Abstract
Seed vigor is an important characteristic of seed quality, and rice cultivars with strong seed vigor are desirable in direct-sowing rice production for optimum stand establishment. In the present study, the quantitative trait loci (QTLs) of three traits for rice seed vigor during the germination stage, including germination rate, final germination percentage, and germination index, were investigated using one recombinant inbred line (RIL) population derived from a cross between japonica Daguandao and indica IR28, and using the multiple interval mapping (MIM) approach. The results show that indica rice presented stronger seed vigor during the germination stage than japonica rice. A total of ten QTLs, and at least five novel alleles, were detected to control rice seed vigor, and the amount of variation (R 2) explained by an individual QTL ranged from 7.5% to 68.5%, with three major QTLs with R 2>20%. Most of the QTLs detected here are likely to coincide with QTLs for seed weight, seed size, or seed dormancy, suggesting that the rice seed vigor might be correlated with seed weight, seed size, and seed dormancy. At least five QTLs are novel alleles with no previous reports of seed vigor genes in rice, and those major or minor QTLs could be used to significantly improve the seed vigor by marker-assisted selection (MAS) in rice.
Keywords: Rice, Recombinant inbred line (RIL) population, Seed vigor, Quantitative trait locus (QTL), Germination
1. Introduction
Seed vigor is an important characteristic of seed quality, reflecting potential seed germination, seedling growth, seed longevity, and tolerance to adversity (Sun et al., 2007). Seeds with strong vigor may significantly improve the speed and uniformity of seed germination and the final percentage of germination, and lead to perfect field emergence, good crop performance, and even high yield under different conditions (Foolad et al., 2007). Seed vigor has been known as a comprehensive characteristic affected by many factors, such as the genetic background, environmental factors during seed development, and storage stages (Sun et al., 2007), which makes the genetic analysis of seed vigor very difficult. With the development of the techniques of DNA molecular marker and genome graphing, quantitative trait locus (QTL) analysis of seed vigor has been reported, mainly focused on limited plants, including Arabidopsis (Clerkx et al., 2004), rice (Cui et al., 2002; Miura et al., 2002; Zhang et al., 2005; Fujino et al., 2008), lettuce (Hayashi et al., 2008), tomato (Foolad et al., 2003; 2007), barley (Mano and Takeda, 1997), and maize (Hund et al., 2004).
Rice is one of the most important food crops in the world, and the cultivars with strong seed vigor are desirable for farmers to get optimum stand establishment in a direct-sowing culture system, which has increased in some economic developed areas in China recently. There are several reports of QTLs for rice seed vigor, mainly focused on seedling growth, seed longevity, and tolerance to adversity. For example, Cui et al. (2002) mapped 31 QTLs for five traits of seedling vigor using a set of 241 F10 recombinant inbred lines (RILs). Zhang et al. (2005) identified a total of 34 QTLs for four traits of seedling vigor using 282 F13 RILs. Miura et al. (2002) found three putative QTLs for seed longevity, qLG-2, qLG-4, and qLG-9, using 98 backcross inbred lines (BILs). Fujino et al. (2004) detected three putative QTLs (qLTG-3-1, qLTG-3-2, and qLTG-4) associated with low-temperature germination using 122 BILs, and qLTG-3-1 was map-based cloned (Fujino et al., 2008). In the evaluation of seed vigor, the germination speed might be more important than the germination rate, since the former usually decreases more quickly than the latter during seed storage (Hayashi et al., 2008). Therefore, the speed of seed germination is a desirable trait for seed vigor testing, such as germination rate and germination index. However, to date there are few QTLs reported on seed vigor during germination stage under optimal conditions.
It has been reported that QTLs associated with germination speed are important for germination, and not specifically affected by stress in tomato (Foolad et al., 1999; 2007) and lettuce (Hayashi et al., 2008), suggesting rapid seed germination controlled by similar genetic mechanisms under different conditions. QTLs for the speed of germination coincided with those for abscisic acid (ABA) sensitivity and salt tolerance in Arabidopsis (Clerkx et al., 2004). In rice, a significant positive correlation was noticed between seed size and seed vigor, probably due to the fact that large-size seed may provide more sugar for seedling growth and fuel rapid early growth (Cui et al., 2002). However, little progress has been made in improving rice seed vigor by conventional breeding methods, such as selecting large grain size. The reason is that rice consumers have a certain requirement for grain size, and increasing the grain size may reduce the popularity of a cultivar (McKenzie et al., 1994). Therefore, identification of QTLs associated with rice seed vigor, not linked to seed size, was needed to improve seed vigor by marker-assisted selection (MAS).
