Abstract
Tandem affinity purification (TAP) coupled with mass spectrometry has become the technique of choice for characterization of multi-component protein complexes. While current TAP protocols routinely provide high yield and specificity for proteins expressed under physiologically relevant conditions, analytical figures of merit required for efficient and in-depth LC-MS analysis remain unresolved. Here we implement a multidimensional chromatography platform, based on two stages of reversed-phase separation operated at high and low pH, respectively. We compare performance metrics for RP-RP and SCX-RP for the analysis of complex peptide mixtures derived from cell lysate, as well as protein complexes purified via TAP. Our data reveal that RP-RP fractionation outperforms SCX-RP primarily due to increased peak capacity in the first dimension separation. We integrate this system with miniaturized LC assemblies to achieve true online fractionation at low (≤5nL/min) effluent flow rates. Stable isotope labeling is used to monitor the dynamics of the multi-component Ku protein complex in response to DNA damage induced by gamma radiation.
Keywords: tandem affinity purification, RP-RP, reversed phase-reversed phase, LC-MS, LC-MS/MS, 2D-LC, two dimensional separation, online fractionation, SCX-RP, mudpit, Orbitrap, Qstar, protein complex, DNA Damage
INTRODUCTION
Normal and aberrant biological phenotypes are driven by numerous interactions of proteins, nucleotides, and small molecule metabolites. As a result, there is growing appreciation that so-called systems level experimental strategies can augment traditional hypothesis-driven approaches for characterization of cellular response to stress, infection, disease, and other biological or environmental perturbations. 1–2 Data derived from large-scale protein characterization studies generally lag behind those for DNA and RNA, both in terms of throughput and comprehensiveness. Generally speaking the technical hurdles that impede progress towards true proteome-scale studies are based on the limited detection and dynamic range available on current mass spectrometry instruments. 3 Hence, faced with the difficulties inherent to the generation of a “complete” proteome catalog, many researchers have opted instead to pursue targeted analyses of protein sub-classes or specific post-translational modifications. Anecdotal evidence suggests that enriched biological materials used in focused studies nevertheless span a wide range of sample complexity, and can often yield a protein and peptide concentration range that exceeds the analytical capabilities of modern mass spectrometry technology. 4
Ongoing efforts to analyze complex biological samples have revealed other analytical deficiencies in LC-MS technologies. For example, it is now generally accepted, although admittedly difficult to quantify, that under typical electrospray conditions, a multitude of co-eluting peptides will compete for charge and effectively suppress the detected signal of low basicity species. 5 In addition, the duty cycle of MS/MS, while steadily improving, 6–7 is nonetheless often overwhelmed by the number of peptides presented simultaneously for analysis. Collectively these data and observations have catalyzed a plethora of work designed to improve overall peak capacity of peptide and protein separations, with the ultimate goal of reducing the number of analytes presented for MS/MS per unit time. Multidimensional fractionation has emerged as the most compelling means to facilitate in-depth analysis of complex peptide mixtures derived from biological extracts. 8 In this approach, two or more separation methods, each based upon different physiochemical properties, are used in tandem to obtain significantly improved peak capacity. To date, the combination of strong cation exchange (SCX) chromatography and reversed phase (RP) chromatography is the most widely used system. 9–12 Despite these successful studies, it is clear that, at the extreme, biological complexity often exceeds the peak capacity of SCX-RP. As a result, numerous other multidimensional separation strategies, including SAX-RP 13–15, gel-based isoelectric focusing coupled with RP 16–20, and CIEF-RP 21–24 have been utilized for shotgun proteomics. More recently Gilar et al. introduced a multidimensional fractionation platform based on two stages of reversed phase operated at high and low pH, respectively. 25 The RP-RP geometry provides high peak capacity in each dimension, while the change in pH between the first and second stages modulates peptide hydrophobicity through a switch in ion pairing at acidic (high buffer pH) and basic (low buffer pH) side chains, respectively. 26–29 Less well understood is the relative benefit of multidimensional fractionation for analysis of samples across the full spectrum of biological complexity (e.g., whole cell lysate, modified peptides, protein complexes, etc.). In principle the analysis of discrete, multi-component protein complexes represents an experimental paradigm that is well-matched to the analytical performance of mass spectrometry. For example, large-scale studies in yeast and human cell lines indicated that protein complexes consist of ~30 proteins on average, 30–34 a level of complexity very amenable to analysis by LC-MS/MS. Moreover, the growing repositories of affinity tag constructs, along with improvements in robotics for cloning, expression, and cell culture, provide a basis for systematic production of reagents suitable for large-scale proteomics studies. 35–39 Indeed, recent reports have described the development and application of new affinity reagents and associated methods designed to improve both yield and specificity of purified protein complexes. 40–45 However, the analytical figures of merit for subsequent LC-MS analysis remain relatively unexplored.
In this report, we compare SCX-RP and RP-RP multidimensional chromatography platforms for analysis of peptides derived from cell lysate and multi-component protein complexes isolated via TAP. A modified Waters 2-D NanoACQUITY system allows convenient use of either SCX or RP in the first dimension, and is readily interfaced with our recently described miniaturized LC electrospray assemblies, 46 to provide for true online, nanoflow (≤5nL/min) multi-dimensional fractionation. Comparison of these separation platforms for the analysis of cell lysate revealed that RP-RP outperformed SCX-RP due largely to improved first dimension peak capacity. We observed similar performance improvements for analysis of tandem affinity purified protein complexes. Finally, we utilize RP-RP in conjunction with iTRAQ-based quantification to monitor changes in protein membership of the multi-component Ku complex in response to DNA damage induced by gamma radiation.
EXPERIMENTAL PROCEDURES
Column Construction
The first dimension column (RP or SCX) consisted of 360 μm O.D. × 100 μm I.D. fused silica capillary tubing capped with 0.5 μm inlet frits (Upchurch, Oak Harbor, WA). Waters 3.5 μm Xbridge C18 silica resin and Sepax Technology (Newark, DE) SCX media were used to pack 10 cm beds for first dimension RP and SCX columns, respectively. Second dimension pre- (PC) and analytical columns were constructed as previously described. 46 Briefly, silicate-based frits were cast in situ within 360 μm O.D. × 100 μm I.D. fused silica capillary tubing. Next, dense slurry of POROS 10R2 resin was prepared in 1 mL of acetonitrile and pressure loaded behind the frit to a final bed length of 5 cm. The AC was constructed from 360 μm O.D. × 25 μm I.D. fused silica capillary tubing, and packed to a bed length of 12 cm with Waters Xbridge 3.5μm dia. silica resin. A laser-based pipette puller (P-2000, Sutter Instruments, Novato, CA) was used to generate an integrated electrospray emitter tip of ≈1 μm dia. at the end of the capillary, ≈2mm from the bed frit.
Our 2D RP-RP platform was based on a Waters NanoACQUITY UHPLC system, equipped with a third, 6-port, 2-position valve (VICI Valco, Houston, TX). The autosampler was used to (i) load samples and (ii) inject first dimension elution buffers, consisting of varying concentrations of either ammonium formate (SCX) or acetonitrile (RP). Discrete first dimension elutions were performed at 1 μL/min with 20mM ammonium formate in 3% acetonitrile or 0.1% formic acid in 10% acetonitrile, for RP-RP and SCX-RP, respectively (Detailed concentrations are listed in Supplementary Table 1.) True nanoflow rates in the second dimension were achieved through use of a passive split located prior to the second dimension PC; the split line was configured in a vented column geometry. 47–48 Flow is blocked in the split line during first dimension elution to ensure that peptides are directed to the second dimension PC. The valve then switches to engage the passive split such that a majority of LC effluent is diverted to waste prior to the second dimension PC.
Cell Culture and Gamma Radiation
HeLa S3 cells that express tandem FLAG and HA peptide epitope tagged Ku86 (FLAG-HA KU86) were grown in BD Falcon 15-cm diameter tissue culture dishes, each containing 25 mL DMEM (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum. Cells were incubated for three days in a 37°C, 5.0% CO2, after which they were supplemented with an additional 3 mL of media, and then incubated again for 24 hrs. A MDS Nordion Gammacell 40 Exactor, equipped with two 137Cs sources, was used for γ-radiation of the HeLa S3 cells. In total twelve, 15 cm dishes of confluent cells were irradiated with 10 Gy, at a central dose rate of 1.1 Gy/min. Cells were then placed back into the incubator, and then removed in groups of 4 plates after 30 minutes, 2 hours, and 5 hours, respectively. Under these conditions we did not observe loss of cell viability or signs of apoptosis. One group of 4 plates (the control) was processed in parallel but not exposed to γ-radiation.
Nuclear Preparation
All purifications were performed from nuclear extracts, prepared as follows: Cells were transferred to 50 mL conical tubes (2 plates/tube). Next, the cells were pelleted at 500×g for 3 minutes at 4°C, in a refrigerated centrifuge (Eppendorf, Hamburg, Germany). The supernatant was discarded, and the cell pellets were re-suspended in 50 mL of 1× PBS solution at 4°C, (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4) and then centrifuged again at 500×g for 3 minutes at 4°C.
