these anti-gp41 antibodies, 2F5 and 4E10, indicate that their epitopes are only exposed or formed on the prehairpin intermediate state during viral entry9. We also find that the hydrophobic CDR H3 loops of these antibodies mediate a reversible attachment to the viral membrane that is essential for their antiviral activities26. These MPER-directed antibodies probably associate with the viral membrane in a required first step and are poised to capture the transient gp41 fusion intermediate9,26.
Gp41 also induces non-neutralizing antibodies which are much more abundant in patients than neutralizing ones. The non-neutralizing antibodies have been classified into two groups based on the location of their epitopes. Cluster I antibodies react with the immunodominant C-C loop of gp41 (residues 590–600), and cluster II antibodies recognize another immunodominant segment (residues 644–663) next to the MPER27. Members in the latter group can bind HIV-1 gp41 with high affinity, but have weak or no neutralizing or antiviral activities28,29. The prototype of this group includes mAbs 98-6, 126-6, 167-D, 1281 and 1379, isolated by immortalizing plasma B cells from HIV-1 positive patients27,30–32. These mAbs appeared to react optimally with a form of gp41 in its postfusion conformation33, but they also bind “monomeric gp41” and oligomer-specific conformations of gp4131,34. As the conformation of these envelope preparations has not been fully assessed, it remains uncertain which conformation(s) of gp41 the cluster II mAbs recognize and why they are incapable of blocking HIV-1 infection, as do the MPER-directed neutralizing antibodies.
Here we set out to investigate the structural basis for the drastic differences between the MPER-directed antibodies and the cluster II antibodies in their ability to neutralize HIV-1 infection. We have produced an improved gp41 construct to mimic its prehairpin intermediate conformation. This protein binds tightly to broadly neutralizing antibodies 2F5, 4E10 and Z13e135. We present biochemical and structural data to demonstrate that anti-HIV-1 gp41 cluster II antibodies show high binding affinity for the postfusion conformation of gp41, and do not bind or bind only weakly to the stable, homogeneous gp41 preparations representing the pre-hairpin intermediate or the prefusion conformation. We propose that these antibodies are non-neutralizing because they target a late step in the viral entry process, when membrane fusion is likely to be complete. They may be induced in HIV-1 infected patients by gp41 antigens in a triggered, postfusion form, which serve as irrelevant decoys to distract the immune system and to contribute to production of ineffective humoral responses. These results could guide rational strategies for HIV-1 gp41-based vaccine design.
Results
Production of GCN4-gp41-inter
In our previous studies, we have produced homogeneous preparations of trimeric HIV-1 envelope protein, derived from a clade A isolate 92UG037.8, to mimic its prefusion (gp140), prehairpin intermediate (gp41-inter) and postfusion (gp41-post) conformations (Figure 1 and ref9). We demonstrated that the different conformational states of gp41 exhibit markedly different antigenic characteristics. In particular, two MPER-directed neutralizing antibodies, 2F5 and 4E10, inhibit HIV-1 infection by targeting the prehairpin intermediate state of gp419,26. To define the conformational state recognized by anti-gp41 cluster II antibodies, we sought to test their reactivity to gp140, gp41-inter and gp41-post. Gp41-inter was designed to capture gp41 in the extended, prehairpin intermediate conformation with the following sequence: (HR2)-linker-(HR1-CCloop-HR2-MPER)-(trimerization foldon tag) (Figure 1). This construct can be pictured as the prehairpin intermediate captured by a covalently linked HR2 peptide, such as T20. When gp41-inter chains trimerize, the N-terminal HR2 segments (T20) form a six-helix bundle with the HR1 segments, while the C-terminal HR2 segments, constrained by the foldon tag, will be unable to form a six-helix bundle. Thus, the two copies of HR2 in gp41-inter are in distinct conformations: the N-terminal HR2 in the six-helix, postfusion state and the C-terminal HR2 mimics the prehairpin intermediate. Using gp41-inter as a reagent to analyze antibodies directed against HR2, such as cluster II mAbs, would complicate data interpretation. We have therefore designed a modified gp41-inter in which the entire six-helix bundle (the segment HR2-linker-HR1) was replaced with a trimeric GCN4-derived coiled-coil to generate GCN4-gp41-inter (Figure 1 and ref36). The heptad repeat of GCN4 needed to be in the same register as the HR1 region of gp41 to avoid any structural distortion. GCN4-gp41-inter was expressed in E. coli and refolded in vitro following the same protocol we developed for gp41-inter (ref9; also see Methods). As expected, purified GCN4-gp41-inter is also a monodisperse trimer and stable after several rounds of gel-filtration chromatography (Supplementary Figure 1).