The purpose of this work was not only to further investigate the genetic dissection of seed vigor, focusing on seed germination under optimum conditions, but also to identify major QTLs for rice breeding through the process of MAS. One RIL population (F10) derived from the cross of indica IR28 and japonica Daguandao rice was employed to map the loci underlying three seed germination traits, including germination rate, final germination percentage, and germination index. The results will provide important information to understand the genetic control of seed vigor in rice, and the major QTLs detected here will be useful for improvement of seed vigor.
2. Materials and methods
2.1. Plant materials
One RIL population was obtained from a cross between two varieties, indica IR28 and japonica Daguandao. The population of 150 F10 RILs was obtained via self-crossing and single seed descent from individual F2 plants. Ten plants of each parent and RIL were grown in the space of 17 cm between plants within a row and 33 cm between rows at the experimental station of Nanjing Agricultural University. The date when 95% of seeds of each plant were yellow-ripe could be defined as maturity date. The maturity dates for parents and RILs were individually recorded. All seeds were harvested at maturity stage for each line, dried at 50 °C for 7 d to break seed dormancy (Jiang et al., 2006), and then stored in dry conditions at −20 °C.
2.2. Evaluation of seed vigor
After breaking down dormancy, a total of 200 healthy grains of each RIL and two parents were soaked in 0.1% (w/v) hydrargyrum chlorination solution for 15 min, and then rinsed three times with sterile distilled water. Fifty seeds were placed in a Petri dish (diameter 9 cm) with two sheets of filter paper, to which 10 ml of distilled water was added. The solution was replaced every 2 d to maintain the distilled water volume. All Petri dishes were placed in an incubator at (30±1) °C for 10 d with a 12-h light/12-h dark photoperiod. Seeds were considered to be germinated when their root length reached the seed length and shoot length half of the seed length. The germinated seeds were observed each day until 10 d, when almost all the seeds were germinated. The percentage of germinated seeds at 3 d was referred to as germination rate (GR) and the percentage of germinated seeds at 10 d was referred to as the final germination percentage (GP). Germination index (GI) was calculated by the method of Cao et al. (2008): GI=∑(Gt/t), where Gt is the number of the germinated seeds on Day t. Four replications were conducted and the mean value was used for data analysis.
2.3. QTL analysis
A set of 167 SSR markers covering most of the rice genetic map at average interval of 11.1 cM was constructed with the MAPMAKER/EXP 3.0 program (Lander et al., 1987). The detection of QTLs for three traits representing seed vigor, germination rate, final germination percentage, and germination index was performed using the QTL Cartographer 2.5 program on the method of multiple interval mapping (MIM) (Churchill and Doerge, 1994). The data of germination percentage were transformed by arcsine transformation into a typical quantitative trait distribution for QTL detection with MIM (limit of detection (LOD) >3.0). In addition, the proportion of observed phenotypic variance explained by each QTL and the corresponding additive effect were estimated. QTL nomenclature followed the method of McCouch et al. (1997).
3. Results
3.1. Phenotypic data
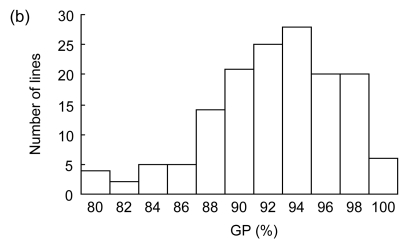
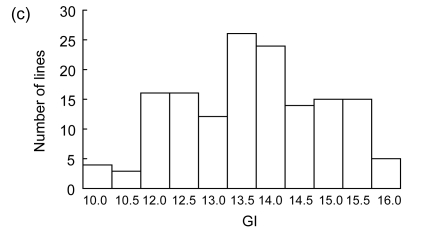
The difference in germination rate and germination index between the two parents was significant at a level of P<0.01, but no statistically significant difference in germination percentage (Table 1). The indica rice IR28 showed a good performance of seed vigor, with higher germination rate and germination index. The RILs also showed statistically significant differences for seed germination (P<0.01). The distributions of three germination traits among the RIL population showed continuous and significantly transgressive segregation with values either larger or smaller than those of the parents, suggesting the involvement of polygenes (Table 1 and Fig. 1). The data of germination percentage were further analyzed by arcsine transformation for QTL detection.