The four-plate equivalents of cells were combined, suspended in 10 mL 1× PBS, and centrifuged at 500×g for 3 minutes at 4°C. The supernatant was discarded and the pellet was suspended in ten times the pellet volume (typically 700 μL for four plates of HeLa S3 cells) of hypotonic buffer (10 mM Tris pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 10 mM β-mercaptoethanol). The suspension was centrifuged at 800×g for 3 minutes at 4°C. The supernatant was discarded and the pellet was suspended in 1 mL of hypotonic buffer and transferred to a 7 mL Dounce tissue homogenizer. Twelve strokes were used to disrupt cells, and then the homogenized lysate was divided equally between two 1.5 mL microcentrifuge tubes. The cell lysates were then centrifuged at 1000×g for 4 minutes at 4°C, yielding a pellet of nuclei with cytoplasm in the supernatant.
After centrifugation, the supernatants were transferred to separate 1.5 mL microcentrifuge tubes. Each nuclear pellet was then re-suspended in 1 mL of hypotonic buffer, and centrifuged at 1000×g for 10 minutes at 4°C. The supernatants were discarded, and remaining nuclei were flash frozen with liquid nitrogen and stored at −80°C.
Tandem Affinity Purification of FLAG-HA tagged Ku86 Complex
Frozen nuclear pellets were re- suspended in 2.5× the pellet volume of lysis buffer (30 mM NaCl, 10% glycerol, 50 mM Tris pH 7.5, 0.2% NP-40, 1 mM EDTA, Roche EDTA-free protease inhibitor cocktail). Solutions were placed on a rotisserie platform (Barnstead-Thermolyne, CA, USA) at 4°C for 30 minutes. The resultant nuclear lysates were centrifuged at 14,000×g for 10 minutes at 4°C. The supernatants were transferred to fresh tubes and remaining pellets were discarded. Supernatants for each time point (0, 30 minutes, 2 hours, and 5 hours) were combined. Agarose beads (80 μL) conjugated with Sigma M2 mouse antiflag antibody were washed three times in 800 μL of lysis buffer and then re-suspended in 80 μL of lysis buffer. Aliquots of bead suspension (25 μL) were added to each of the four nuclei samples (0, 30 minutes, 2 hours, and 5 hours) and the sample tubes were incubated on the rotisserie platform at 4°C for 4 hours.
Beads were washed three times with 200 μL lysis buffer and then once with 200 μL HA buffer (30 mM NaCl, 10% glycerol, 50 mM Tris pH 7.5, 0.05% NP-40, 1 mM EDTA, Roche EDTA-free protease inhibitor cocktail). Ku86 protein complexes were specifically eluted by addition of 18.5 μL HA buffer with 1.5 μL concentrated FLAG peptide (2 mM), followed by vortex at 1200 rpm for 20 minutes at 4°C. FLAG elution was repeated one time; the elutents were combined and filtered through a 0.45 micron PVDF membrane (Millipore, Billerica, MA) at 14,000×g for 30 seconds at 4°C.
Next, 80 μL HA conjugated agarose beads (Santa Cruz Biotechnology) were washed three times in 800 μL HA buffer and re-suspended in 80 μL HA buffer. Aliquots of HA bead suspension (25 μL) were added to each FLAG elution, followed by gentle vortex at 1200 rpm overnight at 4°C. Each sample was specifically eluted two times with 18.5 μL HA buffer and 1.5 μL concentrated HA peptide (2 mM) at 1200 rpm and 4°C. Eluents were filtered as described above and stored at −80°C.
Preparation of iTRAQ labeled Peptides
Frozen protein complex eluates for each of the four time points (four-plate equivalents per time points), were denatured and reduced by addition of 0.1 weight percent Rapigest SM (Waters Corporation, Milford, MA) and 5 mM dithiothreitol (Sigma). The solutions were incubated at 60°C for 30 minutes. After equilibration to room temperature, the denatured protein solutions were then digested with 750 ng sequencing grade trypsin (Promega) at 37°C in a CO2 incubator for 24 hours.
After digestion, the Rapigest was cleaved by addition of 0.5% v/v trifluoroacetic acid. The samples were then incubated at 37°C for 30 minutes in a CO2 incubator. The Rapigest was pelleted by centrifugation (14,000 ×g) at 4°C for 20 minutes. The supernatants were removed and desalted on a reversed phase microelution 96-well plate. Each well was prepared by rinsing once with 400 μL 40% acetonitrile/0.1% TFA, followed by two 400 μL washes with 0.1% TFA. The peptide solution was loaded onto the prepared well, washed twice with 400 μL 0.1% TFA, and eluted with two sequential aliquots of 50 μL 40% acetonitrile/0.1% TFA.
The samples were dried by centrifugal concentration and reconstituted in 14 μL 0.5 M triethylammonium bicarbonate pH 8.5 buffer. Simultaneously, a 40 μL aliquot of Activated Thiol Sepharose 4B beads (GE Healthcare) was prepared by washing approximately 20 μL dry beads five times with 1 mL aliquots of ultrapure, dionized water. The beads were then washed one time with 200 μL 0.5 M TEAB and reconstituted in 40 μL 0.5 M TEAB. From this stock solution of beads, 14 μL were added to each of the four peptide samples and vortexed at 1,400 rpm for one hour at room temperature.
After centrifugation, the supernatants were transferred to fresh tube, and the beads were discarded. At this stage, four channels of iTRAQ reagent [114, 115, 116 and 117] were each reconstituted in 70 μL ethanol yielding a total volume of 90 μL for each channel. To each of the four samples, 40 μL ethanol and 36 μL reconstituted iTRAQ reagent were added using the following scheme: 114 – 0 minutes, 115 – 30 minutes, 116 – 120 minutes, 117 – 300 minutes. The solutions were then incubated for one hour at room temperature.
Sample volumes were reduced to 30 μL each by centrifugal concentration, and then further diluted by addition of 100 μL 0.1% TFA. The four samples were combined and desalted on a reversed phase microelution plate as described above. The combined sample was dried by centrifugal concentration and reconstituted in 65 μL 20 mM ammonium formate pH 10. The sample was then divided into 10 μL aliquots, corresponding to two-plate equivalents of starting cell material, and stored at −70°C until LC-MS.
MS/MS analysis
Both LCQ Decca XP (ThermoFisher Scientific, Waltham, MA) and QSTAR Elite (ABI-SCIEX, Toronoto, Ontario) were used for LC-MS acqusition. The QSTAR Elite was operated in information dependent acquisition (IDA) mode with the following parameters: MS acquisition time = 1 sec.; MS/MS accumulation = 2.5 sec with Smart IDA activated; fragment intensity multiplier value = 5. The five most intense precursors with charge state range of +2 to +4 were selected for CAD with the precursor selection threshold set to 50 counts; selected precursors were excluded for 30 sec. The LCQ Decca XP was operated in data dependent mode with the following parameters: normalized collision energy = 35%; One full-scan mass spectrum was followed by three Data Dependent MS/MS spectra of the three most intense precursor ions. The dynamic exclusion was enabled (Repeat Counts: 2, Repeat Duration: 0.2 minutes, Exclusion Duration: 5 minutes and Exclusion Mass width: 2 Da).
Data processing
MS/MS spectra were searched against a NCBI RefSeq database (released on 04/03/2006) for E.coli. K12 and Uniprot human datadase (release 14.0) for Ku86 complexes using ProteinPilot, v3.0 (Applied Biosystems, Foster City, CA). Peak lists were generated from. RAW and. WIFF files using Mascot Deamon and Protein Pilot, respectively. The number of database entries for E.coli. was 4149 and the number of entries for Uniprot human was 20331. The search parameters were set as follows: Instrument selection was set to “LTQ” for E.coli., and “QSTAR Elite” for analysis of Ku associated proteins. Data were searched in Protein Pilot using the “Thorough” parameter setting. Only those peptide sequences that passed a 1% FDR threshold (based on embedded Protein Pilot “Global FDR” calculation) were considered for the data reported herein. Protein quantification output from Protein Pilot (ratios normalized to 114) was further calibrated according to Ku86 in each iTRAQ channel. Supplementary Tables (see Results) provide complete lists of protein assignments based on parsimonious use of peptide sequences. Further analyses of peptide distributions across RP-RP and SCX-RP fractionation techniques were performed in our multiplierz desktop software environment. 49–50
Permutation test
The frequency of occurrence for amino acids in peptide sequences identified by RP-RP and SCX-RP, respectively, was analyzed by counting the number of each amino acid and then dividing by the total number of residues in each dataset. These data were plotted for each amino acid as the log ratio (base 2) of the frequency of occurrence for amino acid composition in peptide sequences detected in RP-RP and SCX-RP. The significance of each ratio was calculated by combining the peptides detected in RP-RP and SCX-RP and then randomly permuting the assignments such that the number of sequences assigned as (RP-RP) and (SCX-RP) remained constant. After each permutation, the amino acid percent compositions and their log ratios were recalculated as described above, for a total of 10,000 permutations. One-sided p-values were calculated by summing the frequency (probability) of all log ratios equal to or more extreme than the observed log ratios; these values were doubled to yield two-sided p-values. Finally, two-sided p-values were Bonferroni corrected based on the number of amino acids. Frequency of occurrence was considered significant for p-val ≤ 0.01.