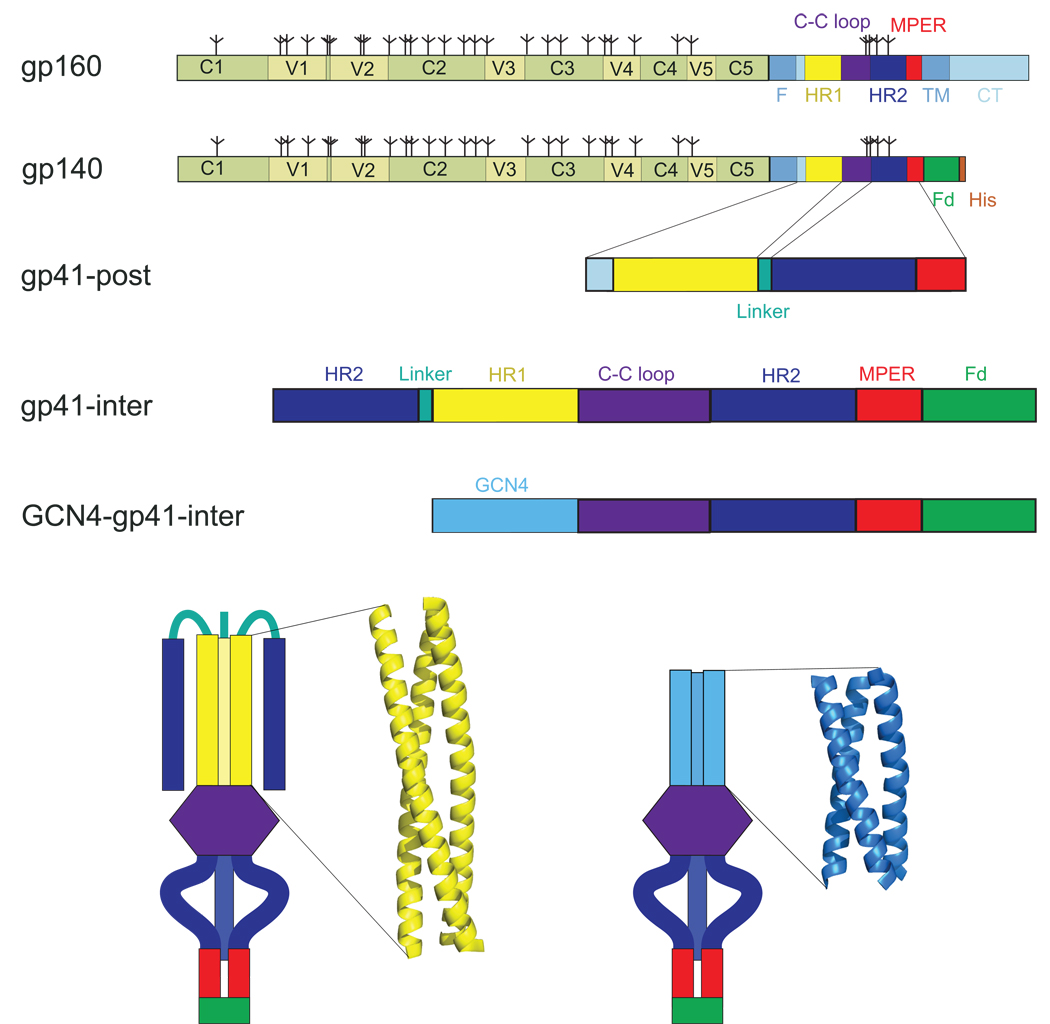
Figure 1. HIV-1 envelope constructs and GCN4-gp41-inter.
Top, schematic representation of HIV-1 envelope glycoprotein gp160, the full-length precursor. Segments of gp120 and gp41 are designated as follows: C1–C5, conserved regions 1–5; V1–V5, variable regions 1–5; F, fusion peptide; HR1, heptad repeat 1; C-C loop, the immunodominant loop with a conserved disulfide bond; HR2, heptad repeat 2; MPER, membrane proximal external region; TM, transmembrane anchor; CT, cytoplasmic tail. Glycans are represented by tree-like symbols. HIV-1 envelope constructs used in this study include gp140, the uncleaved ectodomain of gp160 with a trimerization foldon (Fd) tag and a His-tag at its C-terminus; gp41-post, gp41 in the six helix conformation with partial MPER; gp41-inter, HR2 peptide- and foldon tag-trapped gp41 in the prehairpin intermediate conformation; GCN4-gp41-inter, gp41-inter with the six helix bundle portion replaced with a trimeric GCN4 coiled-coil36 (in light blue). Bottom, diagrams representing 3-D organization of gp41-inter and GCN4-gp41-inter. The trimeric GCN4 with its heptad repeat in the same register as HR1 replaces the HR2-linker-HR1 of gp41-inter. The coordinates of HR111 and GCN436 coiled-coils are shown in yellow and light blue, respectively.
To confirm that replacement of the six-helix bundle with GCN4 does not alter antigenic properties of gp41-inter, we have carried out binding experiments, using surface plasmon resonance (SPR), to assess reactivity of GCN4-gp41-inter to three MPER-directed mAbs 2F5, 4E10 and Z13e1. As shown in Supplementary Figure 2 and Supplementary Table 1, both gp41-inter and GCN4-gp41-inter proteins show the same kinetic profile for binding to the antibodies, indicating that the conformation of MPER is identical in the two constructs. We note that Z13e1, the least potent neutralizing antibody among the three, dissociates from gp41 much more rapidly that do 2F5 and 4E10, consistent with our earlier suggestion that a slow dissociation rate of antibody-gp41 complex may be critical for the neutralizing activity of MPER-directed antibodies26. The interactions of gp41 with the three antibodies fit well to the 1:1 Langmuir binding model (Supplementary Figure 2). Moreover, both gp41-inter and GCN4-gp41-inter form tight complexes with cluster I antibodies, such as 240-D and 246-D27, which could be purified by gel filtration chromatography (data not shown), suggesting that substitution of the six-helix bundle in gp41-inter with GCN4 does not introduce any structural distortion. We conclude that GCN4-gp41-inter is trapped in the same fusion-intermediate conformation as is gp41-inter.