Table 1.
Phenotypic data of seed vigor among parents and RIL population
| Traits | GR (%) | GP (%) | GI | |
| Parentsa | Daguandao | 24.0±0.5 | 97.0±0.0 | 11.8±0.0 |
| IR28 | 80.0±0.1** | 100.0±0.0 | 15.7±0.0** | |
| RILs | Range | 3–90 | 80–100 | 9.7–16.0 |
| Mean | 48.8 | 91.9 | 13.5 | |
| CV (%) | 47.9 | 5.0 | 10.1 | |
| Fb | 2.18## | 1.42 | 2.26## | |
GR: germination rate; GP: germination percentage; GI: germination index; CV: coefficient of variance
Sample size n=50, four replicates
Significant at P<0.01 according to Fisher’s Least Significant Difference (LSD) test
F test of variance among RIL population
Significant at P<0.01, F 0.01(149, 150)=1.47
Fig. 1.
Frequency distributions of germination rate (GR) (a), germination percentage (GP) (b), and germination index (GI) (c) among RILs
3.2. QTL mapping
Three QTLs controlling germination rate were identified on chromosomes 1, 2, and 11, respectively (Table 2 and Fig. 2), accounting for 93.6% of phenotypic variance. Among them, qGR-1 might be one major QTL (R 2=68.5%) with an LOD score of 11.0, and the positive alleles from IR28 enhanced the germination rate by 19.9%. The other two of the minor QTLs (qGR-2 and qGR-11) explained the 11.0% and 14.1% of phenotypic variance, respectively, and the positive alleles originated from Daguandao enhanced the germination rate by 10.0% and 10.5%, respectively.
Table 2.
Chromosome location, coefficient of determination, and additive effect of the putative QTLs conferring seed vigor among the Daguandao/IR28 RIL population
| Trait | Locus | Chra | Marker interval | LOD support interval (cM) | Peak LOD | AEb | R2 (%)c |
| GR | qGR-1 | 1 | RM9-RM7075 | 137.5–156.8 | 11.0 | 19.9 | 68.5 |
| qGR-2 | 2 | RM208-RM240 | 15.7–23.9 | 7.3 | −10.0 | 11.0 | |
| qGR-11 | 11 | RM3668-RM7557 | 8.1–16.0 | 6.3 | −10.5 | 14.1 | |
| GP | qGP-4 | 4 | RM252-RM317 | 102.2–117.5 | 5.7 | 2.5 | 12.7 |
| qGP-6 | 6 | RM528-RM340 | 123.1–132.4 | 5.3 | −3.1 | 24.0 | |
| qGP-8 | 8 | RM22491-RM22694 | 23.4–34.6 | 4.3 | −2.3 | 12.2 | |
| qGP-11 | 11 | RM26632-RM229 | 73.2–84.4 | 3.7 | −2.1 | 7.5 | |
| GI | qGI-1 | 1 | RM306-RM5 | 88.8–111.5 | 3.1 | 0.6 | 16.1 |
| qGI-7 | 7 | RM427-RM1353 | 0–2.0 | 4.3 | −0.6 | 13.9 | |
| qGI-11 | 11 | RM3428-RM6091 | 70.7–74.4 | 5.1 | −2.5 | 54.9 | |
GR: germination rate; GP: germination percentage; GI: germination index; Chr: chromosome; LOD: limit of detection; AE: additive effect; R 2: coefficient of determination
Chromosome on which the QTL was located
Additive effect is the effect of substituting a IR28 allele for a Daguandao allele, and its positive value indicates that IR28 has the positive allele
Variation explained by each putative QTL
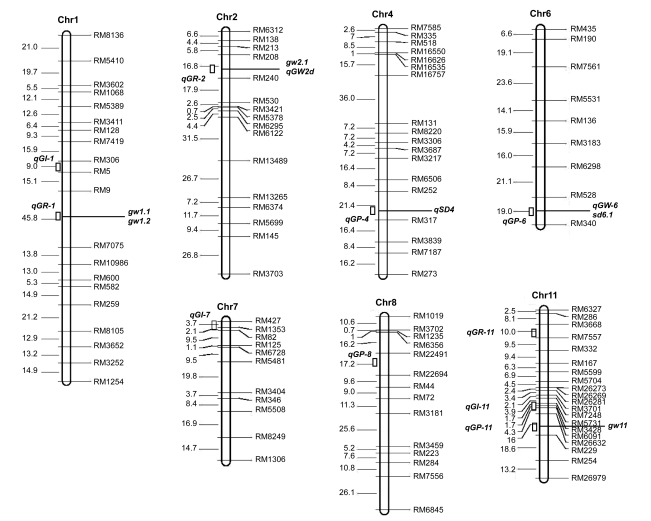
Fig. 2.