Western Blotting
Gels used for Western blot were prepared fresh and consisted of a 10% acrylamide running gel and a 4% acrylamide stacking gel. Each gel was run in a Mini Protean III Cell (BioRad) using SDS electrophoresis buffer (25 mM Tris, 192 mM Glycine, 34.7 mM SDS). The 2 μL aliquots taken from the denatured silver stain aliquots (for each time step) were loaded onto the gel, along with aliquots of Benchmark Protein Ladder solution (Invitrogen), and the gel was run at 150 V (constant voltage mode) with a 40 mA current limit for 60 minutes. Proteins were transferred to a PVDF membrane using a semi-dry apparatus operated at 12 V (constant voltage mode) with a 300 mA current limit for 50 minutes.
After transfer, the membrane was removed and incubated in Ponceau S solution for two minutes and then rinsed twice in water for one minute. Next, the membrane was incubated in a milk solution (2% w/v powdered milk/1× PBS/0.04% Tween 20 v/v) for 30 minutes on an orbital shaker. After blocking, the membrane was incubated overnight in HRP-conjugated α-flag peptide antibody diluted 1:1000 v/v in milk solution on a rotisserie platform at 4°C.
After overnight incubation, the membrane was washed (4×) with 1× PBS/0.04% Tween 20 v/v for 5 minutes on an orbital shaker, with the membrane flipped after each wash. Blots were developed with the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL), and imaged in a luminescence analyzer (Fujifild, LAS-4000). Quantitation of band intensities was performed with Fujifilm Multi Gauge v3.1 software. Load amounts for each gel lane were determined by normalizing the amount of Ku per sample, as measured by the detection of the FLAG epitope.
Validation immunoblots for Ku complex proteins, DDX21, HNRNPC, and RCC2, were performed as described above with the following modifications. Primary stock antibody solutions were diluted 1:2000 v/v (Rabbit α-DDX21, Bethyl Laboratories, Inc.), 1:2000 (Rabbit α-RCC2, Bethyl Laboratories, Inc.), and 1:1000 (Mouse monoclonal α-hnRNP C1/C2, Abcam Inc.). Blots were developed with the SuperSignal West Femto Maximum Sensitivity Substrate kit (Pierce, Rockford, IL), with band imaging and quantification as described above.
Silver Staining
Aliquots, representing approximately 20% of each sample, were reduced and denatured in 4× NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA) with 9% 2-mercaptoethanol at 70°C for 10 minutes. Each was analyzed by anti-FLAG western blot to normalize total protein load for each gel lane. Appropriate volumes of TAP eluted proteins were loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen, Carlsbad, CA). Proteins were separated at 200V (constant voltage mode) for 50 minutes.
After protein separation, the gel was incubated in a 125 mL solution of 40% ethanol v/v and 10% acetic acid v/v for 30 minutes on an orbital shaker. A second 125 mL solution of 40% ethanol/10% acetic acid was added to the gel and it was incubated overnight. After overnight incubation, the gel was incubated in a 125 mL solution of 8 mM sodium thiosulphate/0.83 M sodium acetate/30 % ethanol v/v for 30 minutes. Next, the gel was washed 3× in 250 mL ultrapure deionized water for five minutes. Finally, the gel was then incubated in 125 mL of 15 mM silver nitrate for twenty minutes and washed 2× in 250 mL of water for one minute each. The gel was developed by incubation in 125 mL of 0.236 M sodium carbonate/.0074% formaldehyde w/v for 20 minutes to yield sufficient band intensity. The reaction was quenched by incubation in 125 mL of 40 mM EDTA for 10 minutes, followed by two washes in 250 mL of ultrapure deionized water for 30 minutes each. Gels were imaged with a Fujifilm LAS-1000 CCD camera.
RESULTS AND DISCUSSION
2D RP-RP provides for improved peptide and protein identification from complex cell lysates as compared to 2D SCX-RP
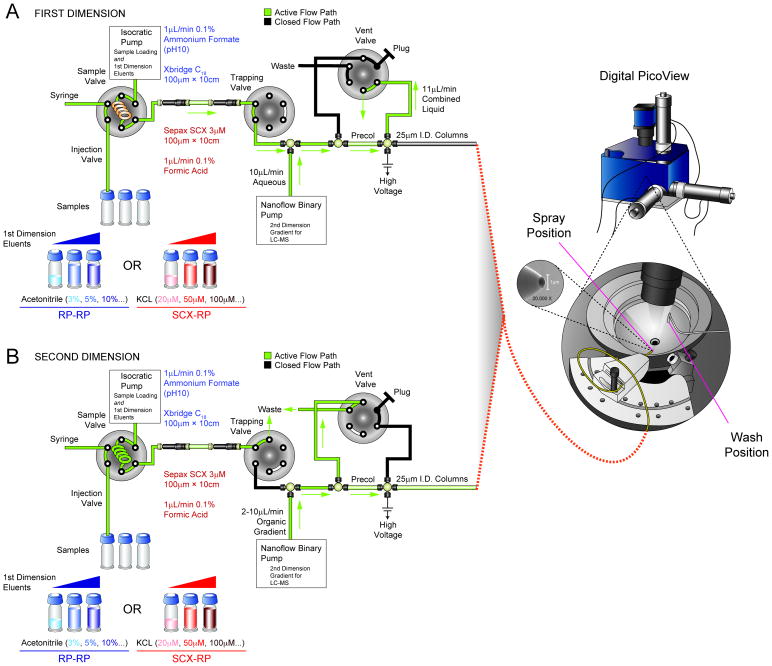
We recently described the fabrication of miniaturized LC-electrospray assemblies that provided significantly improved analytical figures of merit in the context of proteomics-based analyses of tyrosine signaling in embryonic stem cells. 46 The primary challenge in this application was detection of a rare post-translational modification (tyrosine phosphorylation) in primary cells that exhibited relatively low levels of signaling activity. As an extension of these findings, we next asked whether our 25 μm I.D. nanoflow columns could be integrated within an automated, multidimension fractionation platform for analysis of higher complexity biological extracts. We chose RP-RP as our target implementation based on recent reports that suggested the potential for improved performance as compared to other multidimensional fractionation strategies. 25 Figure 1 shows a schematic representation of our 2-D nanoflow system (see experimental section). Briefly, our platform is based on a Waters NanoACQUITY UHPLC modified to accept self-packed columns 46 and reconfigured with an additional two-position, 6-port valve to enable a pre-column effluent split, in a vented column configuration. 47,48 An on-board pump was used in isocratic mode to introduce samples or first dimension eluents loaded from autosampler vials, while a second UHPLC binary pump generated the solvent gradient for LC-MS/MS in the usual manner. This configuration facilitated rapid switching between RP and SCX columns for the first dimension separation, without the need to prepare large quantities of fresh first dimension solvents. Unattended acquisition of RP/SCX-RP MS/MS data was facilitated by use of a computer-controlled electrospray positioning device (Digital PicoView, New Objective, Woburn, MA) that automatically moved the emitter tip beneath a wash station (see Figure 1) for sample loading in-between fractions. This eliminated build-up of salts or other residue on the outer surface of the emitter, that would otherwise degrade performance. Under these conditions and for the sample types analyzed herein, we typically achieved 2–3 months of continuous operation with no observable loss in first- or second-dimension column performance.
Figure 1.
Schematic diagram of automated, online nanoflow RP-RP platform. (A) The autosampler loads peptides and then is used to inject salt or organic eluents to generate first dimension SCX (red) or RP (blue) fractions, respectively. First dimension fractions are diluted (10×) and acidified with reversed phase A solvent (0.1% formic acid, 3% acetonitrile) introduced by an ultra-high pressure binary pump. An additional 6-port, 2-position valve provides a vented second dimension column configuration, and ensures efficient capture of peptides onto the precolumn. (B) The ultra-high pressure binary pump delivers an organic gradient to elute peptides from the second dimension column for MS/MS analysis. The vent valve generates an effluent split of ≈1000:1 to provide a stable column/ESI flow rate of ≈5 nL/min. Active solvent flow paths are highlighted in green. (right) A computer-controlled positioning platform (Digital PicoView) automatically moves the emitter tip beneath a wash station for sample loading in-between fractions.