The postfusion conformation of gp41 is recognized by cluster II antibodies
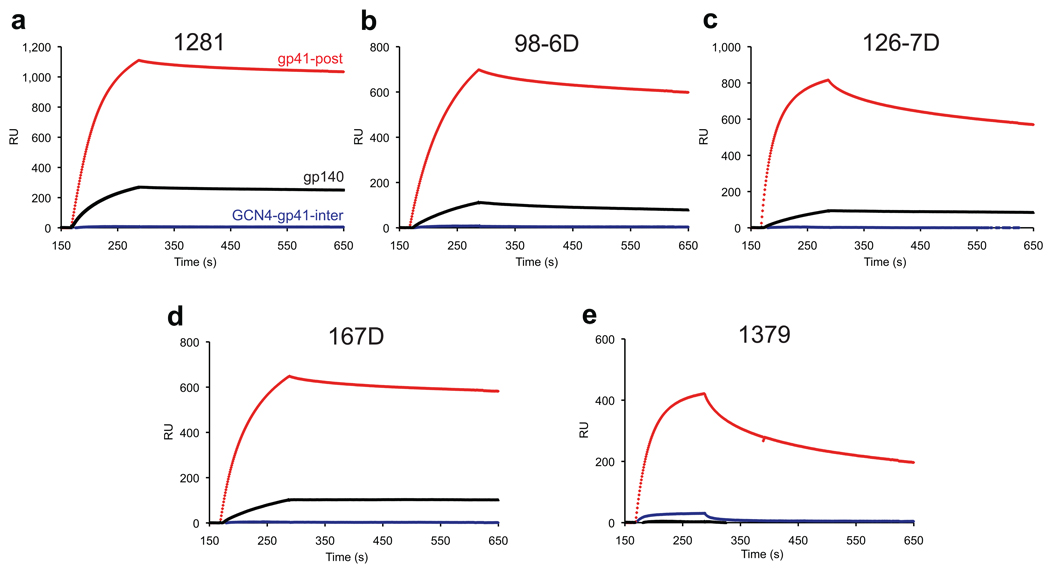
To assess binding specificity of cluster II antibodies, we chose a panel of five human mAbs, including 98-6, 126-6, 167-D, 1281, and 137927,30,31, and tested their reactivity to gp140, GCN4-gp41-inter and gp41-post by SPR. All the antibodies have been shown to bind a full-length recombinant gp41 and oligomeric gp140, as well as a six-helix bundle formed by HR1 and HR2 peptides using ELISA; 98-6 is the only one that reacts with an HR2 peptide alone31,33. As shown in Figure 2 and Supplementary Figure 3, all the antibodies showed tight binding to the postfusion conformation of gp41 with extremely fast on-rates, in agreement with the previous findings33, indicating that the six-helix bundle presents the optimal conformation recognized by these antibodies. In contrast, none of the antibodies shows any binding to GCN4-gp41-inter, which does contain the cluster II epitopes (residues 644–663), suggesting the residues in gp41 critical for interacting with cluster II antibodies are either buried or not correctly configured for antibody recognition in the prehairpin intermediate state. In particular, mAb 98-6, capable of forming a complex with an unconstrained and flexible HR2 peptide, does not show any detectable binding to GCN4-gp41-inter, further confirming that gp41-inter presents a unique conformation that is incompatible with recognition by the non-neutralizing cluster II antibodies. Our stringently characterized gp140 trimer shows only weak binding to four of these mAbs and no binding at all to mAb 1379 (Figure 2 and Supplementary Figure 3). Moreover, all the cluster II antibodies showed barely detectable binding to the same envelope trimer expressed on 293T cell surfaces, just as 2F5, which does not recognize the native, prefusion conformation of gp41 (Supplementary Figure 4). Taken together, the non-neutralizing, anti-HIV-1 gp41 cluster II antibodies only recognize the postfusion conformational state of gp41, which is distinct from the fusion-intermediate conformation targeted by the MPER-directed broadly neutralizing antibodies.
Figure 2. Anti-HIV-1 gp41 cluster II antibodies preferentially bind gp41 in its postfusion conformation.
Human anti-gp41 cluster II mAbs, 1281, 98-6D, 126-7D, 167D and 1379, were analyzed by a surface plasmon resonance (SPR) assay for binding to HIV-1 gp41 constructs: gp140 (sensorgrams in black); GNC4-gp41-inter (blue); and gp41-post (red). GCN4-gp41-inter or gp41-post was immobilized on CM5 chips; gp140 was captured on a Ni-NTA chip. Each IgG at 50 nM was passed over each surface individually. Data with the antibodies immobilized on a Protein A chip are shown in Supplementary Figure 3.
Interaction of gp41-post with the monovalent Fab fragment derived from mAb 1281
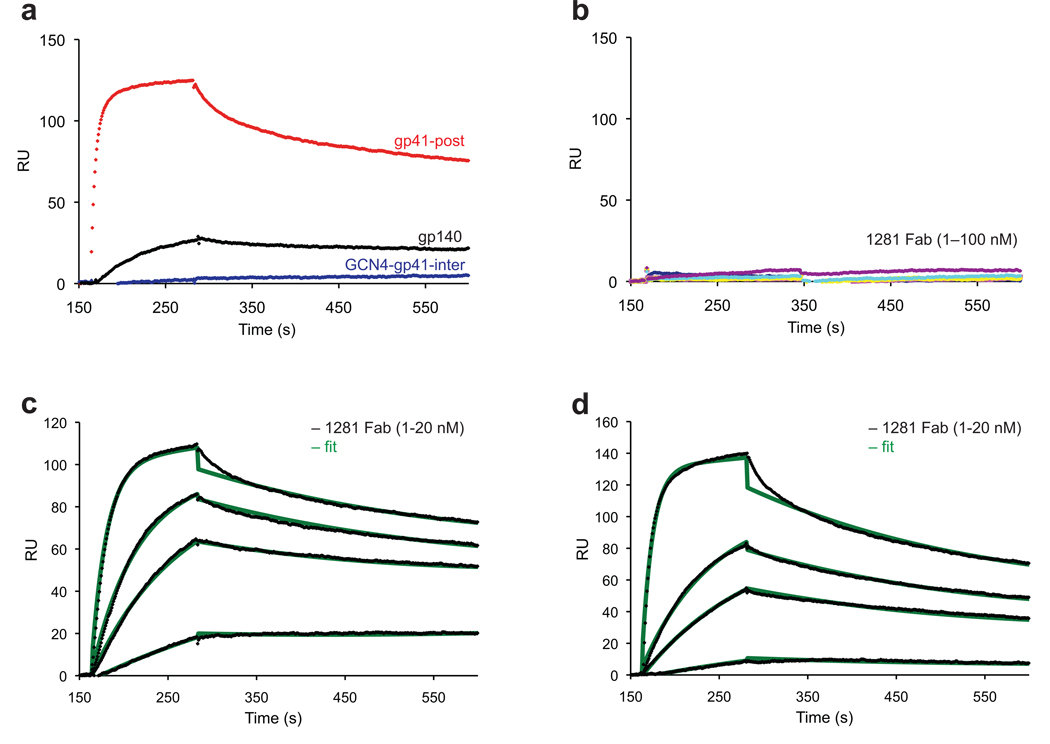
Although bivalent IgG is the physiologically relevant form, intrinsic affinity of its antigen-combining site to target antigens could be masked by avidity effects. To gain further insights into how cluster II antibodies interact with gp41 in its postfusion conformation, we chose mAb 1281 as a representative, and produced the Fab fragment to eliminate avidity effects. As shown in Figure 3a, the 1281 Fab showed the same pattern for binding to the envelope proteins: high affinity to gp41-post, weak affinity to gp140 and no binding to GCN4-gp41-inter, fully consistent with our results using the intact IgG. Additionally, the Fab failed to interact with GCN4-gp41-inter in a wide range of concentrations (1–100 nM; Figure 3b), further confirming the notion that HR2 adopts a completely different conformation in the prehairpin intermediate from the six-helix bundle conformation. We predicted that gp41-inter with the N-terminal HR2 folding back on the HR1 to form a six-helix bundle would bind the 1281 Fab the same way as does gp41-post. Shown in Figure 3c and 3d, the kinetic properties of the Fab binding to gp41-inter and gp41-post are almost identical. We note that the kinetic characteristics of 1281 Fab binding to gp41-post are not that different from those of Z13e1 binding to gp41-inter, suggesting that affinity of an antibody to gp41 alone is not an indicator of its antiviral activity.