Chromosomal positions of QTLs for germination rate, germination percentage, and germination index, and previously mapped QTLs in rice
Map distances (cM) are shown on the left and previously mapped QTLs are shown on the right (Li J.X. et al., 2000; Moncada et al., 2001; Gu et al., 2004; Marri et al., 2005; Wan et al., 2005; Li C. et al., 2006; Bai et al., 2010)
Four QTLs associated with germination percentage were found on chromosomes 4, 6, 8, and 11, respectively (Table 2 and Fig. 2). The phenotypic variance explained by a single QTL ranged from 7.5% to 24.0%, and one major QTL qGP-6 (LOD=5.3) with R 2 of 24.0%. The Daguandao alleles at qGP-6, qGP-8, and qGP-11 increased by 3.1%, 2.3%, and 2.1% of germination percentage, respectively, and IR28 alleles at qGP-4 enhanced germination percentage by 2.5%.
Three QTLs (qGI-1, qGI-7, and qGI-11) were responsible for germination index located on chromosomes 1, 7, and 11, respectively (Table 2 and Fig. 2), explaining the total of 84.9% of phenotypic variance. Among them, a major QTL qGI-11 (LOD=5.1), with a relatively large effect (R 2=54.9%), was detected, and the additive effect of qGI-11 was negative, which showed that the positive alleles from Daguandao contributed to the increase of germination index by 2.5. The positive alleles of QTL qGI-1 and qGI-7 enhanced germination index by 0.6.
However, no significant digenic interaction was detected in this study.
4. Discussion
The most crucial step in a QTL mapping project is the evaluation and screening of the quantitative traits. Seed vigor is defined as “those seed properties which determine the potential for rapid, uniform emergence and development of normal seedlings under a wide range of field conditions” (McDonald, 1994). Consequently, the results of germination percentage determined by standard germination test under optimum conditions usually overestimate field emergence under suboptimal field conditions. Therefore, many additional vigor tests have been suggested, such as conductivity test, accelerated aging test, cold test, cool germination test, complex stressing vigor test, Hiltner test, tetrazolium test, and seedling growth test (Zhang and Hu, 2010). McKenzie et al. (1980) reported that seedling traits measured under controlled laboratory conditions were correlated with seedling vigor measured under field conditions. Cui et al. (2002) indicated that germination rate (speed) and early seedling growth were interrelated in rice. In this study, the traits of seed vigor during the germination stage, including germination rate, germination percentage, and germination index, were determined under laboratory conditions. We considered that indica rice has higher seed vigor during the germination stage under optimum conditions than japonica rice. The frequency distribution of seed vigor among RILs showed continuous segregation, suggesting that seed vigor during the germination stage is a quantitative trait controlled by several genes.
QTLs for germination speed were detected in several different crops (Foolad et al., 1999; Al-Chaarani et al., 2005; Hayashi et al., 2008). Al-Chaarani et al. (2005) detected two QTLs conferring the time to 50% of germination of the population in sunflower, using a population of 84 RILs. Foolad et al. (1999) mapped several QTLs for the mean time to 50% of germination under non-stress, cold-stress, and salt-stress conditions, using a 119 BC1S1 population. Similarly, Hayashi et al. (2008) found 28 QTLs for germination rate in lettuce under different temperatures using a population of 131 F8 RILs. These results show that the speed and uniformity of germination under different conditions share the same basic genetic mechanism. In the present study, we conducted a screen for QTLs of germination under optimum conditions using a population of 150 RILs in rice. Ten QTLs were mapped on chromosomes 1, 2, 4, 6, 7, 8, and 11, respectively. The amount of variation explained by an individual QTL ranged from 7.5% to 68.5%. Among them, three major QTLs (qGR-1, qGP-6, and qGI-11) with R 2>20% were detected. Thus, the performance of rice seed vigor during seed germination might be considered to be controlled by a limited number of major QTLs and several minor QTLs.