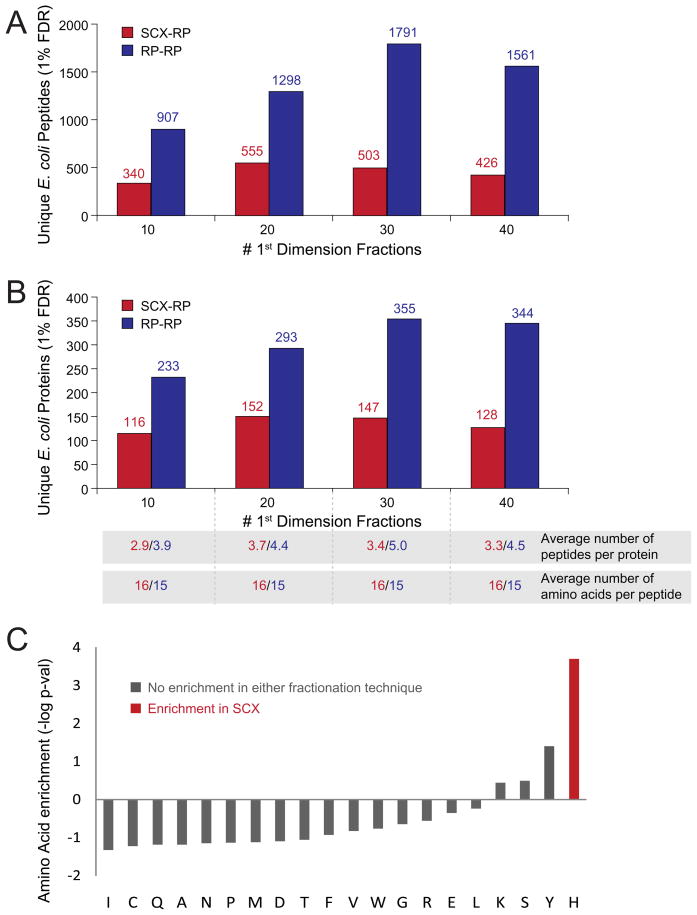
As an initial evaluation for analysis of complex mixtures we chose a commercially prepared tryptic digest of E.coli lysate (see experimental section). First, we performed pilot studies to determine first dimension salt (SCX) and organic (RP) eluent concentrations that provided for uniform distribution of peptide identifications across a complete 2-D LC-MS/MS analysis (data not shown). Next, 200 ng of tryptic peptides derived from E.coli were analyzed by either SCX-RP or RP-RP, using identical mass spectrometry acquisition methods. RP-RP provided for a higher number of unique peptide (Figure 2a) and protein (Figure 2b) identifications across the full range of fractionation conditions analyzed, as compared to SCX-RP (Supplementary Tables 2 and 3 provide a full list of peptide and protein identifications). Consistent with these data, we also observed that RP-RP provided for identification of more peptides per protein (Figure 2b).
Figure 2.
Analysis of tryptic peptides derived from e.coli lysate by SCX-RP (red) and RP-RP (blue) fractionation. RP-RP provided for higher numbers of identified (A) peptides and (B) proteins as compared to SCX-RP for independent experiments spanning 10, 20, 30, and 40 first dimension fractions, respectively. The number of peptides identified per protein was slightly higher for RP-RP, while peptide length was on average shorter by one amino acid (B, bottom). A permutation test (C), revealed that peptides identified by SCX-RP exhibited a statistical bias for histidine (-log[p-val]>2) residues as compared to those identified by RP-RP.
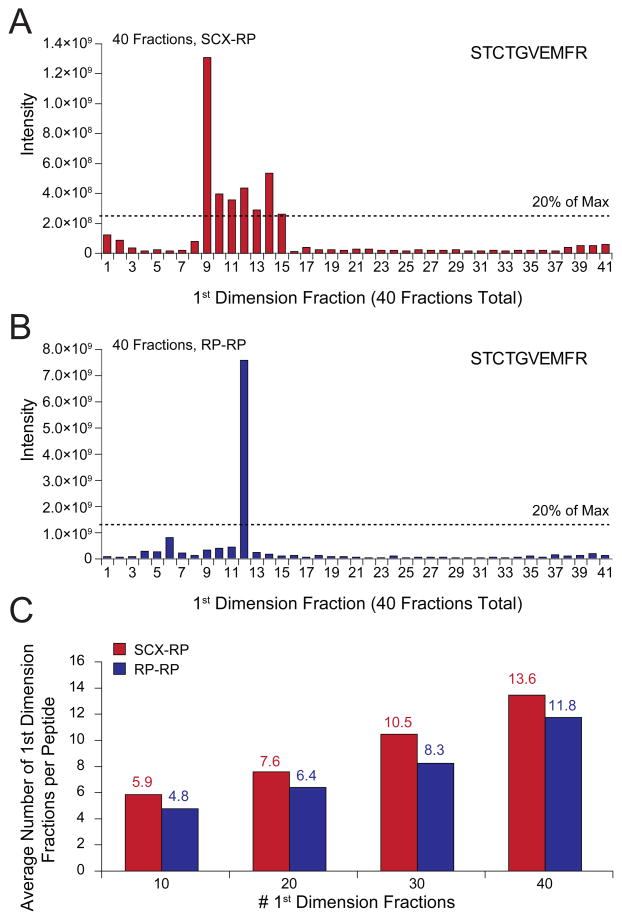
Given the different retention mechanisms of reversed phase and strong cation exchange resins, we next asked if peptides identified by either SCX-RP or RP-RP differed in their respective physio-chemical properties. For example, we observed that the average length of peptides identified remained very consistent at 16 and 15 amino acid residues for SCX-RP and RP-RP, respectively, across all conditions analyzed (Figure 2b). Next we applied a permutation test 51 to decipher any bias present in the amino acid content of peptides identified by each fractionation technique. Figure 2c demonstrates that only histidine was enriched in peptides identified with SCX-RP, while no statistically significant bias was observed for the amino acid content of peptides identified by RP-RP. Data associated with the permutation test is provided in Supplementary Table 4. Collectively the data above suggest that RP-RP provided for an overall increase in peptide identification, rather than improved performance for a particular sub-class of peptides that otherwise would go undetected. These results are consistent with previous work by Dowell et al. 27 in which no significant differences in the physiochemical properties of proteins were observed across various multi-dimensional peptide fractionation techniques. To further elucidate the performance discrepancy between RP-RP and SCX-RP we next analyzed first dimension (SCX and RP) peak capacities for the data in Figure 2. The use of peptide identification to verify the presence of a given peptide in consecutive first dimension fractions was complicated by the stochastic nature of MS/MS. In an effort to mitigate this effect we used a combination of precursor mass tolerance (±1 Da) and second dimension LC elution time (±1 minute) to search for each peptide in first dimension fractions adjacent to that in which the original identification occurred. Precursors within the mass tolerance window, but below 20% relative abundance as compared to the most intense instance of the identified peptide, were not considered (see experimental section). Figure 3a and 3b shows representative data for the peptide, STCTGVEMFR dentified by both techniques, with 40 first dimension fractions. Precursors putatively matching the identified peptide spanned 7 first dimension fractions in SCX-RP, but eluted in only 1 fraction in the RP-RP analysis. Accordingly, the maximum precursor signal intensity observed for STCTGVEMFR was significantly higher for RP-RP as compared to SCX-RP. Overall RP-RP consistently demonstrated superior peak capacity as evidenced by the average number of first dimension fractions spanned by all identified peptides (Figure 3c).
Figure 3.
MS precursor peak intensity (mass tolerance ±1.0 Da, intensity threshold = 20% of max.) for the peptide STCTGVEMFR plotted across first dimension (A) SCX and (B) RP fractions, respectively. (C) Peptides consistently eluted across fewer first dimension fractions in RP-RP (blue) as compared to SCX-RP (red) for independent experiments spanning 10, 20, 30, and 40 first dimension fractions, respectively.
In separate experiments, we used a data analysis approach similar to that described above to explore the reproducibility of RP-RP fractionation. From duplicate, 10 fraction RP-RP experiments (as in Figure 2a, left column), and a stringent threshold that required a peptide identification in both 2-D LC-MS/MS analyses to qualify as “reproduced,” we observed that 78% of all peptides were found in the same first dimension fraction across replicate analyses. As was noted above, the stochastic nature of MS/MS likely makes this result the lower bound of reproducibility. In fact, the use of more permissive criteria as described above (second dimension retention time window ±1 minute and precursor mass tolerance of ±1 Da) demonstrated that potentially 95% of all identified peptides appeared in the same fraction in replicate 2-D RP-RP analyses (data not shown).
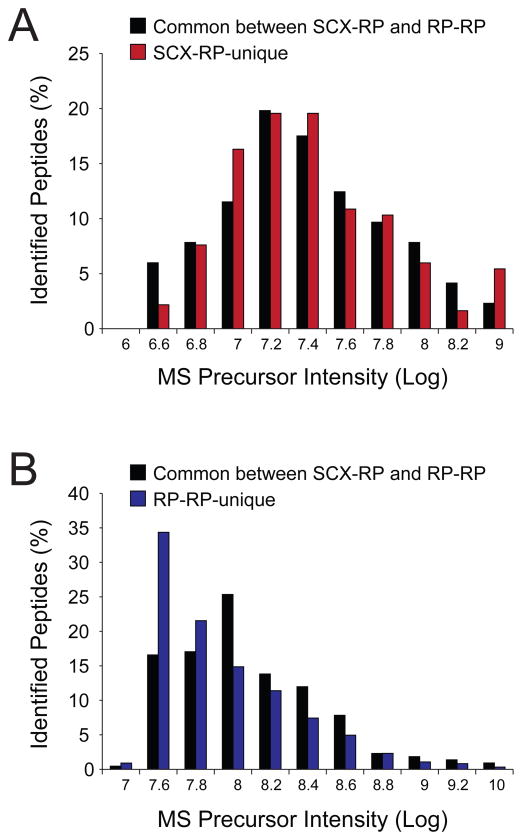
We next asked whether the improved peak capacity of 2-D RP-RP (Figure 3a and 3b) would provide for identification of peptides over a wider dynamic range as compared to 2-D SCX-RP. To address this question we compared precursor intensities of peptides identified in common between SCX-RP and RP-RP as well as those sequences uniquely associated with each fractionation technique. Figure 4 shows histogram plots for the precursor intensities of the commonly detected peptides (black bars) and those identified uniquely by SCX-RP (red bars) and RP-RP (blue bars), respectively. Consistent with the data in Figures 2 and 3, we observed that the distributions of unique- and commonly-identified peptides overlapped in the case of SCX-RP (Figure 4a) while those peptides uniquely identified by RP-RP (Figure 4b, blue bars) systematically clustered towards the low end of the abundance range as compared to peptides identified by both techniques (Figure 4b, black bars).