Figure 3. Analysis of interactions of 1281 Fab with various gp41 constructs.
Fab fragment derived from mAb 1281 was tested by SPR for binding to gp41 constructs. (a) The recorded sensorgram for gp41-post is in red, gp140 in black and GCN4-gp41-inter in blue. (b) To confirm no detectable binding of 1281 Fab to GCN4-gp41-inter, solutions of 1281 Fab at various concentrations were flowed over the GCN4-gp41-inter surface. The sensorgrams are shown in various colors. In c and d, 1281 Fab at various concentrations were passed over the surfaces immobilized with gp41-post, and gp41-inter containing the six-helix bundle, respectively. Binding kinetics were evaluated using a 1:1 Langmuir binding model and binding constants are summarized in Supplementary Table 1. The sensorgrams are shown in black and the fits in green. All injections were carried out in duplicate and gave essentially identical results. Only one of the duplicates is shown.
Crystal structure of the complex of gp41-post and the 1281 Fab fragment
To obtain a structural definition of the cluster II epitopes, we have determined the crystal structure of the complex of gp41-post and 1281 Fab at 3.3 Å resolution. Crystals of the 1281 Fab–gp41-post complex diffract to 3.3 Å resolution and belong to space group R3, with one Fab and one gp41 monomer per crystallographic asymmetric unit. The structure was determined by molecular replacement using HIV-1 gp41 monomer (pdb:1AIK; ref10) and a library of Fab coordinates as search models37. Searches for gp41 and Fab yielded convincing solutions. The constant region of 1281 Fab shares 100% sequence identity with that of the search model, and thus the two should have the same structure. However, density for the constant region remained poor throughout the rebuilding and refinement process (Supplementary Figure 5a and 5b), indicating there is some packing disorder in this domain. The variable region together with gp41 could form a complete lattice in the absence of the constant domain, which may therefore have more than one orientation in the crystal (Supplementary Figure 5c). Complementarity determining regions (CDRs) of both heavy- and light-chains of 1281 were rebuilt iteratively, and the final model was refined with an Rwork of 26.1% and an Rfree of 28.9% (Table 1).
Table 1.
Data collection and refinement statistics
| 1281 Fab–gp41-post | |
|---|---|
| Data collection | |
| Space group | R3 |
| Cell dimensions | |
| a, b, c (Å) | 115.83,115.83,119.54 |
| α, β, γ (°) | 90,90,120 |
| Resolution (Å) | 33.4-3.30(3.39-3.30)* |
| Rsym or Rmerge | 8.7(42.5) |
| I / σI | 16.5(2.3) |
| Completeness (%) | 99.5(99.8) |
| Redundancy | 3.2(3.2) |
| Refinement | |
| Resolution (Å) | 33.4-3.30(3.39-3.30) |
| No. reflections | 8,478(640) |
| Rwork / Rfree | 26.1(37.8)/28.9(43.1) |
| No. atoms | |
| Protein | 3,771 |
| B-factors | |
| Protein | 130.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.743 |
Values in parentheses are for highest-resolution shell.