The exploration of physiological and genetic mechanisms in seed vigor is a highlight of seed science. Comparing the positions of QTLs detected here with other QTLs reported previously (Fig. 2), we found that the region of major QTL qGR-1 on chromosome 1 coincided with gw1.1 and gw1.2 for 1000-seed weight (Moncada et al., 2001). The region of major QTL qGP-6 on chromosome 6 was familiar with the region of qGW-6 for 1000-seed weight (Wan et al., 2005) and sd6.1 for seed dormancy (Li et al., 2006). In addition, the minor QTL qGR-2 was on the similar location of gw2.1 for 1000-seed weight (Marri et al., 2005) and grain width (Bai et al., 2010) on chromosome 2. Similarly, qGP-11 is near to gw11 for 1000-seed weight (Li et al., 2000). Furthermore, qGP-4 located on the same region of qSD4 for seed dormancy on chromosome 4 (Gu et al., 2004). However, other five mapped QTLs (qGI-1, qGI-7, qGP-8, qGR-11, and qGI-11) are novel alleles, with no previous reports of seed vigor genes in rice.
The molecular evidence indicates that de novo transcripts are required for increasing seed vigor, notably those involved in gibberellic acid (GA) biosynthesis and/or sensitivity to GAs during germination (Catusse et al., 2008). In addition, genes related to reserve mobilization and endosperm weakening could possibly affect the speed of germination (Fait et al., 2006; Bethke et al., 2007). Obviously, seed germination rate and vigor are closely related to seed weight, seed size, shell thickness, seed coat, and so on. In this study, we found the regions of major QTLs for seed vigor are likely to coincide with QTLs for seed weight, seed size, and seed dormancy, suggesting that seed vigor, seed weight, seed size, and seed dormancy are partly under the control of the same genetic mechanism. Further study is needed to clarify the function of the major QTLs.
It is extremely difficult to improve rice seedling vigor by conventional strategies, due to undesirable traits associated with seedling vigor, such as large grain size (McKenzie et al., 1994; Cui et al., 2002). Cui et al. (2002) found two QTLs for seedling vigor and α-amylase, not linked to seed weight, which will be useful for MAS to improve seedling vigor. However, with the number of QTLs increasing, the utility of MAS becomes less obvious (Foolad et al., 2003). Therefore, marker-assisted introgression of the three major QTLs detected here is expected to significantly improve seed vigor in rice, especially the QTL qGI-11 independent of seed weight.
Seed vigor is a very complex physiological process with a comprehensive genetic background. In this study, ten QTLs associated with seed vigor were identified, and among them, three were major QTLs and others minor QTLs. To extensively elucidate the mechanism of seed vigor, more studies are needed. The QTLs detected here may be used for developing new varieties with a high level of seed vigor by the MAS method.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31000748), the Natural Science Foundation of Jiangsu Province (No. BK2010452), and the Science and Technology Innovation Foundation of Nanjing Agricultural University (No. KJ09003), China
References
- 1.Al-Chaarani GR, Gentzbittel L, Wedzony M, Sarrafi A. Identification of QTLs for germination and seedling development in sunflower (Helianthus annuus L.) Plant Sci. 2005;169(1):221–227. doi: 10.1016/j.plantsci.2005.03.016. [DOI] [Google Scholar]
- 2.Bai X, Luo L, Yan W, Kovi MR, Zhan W, Xing Y. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7 . BMC Genet. 2010;11(1):16. doi: 10.1186/1471-2156-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007;143(3):1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao DD, Jin H, Huang XX, Wang XJ, Guan YJ, Wang ZF. Relationships between changes of kernel nutritive components and seed vigor during development stages of F1 seeds of sh2 sweet corn. J Zhejiang Univ-Sci B. 2008;9(12):964–968. doi: 10.1631/jzus.B0820084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catusse J, Job C, Job D. Transcriptome- and proteome-wide analyses of seed germination. C R Biol. 2008;331(10):815–822. doi: 10.1016/j.crvi.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clerkx EJ, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SP, Vreugdenhil D, Koornneef M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;135(1):432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui K, Peng S, Xing Y, Xu C, Yu S, Zhang Q. Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor Appl Genet. 2002;105(5):745–753. doi: 10.1007/s00122-002-0908-2. [DOI] [PubMed] [Google Scholar]