Figure 4.
Histogram plots of MS precursor intensities for peptides detected by both fractionation techniques (black bars) and for those peptides identified uniquely by SCX-RP (A, red bars) and RP-RP (B, blue bars), respectively.
Collectively our data suggest that RP-RP provides a reproducible platform for online peptide fractionation that outperforms SCX-RP in terms of total number, and dynamic range, of peptide identifications, primarily due to increased peak capacity in the first dimension.
2D fractionation for proteomic characterization of nuclear multi-component Ku complexes
As described above there is growing appreciation that control of cellular physiology occurs through a delicate balance of protein post-translational modification and protein-protein interactions. As a result, numerous targeted and large-scale studies have attempted to decipher the dynamics of protein complexes in the context of normal physiology, and sought to understand how disruption of protein-protein interactions contributes to human disease. 4, 30 For example, the Ku proteins are involved in a number of cellular functions, including telomere maintenance, transcriptional regulation, apoptosis and antigen-receptor gene rearrangement. In particular the Ku proteins are essential components of the response to DNA damage. 52 Consistent with the involvement of Ku in multiple pathways, a recent mass spectrometry-based study identified some 90 protein groups in association with Ku, immunoaffinity purified from HEK 293 whole cell lysate.45 Moreover, several reports have described how disruption of Ku protein complexes contributes to aberrations in multiple cellular processes. 53
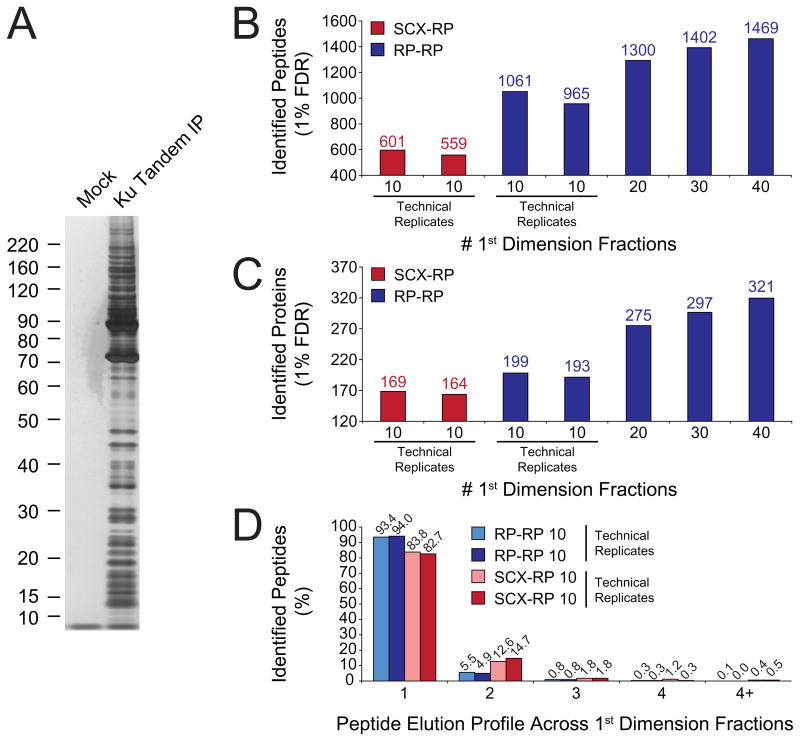
Based on the apparent molecular complexity and functional diversity of Ku, we asked whether proteomics analysis based on RP-RP fractionation would provide additional molecular resolution for the Ku complex. Towards this end we performed tandem affinity purification of FLAG-HA Ku86 from the nuclear fraction of FLAG-HA Ku86 HeLa S3 cells. As a negative control, TAP was performed in parallel from parental HeLa S3 cells. After elution of the complex with HA peptide, a small aliquot corresponding to approximately 20% of the total eluate was resolved by SDS-PAGE and visualized with silver stain. Figure 5a shows that the Ku complex consists of a very large number of proteins spanning a wide range of stoichiometry and molecular weight. The absence of detected proteins in the control lane suggested that the majority of protein bands resolved by SDS-PAGE represented specific protein interactions within the Ku complex.
Figure 5.
Analysis of tandem affinity purified FLAG-HA Ku86. (A) SDS-PAGE followed by silver stain revealed that FLAG-HA Ku86 interacts specifically (compare Mock and Ku Tandem IP lanes) with a diverse set of proteins that span a wide molecular weight range. (B and C) LC-MS/MS analysis of technical replicates, 10 fractions each, demonstrated that RP-RP (blue bars) identified a higher number of peptides (B) and proteins (C) as compared to SCX-RP (red bars). Additional RP-RP fractionation (B and C – 20, 30, 40 first dimension fractions) provided a continual increase in peptide (B) and protein (C) coverage for the Ku86 multi-component complex. (D) Peptide elution profile across first dimension fractions illustrates that RP-RP provides improved peak capacity as compared to SCX-RP.
Next, we performed replicate LC-MS/MS analyses for both SCX-RP and RP-RP, collecting 10 first dimension fractions for each. Figure 5b and 5c show that, although each technique provided reproducible fractionation, RP-RP provided for detection of significantly more unique peptides and proteins as compared to SCX-RP (Supplementary Table 5 provides information for all peptide and protein identifications). Consistent with the data in Figures 2 and 3, we observed that ≈ 94% of all peptides detected by RP-RP were constrained within a single, first dimension fraction (Figure 5d), while in the corresponding SCX-RP experiment, only ≈ 83% of identified peptides met this criteria. Given these data we next asked whether further RP-RP fractionation depth would provide for identification of additional proteins in the Ku complex. In subsequent RP-RP experiments, equal aliquots of Ku complex were analyzed by acquisition of 20, 30, and 40 first dimension fractions, respectively. Figure 5b and 5c, show that in fact we observed a continual increase in the number of peptide and protein identifications as a function of fractionation depth, again confirming the high peak capacity afforded by RP-RP fractionation.
In the context of these experiments, we observed many proteins previously identified as partners of Ku 70/86, including: Werner syndrome associated protein (WRN) the poly(ADP-ribose) polymerase I (PARP-1), YY1, the RNA-dependent Helicase A/Nuclear DNA-dependent Helicase II/(RHA/NDH-II/DDX9), and several heterogeneous nuclear proteins (hnRNPs). 45 Collectively, our results add significantly to the repertoire of known Ku complex members. (see Supplementary table 5)
As we intended to use iTRAQ-based quantification in the course of these studies, we next explored the potential for reporter ion scrambling due to co-elution of peptides with similar mass-to-charge ratios. Using an approach similar to that described in Figure 3, we queried all MS scans preceding each MS/MS event that yielded a peptide identification, and recorded the number of other peptide precursors that appeared within a m/z range of −0.5 to +1.0 Da with respect to the identified peptide. As before, co-eluting species were scored as potential interferents when their precursor intensity exceeded 20% relative to that of the peptide of interest. By this relatively crude measure, we observed potential contaminants associated with 12.7% of all peptides identified in the 10 fraction SCX-RP experiment. In the corresponding 10 fraction RP-RP analysis, only 7.4% of identified peptides co-eluted with a potential interfering species. Consistent with our observations for improved peak capacity associated with RP-RP, the potential precursor contamination rate dropped to 2.4% at the highest fractionation depth (data not shown).
Ku multi-component protein complex is dynamically remodeled in response to gamma radiation
While Ku participates in many cellular processes it is primarily associated with the DNA damage response. 54 An emerging body of work has revealed that Ku along with several protein interaction partners, including SSRP1, WRN, and nucleolin, re-localize within the nucleus or are otherwise activated, via post-translational modification for example, in response to genotoxic stress. Moreover recent reports suggest that Ku/DNA-PK and SSRP1 55 may also play a role in sensitizing tumor cells to chemotherapeutic agents. 18, 56
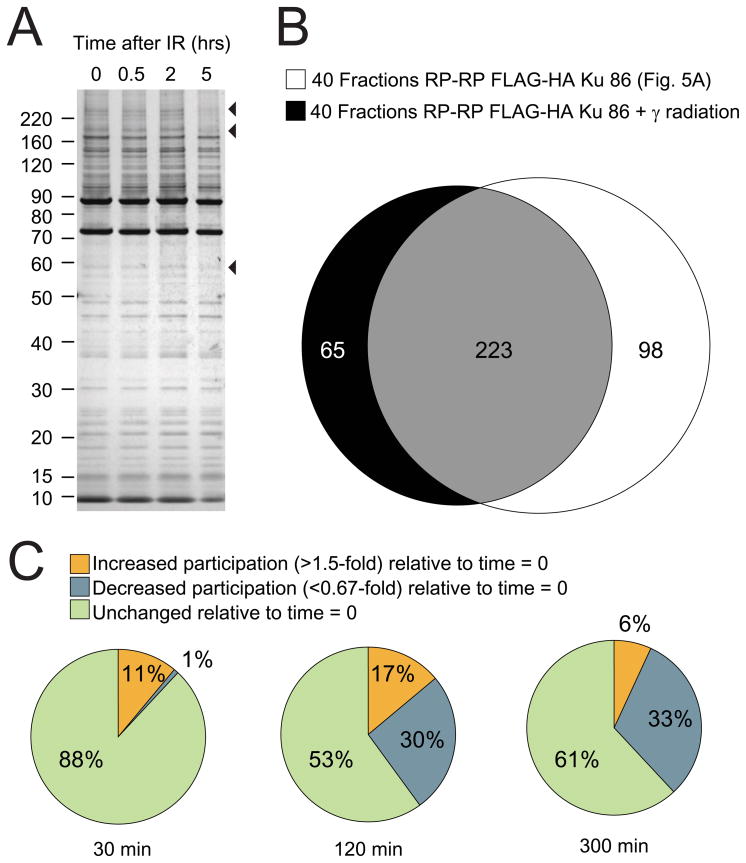
Given these intriguing results and the underlying complexity of DNA repair pathways 53, we next asked whether RP-RP fractionation combined with stable isotope labeling would provide sufficient peak capacity to decipher the molecular dynamics of Ku protein complexes in response to DNA damage. Towards this end we used a model system in which HeLa S3 cells that expressed FLAG-HA Ku86 were subjected to gamma radiation to induce DNA damage. Tandem affinity purification was performed at varying time intervals after treatment, and a small aliquot of each HA eluate was resolved on SDS-PAGE. Comparison of the gel lanes (Figure 6a) revealed that changes in protein membership of the Ku complex under these conditions were subtle at best. Next, the remaining portions of each TAP elution were digested with trypsin, labeled with iTRAQ reagents, and analyzed by 40 fraction RP-RP MS/MS as in Figure 5.