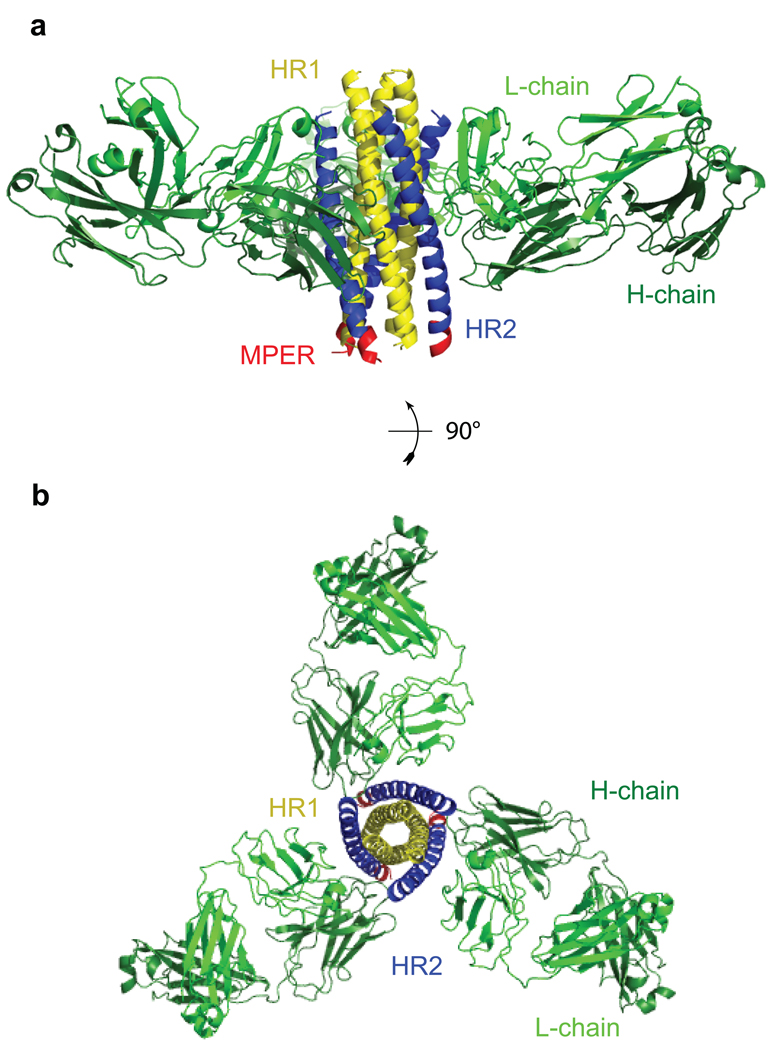
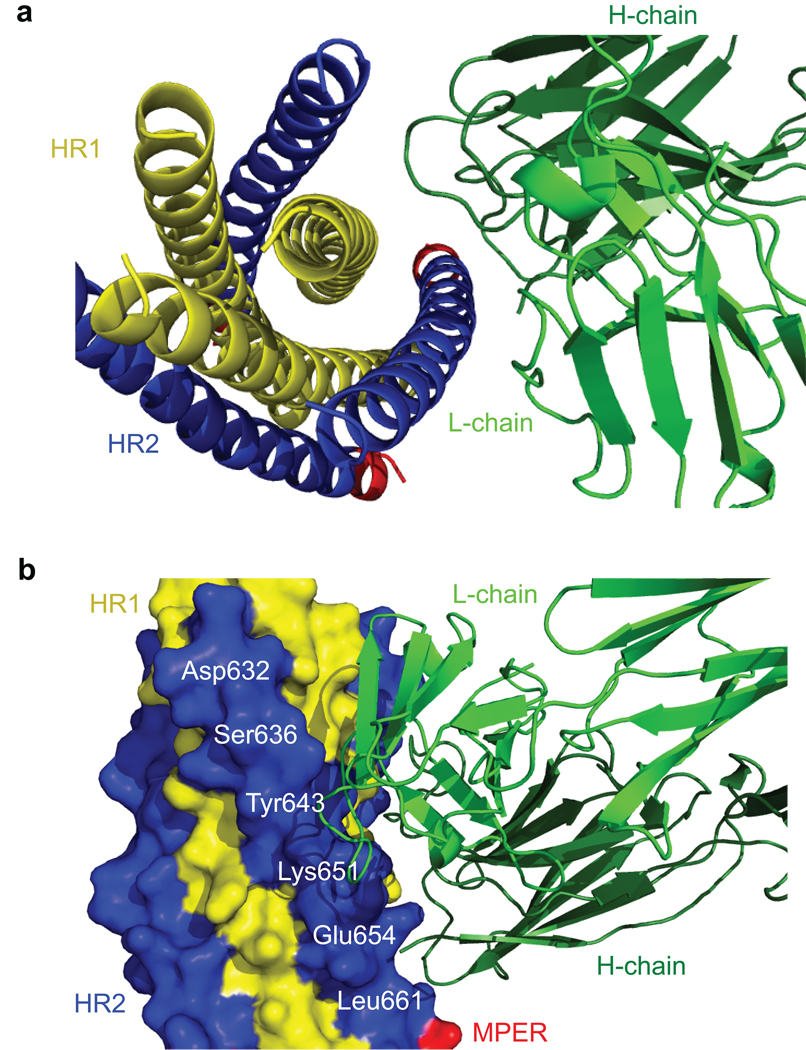
As expected, gp41-post derived from the clade A isolate 92UG037.8 forms a six-helix bundle similar to the structures of gp41 determined for HIV-1 clade B isolates and SIV (simian immunodeficiency viruses)10,11,38,39. The 2F5 epitope in the MPER of gp41-post adopts an α-helical conformation (Figure 4) distinct from that in the crystal structure of the 2F5-gp41 peptide complex, where the epitope stretches into an extended conformation with two overlapping type I β turns40. The CDR loops from both the heavy- and light- chains of 1281 Fab make extensive interactions with the six-helix bundle. The CDR H1 and L2 loops contact the HR2 helix in gp41-post exclusively, while the CDR H3 interacts with both the HR1 and HR2 helices (Figure 4). Two segments (residues 97–100 and 100C–100E) of CDR H3 pack against the residues 560–564 in HR1 (Figure 5a). The footprint of the antibody covers residues 643–661 of HR2 (Figure 5b), fully consistent with previous epitope-mapping data27,33,34. The observation that 1281 Fab makes direct contacts with the HR1 helix is in agreement with the previous findings that most cluster II antibodies interact with the six-helix bundle of gp41, but not with HR1 or HR2 peptide alone33. Binding of 1281 Fab does not induce major structural rearrangements in gp41, as judged by comparison of gp41-post with the other six-helix bundle structures10,11. The distance between the centers of the 2F5 and 1281 epitopes is ~ 30 Å, confirming their spatial closeness on gp41 (Figures 4a and 5b).
Figure 4. Crystal structure of the complex of gp41-post and the Fab fragment of cluster II antibody 1281.