- 9.Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006;142(3):839–854. doi: 10.1104/pp.106.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foolad MR, Lin GY, Chen FQ. Comparison of QTLs for seed germination under non-stress, cold stress and salt stress in tomato. Plant Breed. 1999;118(2):167–173. doi: 10.1046/j.1439-0523.1999.118002167.x. [DOI] [Google Scholar]
- 11.Foolad MR, Zhang LP, Subbiah P. Genetics of drought tolerance during seed germination in tomato: inheritance and QTL mapping. Genome. 2003;46(4):536–545. doi: 10.1139/g03-035. [DOI] [PubMed] [Google Scholar]
- 12.Foolad MR, Subbiah P, Zhang L. Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int J Plant Genomics. 2007;2007:97386. doi: 10.1155/2007/97386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin SY, Yano M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.) Theor Appl Genet. 2004;108(5):794–799. doi: 10.1007/s00122-003-1509-4. [DOI] [PubMed] [Google Scholar]
- 14.Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. PNAS. 2008;105(34):12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu XY, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166(3):1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi E, Aoyama N, Still DW. Quantitative trait loci associated with lettuce seed germination under different temperature and light environments. Genome. 2008;51(11):928–947. doi: 10.1139/G08-077. [DOI] [PubMed] [Google Scholar]
- 17.Hund A, Fracheboud Y, Soldati A, Frascaroli E, Salvi S, Stamp P. QTL controlling root and shoot traits of maize seedlings under cold stress. Theor Appl Genet. 2004;109(3):618–629. doi: 10.1007/s00122-004-1665-1. [DOI] [PubMed] [Google Scholar]
- 18.Jiang L, Liu S, Hou M, Tang J, Chen L, Zhai H, Wan J. Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.) Field Crops Res. 2006;98(1):68–75. doi: 10.1016/j.fcr.2005.12.015. [DOI] [Google Scholar]
- 19.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Zhou A, Sang T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara . New Phytol. 2006;170(1):185–194. doi: 10.1111/j.1469-8137.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- 21.Li JX, Yu SB, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q. Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor Appl Genet. 2000;101(1-2):248–254. doi: 10.1007/s001220051476. [DOI] [Google Scholar]
- 22.Mano Y, Takeda K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.) Euphytica. 1997;94(3):263–272. doi: 10.1023/A:1002968207362. [DOI] [Google Scholar]
- 23.Marri PR, Sarla N, Reddy LV, Siddiq EA. Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon . BMC Genet. 2005;6(1):33. doi: 10.1186/1471-2156-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T. Report on QTL nomenclature. Rice Genet Newslett. 1997;14:11–13. [Google Scholar]
- 25.McDonald MB. The history of seed vigor testing. J Seed Technol. 1994;17:93–101. [Google Scholar]
- 26.McKenzie KS, Rutger JN, Peterson ML. Relation of seedling vigor to semidwarfism, early maturity, and pubescence in closely related rice lines. Crop Sci. 1980;20(2):169–172. doi: 10.2135/cropsci1980.0011183X002000020005x. [DOI] [Google Scholar]
- 27.McKenzie KS, Johnson CW, Tseng ST, Oster JJ, Brandon DM. Breeding improved rice cultivars for temperate regions–a case study. Aust J Exp Agric. 1994;34(7):897–905. doi: 10.1071/EA9940897. [DOI] [Google Scholar]
- 28.Miura K, Lin S, Yano M, Nagamine T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.) Theor Appl Genet. 2002;104(6-7):981–986. doi: 10.1007/s00122-002-0872-x. [DOI] [PubMed] [Google Scholar]
- 29.Moncada P, Martinez CP, Borrero J, Châtel M, Gauch H, Guimaraes EP, Tohmé J, McCouch SR. Quantitative trait loci for yield and yield components in an Oryza sativa×Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet. 2001;102(1):41–52. doi: 10.1007/s001220051616. [DOI] [Google Scholar]
- 30.Sun Q, Wang JH, Sun BQ. Advances on seed vigor physiological and genetic mechanisms. Agric Sci China. 2007;6(9):1060–1066. doi: 10.1016/S1671-2927(07)60147-3. [DOI] [Google Scholar]
- 31.Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet. 2005;110(7):1334–1346. doi: 10.1007/s00122-005-1976-x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang HS, Hu J. Seed Science. Beijing, China: Science Press; 2010. pp. 165–170. (in Chinese) [Google Scholar]
- 33.Zhang ZH, Yu SB, Yu T. Mapping quantitative trait loci (QTLs) for seedling-vigor using recombinant inbred lines of rice (Oryza sativa L.) Field Crops Res. 2005;91(2-3):161–170. doi: 10.1016/j.fcr.2004.06.004. [DOI] [Google Scholar]