Figure 6.
The FLAG-HA Ku86 multi-component complex is remodeled in response to DNA damage. HeLa S3 cells that expressed FLAG-HA Ku86 were subjected to gamma radiation, with complexes purified at 0, 0.5, 2, and 5 hours after exposure. (A) Aliquots of eluted proteins were resolved by SDS-PAGE and visualized by silver stain; arrows indicate molecular weight regions at which subtle changes in complex membership were observed. (B) Comparison of proteins identified in TAP-purified FLAG-HA Ku86 complexes from gamma-treated HeLa S3 cells with those found in the native complex (Figure 5, 40 frxn) revealed ≈80% reproducibility for RP-RP analysis. Analysis of iTRAQ ratios (C) for proteins identified in the context of gamma radiation indicated that protein membership in the complex decreased by ≈30% (relative to the native complex) over a time course of 6 hours after gamma irradiation.
ProteinPilot 57 was used to facilitate analysis (e.g., identification and quantification) of all identified peptides and proteins. In total we detected and quantified 274 proteins within the Ku complex at time points, 0, 0.5, 2, and 5 hours, relative to treatment with gamma radiation. We observed several proteins known to interact with Ku including DNA-PKcs, WRN, NCL, 45 and H2AX. 58 Encouragingly, the overlap in protein identifications between this experiment and our analysis of the basal complex (Figure 5, 40 RP-RP fractions) was 82%, again illustrating the reproducibility of TAP and RP-RP fractionation (Figure 6b). To better understand global changes in Ku complex membership, we next normalized all peptide ratios to those of FLAG-HA Ku as an internal control, and then plotted the fraction of protein partners whose membership in the complex was modulated across the time course analyzed. Figure 6c shows that protein membership in the complex generally decreased as a result of DNA damage.
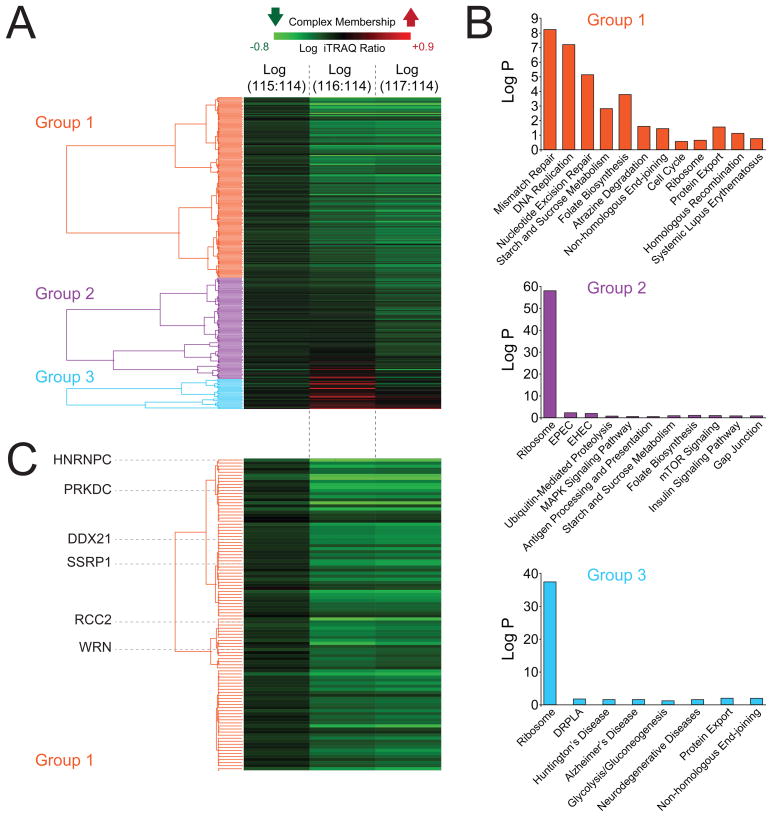
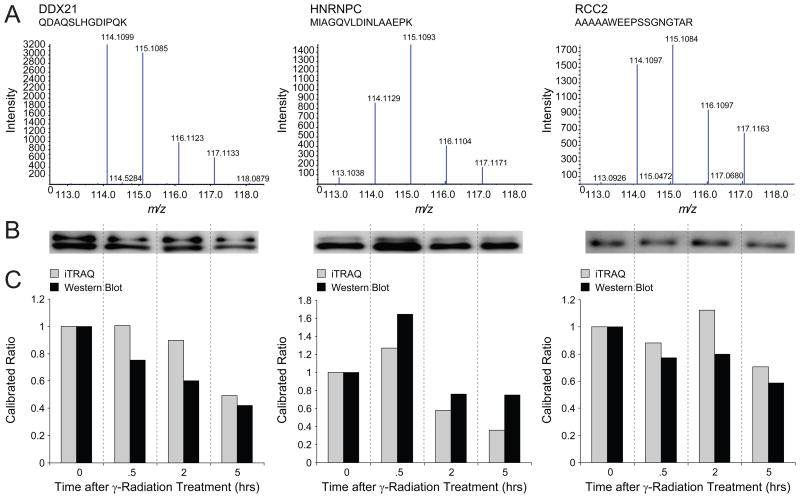
Given the large number of detected proteins, we next applied hierarchical clustering to group proteins based on their coordinate response to gamma radiation. Figure 7a shows that protein members of the Ku complex cluster into 3 primary groups (see also Supplementary Table 6); further analysis based on KEGG pathways revealed that groups 2 and 3 were highly enriched for ribosomal proteins (Figure 7b). Interestingly, several proteins reported to play a role in DNA repair, including DDX21, SSRP1, DNA-PKcs, along with several heterogeneous nuclear ribonucleoproteins, 59–60 clustered in group 1 (Figure 7c). Enrichment of group 1 for proteins involved in DNA repair was further corroborated by KEGG pathway analysis (Figure 7b). Given these intriguing functional links, we chose three proteins distributed across group 1 for further validation. Figure 8 shows (8a) our original iTRAQ data and (8b) western blots for the group 1 proteins DDX21, HNRNPC, and RCC2, isolated via TAP from an independent culture of HeLa S3 cells exposed to gamma radiation as in Figure 6a. To evaluate the agreement between these data types we normalized iTRAQ reporter ion signal and western blot intensities to their respective values at time point zero (Figure 8c). Taken together with our analysis of potential reduction in precursor contamination (Figure 5) these data suggest that RP-RP fractionation combined with iTRAQ-based stable isotope labeling provides for accurate quantification of subtle changes in multi-component protein complex membership.
Figure 7.
(A) Hierarchical clustering of iTRAQ ratios, relative to the zero time point (Figure 6A, left lane), partitioned the response of FLAG-HA Ku86 associated proteins to gamma irradiation into three major groups. Functional analysis of each group based on KEGG Pathways (B) revealed that groups 2 and 3 were highly enriched for ribosomal proteins, while multiple DNA repair pathways were overrepresented in group 1 (B - Top, and C). Several proteins previously demonstrated to interact with Ku (DNA-PK, SSRP1, WRN, and HNRNPC), along with novel complex members (DDX21 and RCC2) validated in this study (Figure 8) are indicated for group 1.
Figure 8.
Biochemical validation of FLAG-HA Ku86 associated proteins quantified by iTRAQ RP-RP MS/MS. (A) iTRAQ reporter ion region for representative peptides from DDX21, HNRNPC, and RCC2, identified in a 40 fraction RP-RP MS/MS analysis (Figure 6A). (B) Immunoblot of the same proteins after tandem affinity purification of FLAG-HA Ku86 from an independent culture of HeLa S3 cells subjected to gamma radiation as in Figure 6A. (C) iTRAQ and western blot data for DDX21, HNRNPC, and RCC2 replotted with both data types normalized to their respective values at time point zero.