Side (in a) and top (in b) views of the overall structure of the postfusion conformation of HIV-1 gp41 in complex with the Fab derived from an anti-gp41 cluster II mAb 1281 are shown in ribbon representation. The heavy chain of the antibody is in dark green and the light chain in light green; HR1 of gp41 in yellow, HR2 in blue and the part of MPER in red. The Fab mainly grips HR2, but also makes direct contacts with HR1 by CDR loops from both the heavy- and light- chains, suggesting the six-helix bundle conformation of gp41 is critical for 1281 binding. The MPER part in red contains the 2F5 epitope (residues 663–669), which is α-helical in the postfusion conformation.
Figure 5. Close-up of major contacts between gp41 and 1281 Fab.
Gp41 and1281 Fab are both shown in ribbon diagram in a; gp41 in surface representation and the Fab in ribbon diagram in b. The heavy chain of the antibody is in dark green and the light chain in light green; HR1 of gp41 in yellow, HR2 in blue and the part of MPER in red; surface-exposed residues in HR2 are labeled in white. The CDR H1 and L2 loops of the antibody contact the HR2 helix in gp41-post; the CDR H3 reaches out and interacts with both the HR1 and HR2 helices. The footprint of the antibody covers residues 643–661, consistent with the previous epitope-mapping data27,33,34. The 2F5 in red is spatially close to the cluster II epitope.
Discussion
Developing a safe and effective vaccine that durably blocks HIV-1 infection is one of the highest priorities for global health. Conventional strategies based on empirical approaches have failed to provide adequate protection against HIV-1 infection in clinical trials41,42. Innovative approaches are urgently needed. The HIV-1 envelope glycoprotein has evolved to undergo large structural rearrangements with very different conformational states during viral entry and each state exhibits distinct antigenic characteristics. Rational design of an effective envelope-based immunogen will likely require a deeper understanding of the structural correlates of envelope antigenicity and immunogenicity. We sought to study the structural basis for the drastic differences between the MPER-directed antibodies and the cluster II antibodies in their ability to neutralize HIV-1 infection, despite their equivalently high affinity for HIV-1 gp41. We have previously reported that the broadly neutralizing mAbs, 4E10 and 2F5, do not bind the prefusion form of gp41, but rather target only the prehairpin intermediate conformation9. Gp41-inter, a gp41 design to mimic the prehairpin intermediate, was instrumental in that study. 4E10 and 2F5 bind almost irreversibly to gp41-inter, while their complexes with the soluble peptide epitopes dissociate much more rapidly9, consistent with the notion that the very slow dissociation rate of the antibody-gp41 complex may be critical for targeting a fusion-intermediate, as dissociation could allow gp41 to proceed toward fusion. Moreover, addition of gp41-inter could efficiently block neutralization by 4E10 even when the antibody was preincubated with the virus26, while the 4E10 epitope peptide was ineffective under the same setting (M. Alam and B. Chen, unpublished data), suggesting that gp41-inter is in a conformation relevant to both membrane fusion and antibody neutralization. To examine conformational specificity of the cluster II mAbs, it was necessary to modify our original gp41-inter design by replacing the postfusion component with an unrelated trimeric GCN4 (Figure 1; ref9). We demonstrate that anti-HIV-1 gp41 cluster II antibodies, which recognize a segment adjacent to the neutralizing epitopes in the MPER, show high affinity only to gp41 in the postfusion conformation. These antibodies are ineffective in preventing HIV-1 infection as they target a late step in the viral entry process, when membrane fusion is likely to be complete. We propose that the stable postfusion conformation of gp41 probably serves as a decoy to help HIV-1 evade the immune system and induce ineffective antibody responses in infected patients. Rational design of gp41-based immunogens would require strategies to constrain gp41 and prevent it from folding into the six-helix bundle conformation.
Neutralizing antibodies against gp41 are rare, while gp41-specific non-neutralizing antibodies are often quite abundant27. Cluster I antibodies directed at the C-C loop are not neutralizing, either because these epitopes are not readily accessible on the functional envelope trimer or because antibody binding to the C-C loop does not impede the envelope function28,43,44. The reason why cluster II antibodies are non-neutralizing has been puzzling, however, especially, since their epitopes are near the broadly neutralizing epitopes in the MPER. Our previous studies on the neutralization mechanism by MPER-directed mAbs show that 4E10 and 2F5 target the gp41 prehairpin intermediate9,26. Furthermore, the ability of these antibodies to interact with HIV-1 membrane is critical for them to capture their MPER target presented in the transient intermediate state45,46. Thus, it appears that neutralizing activity for an anti-gp41 antibody correlates with its capacity to bind viral membrane and gp41 in the fusion-intermediate conformation. Some cluster II mAbs, including 126-6, 167-D and 1281, can also interact with membrane lipids (M. Alam and B. F. Haynes, Duke University, personal communication). In addition, the cluster II epitopes are more membrane-distal than the MPER, and these mAbs would need a much longer hydrophobic CDR loop to bind both gp41 and the membrane simultaneously. Thus, membrane-binding properties are unlikely to be the reason why the cluster II mAbs are not neutralizing. Our results clearly demonstrate that the postfusion conformation of gp41 is the high-affinity target of the cluster II antibodies. The only two opportunities when this conformational state of gp41 is accessible to antibodies during viral entry are either when gp120 dissociates prematurely (gp120 shedding) to leave nonfunctional gp41 “stumps” on the surface of virion47, or when membrane fusion is complete. In both cases, binding by cluster II mAbs would not obstruct the function of gp41 and thus would have no impact on HIV-1 entry.
Cluster II antibodies could mediate HIV-1 specific antibody-dependent cellular cytotoxicity (ADCC) and other Fc-mediated antiviral activities29,48–50. Our data indicate that these antibodies only bind with high affinity to the triggered form of gp41 on the surface of virion, not the native envelope spikes. For any given HIV-1 envelope, ADCC that targets the cluster II epitopes would have to depend on how much gp120 sheds spontaneously and how many nonfunctional gp41 stumps are present on the viral membrane surface. Thus, cluster II mAb-mediated ADCC would be more effective against isolates which shed gp120 readily, while those with much more stable (gp120–gp41)3 complex would be more resistant.
Cluster II epitopes are very immunogenic in vivo27 and could help HIV-1 evade the immune system by triggering production of non-neutralizing antibody responses. A recent study to clone anti-HIV antibodies against HIV-1 gp41 from the memory B-cell compartment of HIV-1 infected individuals has shown that unique B-cell clones targeting cluster II epitopes account for 49% of all anti-gp41-reactive B cells51. The crystal structure of 1281 Fab in complex of gp41-post demonstrates that the antibody makes direct contacts with both HR1 and HR2, suggesting that the six-helix bundle is likely the immunogen that induces this type of antibody responses in HIV-1 infected patients. The gp120-depleted gp41 stumps, which do interact with cluster II antibodies47, observed on the surface of virion, are probably in the triggered, six helix bundle form and may be the major source of gp41 immunogens responsible for this type of antibody responses. HIV-1 may thereby exploit the envelope stability as one of immune evasion tactics to distract the immune system from the native, functional trimers.
HIV-1 envelope-based immunogens often induce high ELISA-titer antibody responses with limited neutralizing activity or breadth. Most envelope immunogens containing gp41 are not rigorously characterized, particularly, in their conformational homogeneity. For example, HIV-1 gp140 preparations are often a mixture of monomers, dimers, trimers and aggregates, and it is difficult to discern what conformation each of these species represents and whether they are physiologically relevant. Gp41 could adopt the most stable, postfusion conformation in some of these irrelevant forms and expose immunodominant, non-neutralizing epitopes, such as those recognized by the cluster II antibodies. These preparations could lead to misinterpretation of the antigenic and immunogenic properties of the envelope protein, and hence misguide the effort for immunogen design. Another important and understudied aspect of protein-based immunogen design is the potential impact that adjuvant formulation may have on immunogen structure. For instance, emulsions using oil-based adjuvants could potentially disrupt protein structural integrity and trigger conformational changes in gp41. Empirical approaches to develop gp41-based immunogens that overlook these structural details of immunogens might primarily induce non-neutralizing antibody responses. For a genuinely rational immunogen design, steps must be taken to prevent gp41 from folding into the postfusion conformation.
Methods
Expression and refolding of gp41 proteins
Details of the production of gp41 proteins are described in Supplementary Methods.
Antibody and Fab production
Human anti-HIV-1 gp41 cluster II monoclonal antibodies, 98-6, 126-6, 167-D, 1281 and 1379, were produced as described27,30,31. Fab fragments of 2F5, 4E10 and Z13e1 were expressed in insect cells and purified by Gamma Bind Plus-Sepharose beads (GE Healthcare). Recombinant baculoviruses containing the heavy chain or the light chain of Fab were generated separately and mixed at a volume ratio of 1:1. Typically, 12 L of Sf9 cells were infected with recombinant baculoviruses at a multiplicity of infection of 2.5. The cell supernatants were harvested 72 h post-infection by centrifugation, concentrated and loaded onto a Gamma Bind Plus-Sepharose column. Bound Fab was eluted by 100 mM glycine, pH 2.5 and further purified by gel filtration chromatography on a Superdex 200 (GE Healthcare). The Fab fragment of mAb 1281 was produced by papain (Sigma) digestion at 37°C for 4 hours with an enzyme to antibody ratio of 1:1,000 by weight, and then purified by protein A affinity and gel-filtration chromatography.
SPR binding assays
All experiments were performed in duplicate with a Biacore 3000 instrument (Biacore Inc, Piscataway NJ) at 20°C, with immobilization levels between 300 and 400 RU to avoid rebinding events. The experiments were run with a flow rate of 50 µl min−1 with a 2 min association phase and a 10 min dissociation phase. HBS-EP (10 mM HEPES pH 7.4, 150 mM NaCl and 3 mM EDTA) was the running buffer for experiments using CM5 chips; and a buffer containing 10 mM HEPES pH7.4, 150 mM NaCl, 50 µM EDTA and 0.005% (v/v) P20 for those using Ni-NTA chips.
Binding of 2F5, 4E10 or Z13e1 Fab to GCN4-gp41-inter and gp41-inter were performed as follows. GCN4-gp41-inter or gp41-inter was coupled to a CM5 chip at ~ 300–400 RU for 2F5 and 4E10 binding and at ~100 RU for Z13e1 binding using a standard amine coupling procedure. Sensorgrams were recorded by passing each Fab at various concentrations over the ligand surface. The surface was regenerated between each experiment by a single injection (3 sec) of 35 mM NaOH and 1.3 M NaCl at a flow rate of 100 µl min−1. 1281 Fab and cluster II IgG’s binding to gp41-post and GCN4-gp41-inter were performed with gp41-post or GCN4-gp41-inter immobilized to a CM5 chip as described above. The chip surface was regenerated between each experiment using a single injection (3 sec) of 35 mM NaOH and 1.3 M NaCl at a flow rate of 100 µl min−1. Binding of 1281 Fab to the gp140 trimer was done by capturing the his-tagged gp140 on a Ni-NTA chip and 1281 Fab at various concentrations were passed over the chip surface. The surface was regenerated between each experiment with 10 mM HEPES, pH 8.3, 150 mM NaCl, 350 mM EDTA and 0.005% P20. To avoid potential artifacts introduced by protein immobilization to a CM5 chip, Protein A was first immobilized to a CM5 chip at ~1000 RU using the standard procedure. Each IgG was then captured to the Protein A surface at ~400 RU. Each of gp140, GCN4-gp41-inter or gp41-post at 50 nM was passed over each antibody surface individually. The surface was regenerated using 10 mM HCl. Binding kinetics were analyzed by BiaEvaluation software (Biacore) using a 1:1 Langmuir binding model. All injections were carried out in duplicate and gave essentially identical results.