CONCLUSIONS
Successful application of SCX-RP in the context of multiple, disparate studies 8–12 provides unequivocal evidence that multi-dimensional fractionation improves data quality, and concomitantly augments researchers’ ability to place proteomics data into appropriate biological context. Simultaneously these studies reveal that still higher performance fractionation is required to probe the proteome in further depth, even in the case of studies targeted to sub-classes of proteins or peptides. Our data herein demonstrate that RP-RP outperforms SCX-RP primarily due to greater peak capacity in the first dimension separation. Collectively, our results indicate that the analytical figures of merit for RP-RP translate directly into greater numbers of peptide and protein identifications as well as improved dynamic range, as compared to SCX-RP, for analysis of both complex cell lysates and multi-component protein complexes. Moreover, addition of a 6-port valve to a Waters NanoACQUITY UHPLC system allowed us to utilize our recently described miniaturized LC-ESI assemblies in the second dimension, hence enabling RP-RP fractionation in a true nanoflow format. 61 As noted previously, 18, 27 offline fractionation schemes may be prone to low yield as a result of additional sample handling, lyophilization, and non-specific adsorption to tube surfaces. Not surprisingly the relative impact of these issues on peptide recovery typically scales inversely with total sample quantity. In contrast to previous reports, 16–18, 25, 27–29, 62 our system provides an automated, online platform that leverages the combined advantages of RP-RP fractionation and high efficiency electrospray ionization achieved at ultralow effluent flow rates, yielding improved performance as compared to SCX-RP for samples that span a wide range of protein complexity (e.g., whole cell lysates and multi-component protein complexes).
Despite growing interest in the analysis of multi-component protein complexes by mass spectrometry, the performance metrics for efficient fractionation remain somewhat unexplored. We used HeLa S3 cells that expressed FLAG-HA Ku86 as a model system to compare analytical figures of merit for multi-dimensional fractionation coupled to mass spectrometry analysis. Consistent with our analysis of complex cell lysates, we observed that RP-RP provided for identification of significantly more unique peptides and proteins as compared to SCX-RP, for an equivalent fractionation depth. Moreover, we observed a continual increase in the number of proteins identified as a function of fractionation depth, suggesting that efforts aimed at systematic analysis of discrete protein complexes will benefit from higher peak capacity separation strategies. Our data add some 175 putatively novel members to the repertoire of known proteins in the multi-component Ku complex.
To further explore the utility of RP-RP fractionation for the analysis of protein complexes, we sought to characterize changes in the Ku complex in response to DNA damage induced by gamma radiation. Although Ku86 and Ku70 are known to play important roles in DNA repair, we observed only modest modulation of the associated complex at the level of a silver-stained gel, over a time course of 5 hours after exposure of cells to gamma radiation. To gain additional insight, we utilized iTRAQ stable isotope labels in conjunction with RP-RP MS to decipher Ku protein complex dynamics under these conditions. Hierarchical clustering revealed coordinate participation of many proteins known to associate with Ku, including WRN, 63 DNA-PK, 55, 64 and H2AX, 58 in addition to a wide range of proteins not previously reported as members of the multi-component Ku complex (see Supplementary Table 6). For example, our data significantly expand the number of ribosomal proteins that may participate in the Ku86 complex. When considered collectively with previous work 45, our results raise the intriguing possibility that Ku86 regulates translation of specific mRNAs via recruitment of ribosomal components to their site of synthesis. Consistent with this hypothesis Ku86 and Ku70, among other proteins, have been reported to interact with the 5′UTR sequence of p53. Indeed, this DNA sequence is critical for the increased expression of p53 upon DNA damage.65 We used western blot to validate the response of HNRNPC, DDX21, and RCC2 to gamma radiation; the agreement between our immunoblot and proteomics data suggest that membership of many proteins within the Ku complex are dynamically modulated in response to DNA damage. Moreover, our ability to monitor modest ratios for members of the Ku complex suggests that our online RP-RP fractionation platform provides sufficient peak capacity to minimize deleterious scrambling 66–67 of iTRAQ reporter ions. In the specific cases of RCC2, 68 HNRNPC, 59 and DDX21, their roles in DNA repair have been linked to different protein kinases, ATM/ATR, 68 DNA-PKcs, 69 and JNK, 60 respectively. Collectively these observations highlight the complex interplay between protein-protein interactions and phosphorylation in regulation of cellular response to DNA damage.
Given our results above, along with recent work that the highlighted the complex nature of DNA repair pathways 68, it is not surprising to find that the molecular response to DNA damage, or other genotoxic insult, is observed across sub-nuclear compartments. For example, re-localization of WRN, SSRP1 and Ku86 between the nucleolus and nucleoplasm has been linked with different types of cellular stress 55. It is intriguing to note that many proteins identified in our study, particularly those in group 1 (Figure 7c), shuttle between different sub-nuclear compartments as a result of DNA damage. While the mechanistic impact of protein re-localization, for example DNA-PK, SSRP1, and DDX21 in our work, or WRN, 63 Spt16 70 and TRF2 71 as described elsewhere, remains unclear, our data provide an expanded set of proteins, putatively involved in DNA repair pathways, that are viable candidates for further biochemical and functional characterization. In this context, immunohistochemical-based assays would provide orthogonal validation of protein-protein interactions in addition to exquisite temporal and sub-cellular localization data. These assays, when combined with proteomics-based analysis of protein complexes involved in DNA repair, will provide a powerful means to decipher the molecular response to therapeutic strategies that rely on induction of genotoxic stress.
Supplementary Material
Acknowledgments
The authors thank Eric Smith for preparation of figures and valuable discussion. Generous support for this work was provided by the Dana-Farber Cancer Institute and the National Institutes of Health, NHGRI (P50HG004233), and NINDS (P01NS047572).
References
- 1.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405 (6788):837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422 (6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Hille JM, Freed AL, Watzig H. Possibilities to improve automation, speed and precision of proteome analysis: A comparison of two-dimensional electrophoresis and alternatives. Electrophoresis. 2001;22 (19):4035–4052. doi: 10.1002/1522-2683(200111)22:19<4035::AID-ELPS4035>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4 (10):807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 5.Sterner JL, Johnston MV, Nicol GR, Ridge DP. Signal suppression in electrospray ionization Fourier transform mass spectrometry of multi-component samples. J Mass Spectrom. 2000;35 (3):385–391. doi: 10.1002/(SICI)1096-9888(200003)35:3<385::AID-JMS947>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 6.Proteomics on a thermo finnigan LTQ-FTICR mass spectrometer. Mol Cell Proteomics. 2005;4 (8):S442–S442. [Google Scholar]
- 7.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-Pressure Linear Ion Trap Mass Spectrometer Improving the Analysis of Complex Protein Mixtures. Analytical Chemistry. 2009;81 (18):7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19 (3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 9.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, Van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419 (6906):512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 10.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu YM, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419 (6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 11.Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, Wolters D, Yates JR. Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci U S A. 2002;99 (18):11969–11974. doi: 10.1073/pnas.172183199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Multidimensional separations-based shotgun proteomics. Chem Rev. 2007;107 (8):3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 13.Mawuenyega KG, Kaji H, Yamauchi Y, Shinkawa T, Saito H, Taoka M, Takahashi N, Isobe T. Large-scale identification of Caenorhabditis elegans proteins by multidimensional liquid chromatography - Tandem mass spectrometry. J Proteome Res. 2003;2 (1):23–35. doi: 10.1021/pr025551y. [DOI] [PubMed] [Google Scholar]
- 14.Taoka M, Yamauchi Y, Shinkawa T, Kaji H, Motohashi W, Nakayama H, Takahashi N, Isobe T. Only a small subset of the horizontally transferred chromosomal genes in Escherichia coli are translated into proteins. Mol Cell Proteomics. 2004;3 (8):780–787. doi: 10.1074/mcp.M400030-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Nagano K, Taoka M, Yamauchi Y, Itagaki C, Shinkawa T, Nunomura K, Okamura N, Takahashi N, Izumi T, Isobe T. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 2005;5 (5):1346–1361. doi: 10.1002/pmic.200400990. [DOI] [PubMed] [Google Scholar]