Crystallization and structure determination
Crystals of the complex of gp41-post and 1281 Fab were obtained using hanging drop vapor diffusion method. Briefly, 1 µl of protein solution (15 mg ml−1) was mixed with 1 µl of mother liquor (0.1 M Tris-HCl, 15% (w/v) PEG 4K) and allowed to equilibrate at 25°C. Crystals were flash frozen in liquid N2 using 20% (v/v) glycerol in the mother liquor as a cryoprotectant. X-ray diffraction data were collected at 100° K at beamline 24-ID, Advanced Photon Source (Argonne National Laboratory, IL). The best crystals diffracted to a Bragg spacing of 3.3 Å with space group R3 (a=115.8, b=115.8, c=119.53). Initial phases were obtained by molecular replacement using the ectodomain of gp41 as a search model10. The top solution of this search was fixed and a second search against a library of 244 antibody fragment structures was performed using MOLREP and Phaser37,52,53, yielding a single solution. The three-fold axis of the gp41 trimer coincides with the crystallographic three-fold and there are one monomer of gp41 and one Fab in an asymmetric unit. Mild anisotropy in the data was observed and subsequently corrected using a diffraction anisotropy server prior to refinement54. Electron density for the constant domain of the Fab fragment was poor. A complete lattice can form by the variable region of Fab and gp41 in absence of the constant domain, suggesting that this domain may have more than one orientation in the lattice. The R factors are ~32% when the constant region was excluded for refinement. Model building was performed iteratively in O and Coot and refinement in Phenix and Refmac55–58. The final model was refined with an Rwork of 26.1% and an Rfree of 28.9%. Analyzed by Procheck59, 89.7% of residues are in most favoured regions of the Ramachandran plot; 8.9%, in additional allowed regions; 1.3%, in generously allowed regions; and none in disallowed regions. All the structure figures are made in PyMOL60.
Supplementary Material
Acknowledgments
We thank Stephen Harrison, Hanqin Peng, Andrea Carfi, Antu Dey, Munir Alam and Barton Haynes for generous advice and assistance, the staff of the Northeastern Collaborative Access Team at Advanced Photon Source, Argonne National Laboratory for assistance with x-ray data collection. We acknowledge support from NIH grants GM083680 (to B.C.), AI084794 (to B.C. and Dan H. Barouch), AI36085 (to S.Z-P.) a Collaboration for AIDS Vaccine Discovery (CAVD) grant (to Barton F. Haynes) from the Bill and Melinda Gates Foundation, the Center for HIV/AIDS Vaccine Immunology (to Barton F. Haynes), and research funds from the Department of Veterans Affairs. J.C. is supported by a fellowship from the Ragon Institute of MGH, MIT and Harvard.
Footnotes
Accession codes
Coordinates and structure factors have been deposited in Protein Data Bank with accession code 3P30.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Allan JS, et al. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 4.Veronese FD, et al. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 5.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Advances in Virus Research. 2005;64:231–259. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 7.Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 8.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 12.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 14.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 15.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009 doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010 doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010 doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 21.Hioe CE, et al. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One. 2010;5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10:527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiegler G, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 24.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwick MB, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hioe CE, et al. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int Immunol. 1997;9:1281–1290. doi: 10.1093/intimm/9.9.1281. [DOI] [PubMed] [Google Scholar]
- 29.Holl V, et al. Nonneutralizing antibodies are able to inhibit human immunodeficiency virus type 1 replication in macrophages and immature dendritic cells. J Virol. 2006;80:6177–6181. doi: 10.1128/JVI.02625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci U S A. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. Effects of oligomerization on the epitopes of the human immunodeficiency virus type 1 envelope glycoproteins. Virology. 2000;267:220–228. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- 32.Pinter A, et al. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorny MK, Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74:6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan W, et al. Oligomer-specific conformations of the human immunodeficiency virus (HIV-1) gp41 envelope glycoprotein ectodomain recognized by human monoclonal antibodies. AIDS Res Hum Retroviruses. 2009;25:319–328. doi: 10.1089/aid.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson JD, et al. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol. 2007;81:4033–4043. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harbury PB, Kim PS, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 37.Aoki ST, et al. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science. 2009;324:1444–1447. doi: 10.1126/science.1170481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caffrey M, et al. Three-dimensional solution structure of the 44kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang ZN, et al. The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution. J Struct Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 40.Ofek G, et al. Structure and Mechanistic Analysis of the Anti-Human Immunodeficiency Virus Type 1 Antibody 2F5 in Complex with Its gp41 Epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 42.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009 doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 43.Nyambi PN, et al. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol. 2000;74:7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulke N, et al. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steger HK, Root MJ. Kinetic dependence to HIV-1 entry inhibition. J Biol Chem. 2006;281:25813–25821. doi: 10.1074/jbc.M601457200. [DOI] [PubMed] [Google Scholar]
- 47.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsmadi O, Tilley SA. Antibody-dependent cellular cytotoxicity directed against cells expressing human immunodeficiency virus type 1 envelope of primary or laboratory-adapted strains by human and chimpanzee monoclonal antibodies of different epitope specificities. J Virol. 1998;72:286–293. doi: 10.1128/jvi.72.1.286-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyler DS, et al. Identification of sites within gp41 that serve as targets for antibody-dependent cellular cytotoxicity by using human monoclonal antibodies. J Immunol. 1990;145:3276–3282. [PubMed] [Google Scholar]
- 50.Forthal DN, Landucci G, Gorny MK, Zolla-Pazner S, Robinson WE., Jr Functional activities of 20 human immunodeficiency virus type 1 (HIV-1)-specific human monoclonal antibodies. AIDS Res Hum Retroviruses. 1995;11:1095–1099. doi: 10.1089/aid.1995.11.1095. [DOI] [PubMed] [Google Scholar]
- 51.Pietzsch J, et al. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J Virol. 2010;84:5032–5042. doi: 10.1128/JVI.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 1997;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods in Enzymology. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 59.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 60.DeLano WL. The PyMOL User's Manual. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.