- 16.Essader AS, Cargile BJ, Bundy JL, Stephenson JL. A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5 (1):24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 17.Cargile BJ, Talley DL, Stephenson JL. Immobilized pH gradients as a first dimension in shotgun proteomics and analysis of the accuracy of pI predictability of peptides. Electrophoresis. 2004;25 (6):936–945. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 18.Slebos RJC, Brock JWC, Winters NF, Stuart SR, Martinez MA, Li M, Chambers MC, Zimmerman LJ, Ham AJ, Tabb DL, Liebler DC. Evaluation of Strong Cation Exchange versus Isoelectric Focusing of Peptides for Multidimensional Liquid Chromatography-Tandem Mass Spectrometry. J Proteome Res. 2008;7 (12):5286–5294. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455 (7217):1251–U60. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 20.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full Dynamic Range Proteome Analysis of S. cerevisiae by Targeted Proteomics. Cell. 2009;138 (4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balgley BM, Wang WJ, Song T, Fang XP, Yang L, Lee CS. Evaluation of confidence and reproducibility in quantitative proteomics performed by a capillary isoelectric focusing-based proteomic platform coupled with a spectral counting approach. Electrophoresis. 2008;29 (14):3047–3054. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 22.Fang XP, Balgley BM, Wang WJ, Park DM, Lee CS. Comparison of multidimensional shotgun technologies targeting tissue proteomics. Electrophoresis. 2009;30 (23):4063–4070. doi: 10.1002/elps.200900367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YJ, Rudnick PA, Evans EL, Li J, Zhuang ZP, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected tumor tissue using a capillary isoelectric focusing-based multidimensional separation platform coupled with ESI-tandem MS. Analytical Chemistry. 2005;77 (20):6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 24.Chen JZ, Balgley BM, DeVoe DL, Lee CS. Capillary isoelectric focusing-based multidimensional concentration/separation platform for proteome analysis. Analytical Chemistry. 2003;75 (13):3145–3152. doi: 10.1021/ac034014+. [DOI] [PubMed] [Google Scholar]
- 25.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28 (14):1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 26.Delmotte N, Lasaosa M, Tholey A, Heinzle E, Huber CG. Two-dimensional reversed-phase x ion-pair reversed-phase HPLC: an alternative approach to high-resolution peptide separation for shotgun proteome analysis. J Proteome Res. 2007;6 (11):4363–73. doi: 10.1021/pr070424t. [DOI] [PubMed] [Google Scholar]
- 27.Dowell JA, Frost DC, Zhang J, Li LJ. Comparison of two-dimensional fractionation techniques for shotgun proteomics. Analytical Chemistry. 2008;80 (17):6715–6723. doi: 10.1021/ac8007994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77 (19):6426–34. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura T, Kuromitsu J, Oda Y. Evaluation of comprehensive multidimensional separations using reversed-phase, reversed-phase liquid chromatography/mass spectrometry for shotgun proteomics. J Proteome Res. 2008;7 (3):1007–1011. doi: 10.1021/pr7005878. [DOI] [PubMed] [Google Scholar]
- 30.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:17. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415 (6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 32.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 33.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440 (7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 34.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40 (4):303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolfs A, Hu Y, Ebert L, Hoffmann D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, Shen B, Baqui MMA, Pearlberg J, Taycher E, DeLoughery C, Hoerlein A, Korn B, LaBaer J. A Biomedically Enriched Collection of 7000 Human ORF Clones. PLoS ONE. 2008;3 (1):e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposito D, Garvey LA, Chakiath CS. Gateway Cloning for Protein Expression. 2009. pp. 31–54. [DOI] [PubMed] [Google Scholar]
- 37.Forstner M, Leder L, Mayr LM. Optimization of protein expression systems for modern drug discovery. Expert Rev Proteomics. 2007;4 (1):67–78. doi: 10.1586/14789450.4.1.67. [DOI] [PubMed] [Google Scholar]
- 38.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Meth. 2008;5 (6):535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, Hu Y, Murthy TVS, Vannberg F, Shen B, Rolfs A, Hutti JE, Cantley LC, LaBaer J, Harlow E, Brizuela L. Building a human kinase gene repository: Bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci U S A. 2005;102 (23):8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang SF, Yu YB, Yang PY, Gu S, Xue Y, Chen X. Analysis of the protein complex associated with 14-3-3 epsilon by a deuterated-leucine labeling quantitative proteomics strategy. J Chromatogr B. 2009;877 (7):627–634. doi: 10.1016/j.jchromb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Yang MZ, Wang XL, Zhang L, Yu CY, Zhang BB, Cole W, Cavey G, Davidson P, Gibson G. Demonstration of the interaction of transforming growth factor beta 2 and type X collagen using a modified tandem affinity purification tag. J Chromatogr B. 2008;875 (2):493–501. doi: 10.1016/j.jchromb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Leene J, Witters E, Inze D, De Jaeger G. Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 2008;13 (10):517–520. doi: 10.1016/j.tplants.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila - The advantages of the GS-TAP system. Fly. 2008;2 (4):229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- 44.Gregan J, Riedel CG, Petronczki M, Cipak L, Rumpf C, Poser I, Buchholz F, Mechtler K, Nasmyth K. Tandem affinity purification of functional TAP-tagged proteins from human cells. Nat Protoc. 2007;2 (5):1145–1151. doi: 10.1038/nprot.2007.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3 (12):1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 46.Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, Smith ED, Boyer L, Schlaeger TM, Luckey CJ, Marto JA. Improved Electrospray Ionization Efficiency Compensates for Diminished Chromatographic Resolution and Enables Proteomics Analysis of Tyrosine Signaling in Embryonic Stem Cells. Analytical Chemistry. 2009;81 (9):3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 47.Licklider LJ, Thoreen CC, Peng JM, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectromentry with a vented column. Analytical Chemistry. 2002;74 (13):3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 48.van der Heeft E, ten Hove GJ, Herberts CA, Meiring HD, van Els CA, de Jong AP. A microcapillary column switching HPLC-electrospray ionization MS system for the direct identification of peptides presented by major histocompatibility complex class I molecules. Anal Chem. 1998;70 (18):3742–51. doi: 10.1021/ac9801014. [DOI] [PubMed] [Google Scholar]
- 49.Askenazi M, Parikh JR, Marto JA. mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods. 2009;6 (4):240–242. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parikh JR, Askenazi M, Ficarro SB, Cashorali T, Webber JT, Blank NC, Zhang Y, Marto JA. multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics. 2009;10:364. doi: 10.1186/1471-2105-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ficarro SB, Parikh JR, Blank NC, Marto JA. Niobium(V) oxide (Nb2O5): Application to phosphoproteomics. Analytical Chemistry. 2008;80 (12):4606–4613. doi: 10.1021/ac800564h. [DOI] [PubMed] [Google Scholar]
- 52.Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97 (5):621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 53.Downs JA, Jackson SP. A means to a DNA end: The many roles of Ku. Nat Rev Mol Cell Biol. 2004;5 (5):367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 54.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8 (3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 55.Dejmek J, Iglehart JD, Lazaro JB. DNA-Dependent Protein Kinase (DNA-PK)-Dependent Cisplatin-Induced Loss of Nucleolar Facilitator of Chromatin Transcription (FACT) and Regulation of Cisplatin Sensitivity by DNA-PK and FACT. Mol Cancer Res. 2009;7 (4):581–591. doi: 10.1158/1541-7786.MCR-08-0049. [DOI] [PubMed] [Google Scholar]
- 56.Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15 (20):6314–20. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- 57.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6 (9):1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Du YC, Gu S, Zhou JH, Wang TY, Cai H, MacInnes MA, Bradbury EM, Chen X. The dynamic alterations of H2AX complex during DNA repair detected by a proteomic approach reveal the critical roles of Ca2+/calmodulin in the ionizing radiation-induced cell cycle arrest. Mol Cell Proteomics. 2006;5 (6):1033–1044. doi: 10.1074/mcp.M500327-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Christian KJ, Lang MA, Raffalli-Mathieu F. Interaction of heterogeneous nuclear ribonucleoprotein C1/C2 with a novel cis-regulatory element within p53 mRNA as a response to cytostatic drug treatment. Mol Pharmacol. 2008;73 (5):1558–1567. doi: 10.1124/mol.107.042507. [DOI] [PubMed] [Google Scholar]
- 60.Mialon A, Thastrup J, Kallunki T, Mannermaa L, Westermarck J, Holmstrom TH. Identification of nucleolar effects in JNK-deficient cells. FEBS Lett. 2008;582 (20):3145–3151. doi: 10.1016/j.febslet.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, Smith ED, Boyer L, Schlaeger TM, Luckey CJ, Marto JA. Improved Electrospray Ionization Efficiency Compensates for Diminished Chromatographic Resolution and Enables Proteomics Analysis of Tyrosine Signaling in Embryonic Stem Cells. Analytical Chemistry. 0(0) doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 62.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 2006;5 (12):3368–75. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 63.Sakamoto S, Nishikawa K, Heo SJ, Coto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6 (5):421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 64.Shi MG, Vivian CJ, Lee KJ, Ge CM, Morotomi-Yano K, Manzl C, Bock F, Sato S, Tomomori-Sato C, Zhu RH, Haug JS, Swanson SK, Washburn MP, Chen DJ, Chen BPC, Villunger A, Florens L, Du CY. DNA-PKcs-PIDDosome: A Nuclear Caspase-2-Activating Complex with Role in G2/M Checkpoint Maintenance. Cell. 2009;136 (3):508–520. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123 (1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 66.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. iTRAQ Underestimation in Simple and Complex Mixtures: “The Good, the Bad and the Ugly”. J Proteome Res. 2009;8 (11):5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 67.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao ZM, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316 (5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 69.Zhang SS, Schlott B, Gorlach M, Grosse F. DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 2004;32 (1):1–10. doi: 10.1093/nar/gkg933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heo K, Kim HJ, Choi SH, Choi JY, Kim KW, Gu JF, Lieber MR, Yang AS, An WJ. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30 (1):86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 71.Buscemi G, Zannini L, Fontanella E, Lecis D, Lisanti S, Delia D. The Shelterin Protein TRF2 Inhibits Chk2 Activity at Telomeres in the Absence of DNA Damage. Curr Biol. 2009;19 (10):874–879. doi: 10.1016/j.cub.2009.03.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.