Abstract
In rapidly growing tumors, hypoxia commonly develops due to the imbalance between O2 consumption and supply. Hypoxia Inducible Factor (HIF)-1α is a transcription factor responsible for tumor growth and angiogenesis in the hypoxic microenvironment; thus, its inhibition is regarded as a promising strategy for cancer therapy. Given that CamKII or PARP inhibitors are emerging anticancer agents, we investigated if they have the potential to be developed as new HIF-1α-targeting drugs. When treating various cancer cells with the inhibitors, we found that a CamKII inhibitor, KN-62, effectively suppressed HIF-1α specifically in hepatoma cells. To examine the effect of KN-62 on HIF-1α-driven gene expression, we analyzed the EPO-enhancer reporter activity and mRNA levels of HIF-1α downstream genes, such as EPO, LOX and CA9. Both the reporter activity and the mRNA expression were repressed by KN-62. We also found that KN-62 suppressed HIF-1α by impairing synthesis of HIF-1α protein. Based on these results, we propose that KN-62 is a candidate as a HIF-1α-targeting anticancer agent.
Keywords: CaMKII, HIF1-α, Hepatocellular carcinoma, Hypoxia, KN-62
INTRODUCTION
Solid tumors inevitably encounter hypoxia because of outgrowth of the cell mass over vessels. To survive under hypoxic conditions, tumor cells must run numerous adaptive processes including glycolysis, glucose uptake, and up-regulation of survival factors [1]. HIF-1 was first identified as a transcription factor that mediates hypoxia-inducible activity of the erythropoietin 3' enhancer [2]. Many lines of evidence have since demonstrated that HIF-1 is a master regulator that induces over 70 genes in response to hypoxia. HIF-1 exists as a heterodimeric complex consisting of HIF-1α and HIF-1β. In normoxia, HIF-1α is rapidly hydroxylated at its two proline residues by prolyl-hydroxylase domain proteins (PHDs), then ubiquitinated by the von Hippel-Lindau protein (pVHL)/E3 ligase components, and finally degraded through the 26S proteasome [3,4]. In hypoxia, however, HIF-1α hydroxylation is limited, and HIF-1α protein accumulates [5]. To ensure the transcriptional activity of HIF-1α, p300/CBP, steroid receptor co-activator-1 (SRC-1), and transcription intermediary factor-2 (TIF-2) bind to the C-terminal transactivation domain (C-TAD) of HIF-1α and function as transcriptional coactivators [6,7]. HIF-1α is also regulated at the translational level by the AKT-mTOR pathway. AKT phosphorylates mTOR, and the activated mTOR in turn phosphorylates and inhibits S6K and 4E-BP1. Then, the translation-initiating factors aggregate to form the translational complex and promote the translation of HIF-1α, which constitutes the so-called "5' cap-dependent translation [8,9]."
Calcium/calmodulin-dependent protein kinase II (CaMKII) is a multifunctional calcium/calmodulin-dependent serine/threonine protein kinase. Recent studies suggest that CaMKII plays important roles in cell cycle progression and cell proliferation [10,11]. To date, several CaMKII inhibitors have been developed that interfere with calcium/calmodulin binding to CaMKII or its catalytic activity. Previous studies showed that CaMKII inhibitors KN-62 and KN-93 induce cell cycle arrest, proliferation inhibition, and apoptosis in cancer cells [12,13]. However, whether CaMKII inhibitors deregulate HIF-1 or not remains controversial. It has been reported that calcium increase within cells positively regulates the translation of HIF-1α by activating cPKC-α and mTOR in PC12 and HEK293 cells [14]. Moreover, calcineurin, which facilitates calcium/calmodulin signaling, has been shown to activate the recruitment of p300 by MEF-2 in T-cells [15] and myocytes [16]. As mentioned previously, given that p300 plays a critical role in HIF-1-driven gene expression, it is plausible that disrupting calcium signaling by CaMKII inhibition would affect HIF-1α expression and activity.
Poly (ADP-ribose) polymerases (PARPs) function as DNA nick sensors and provide nuclear targets for various signaling pathways. PARPs bind to damaged DNA and are activated to conjugate ADP-ribose units to DNA and various acceptor proteins. PARPs are known to regulate diverse cellular processes, such as replication, transcription, differentiation, protein degradation, and mitotic spindle maintenance [17]. Interestingly, the elevation of intracellular calcium is among the wide array of PARP-activating stimuli [18,19]. Moreover, the genetic or pharmacological inhibition of PARP1 attenuates the hypoxic induction of HIF-1α and other hypoxia-induced genes [20-23].
Given that CaMKII and PARP inhibitors are emerging as new drugs for molecular target cancer therapy, we investigated whether they inhibit the tumor response to hypoxia by targeting HIF-1α. We found that the CaMKII inhibitor KN-62, but not PARP inhibitors, effectively suppressed the hypoxic expression and activation of HIF-1α, specifically in hepatocellular carcinoma cells. Moreover, the HIF-1α suppression by KN-62 may be attributed to impaired translation of HIF-1α due to Akt inactivation.
METHODS
Cell culture and chemicals
Hep3B, MCF7 and SK-N-Mc cells were maintained in Dulbecco's modified Eagle's medium from Gibco, and HepG2 cells were maintained in RPMI media supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), antibiotics and L-Glutamine (Invitrogen). The oxygen tension in a CO2 incubator (Vision Science, Seoul, Korea) was 20% (normoxic) or 1% (hypoxic). Cells were pretreated with drugs 1 hr before being subjected to hypoxia and further incubated for 8 or 16 hrs. MG132 was purchased from Alexis Biochemicals (Lausen, Switzerland). All other chemicals were from Sigma-Aldrich (St. Louis, MO).
Western blotting
Equal amounts of total protein were loaded onto an 8% SDS/PAGE gel and transferred to an Immobilon-P (Millipore) membrane. After blocking with PBST (1× PBS with 0.05% Tween 20) plus 5% skim milk at room temperature for 1 hr, the membrane was incubated with primary antibody overnight at 4℃ and then with horseradish peroxidase (HRP)-coupled secondary antibody for 1 hr at room temperature. Immune complexes were visualized using Enhanced Chemiluminescence plus western blotting detection kit (GE-Healthcare, Piscataway, NJ). Anti-HIF-1α antiserum was generated in rabbits against amino acids 418~698 of human HIF-1α, as described previously [24]. Anti-phospho (S473)Akt and anti-Akt antibodies were purchased from Cell Signaling (Danvers, MA), and anti-β-tubulin and anti-rabbit IgG Horseradish peroxidase-conjugated antibody were from SantaCruz Biotechnologies (Santa Cruz, CA).
Reverse Transcription and Quantitative Real-Time PCR
Total mRNA was isolated using Trizol reagent (Invitrogen). Reverse transcription was performed with a MultiScribe Reverse Transcriptase kit (Applied Biosystems). Real-time PCR was performed in triplicates with a total volume of 20 µl using a SYBR Green JumpStart Taq ReadyMix (Sigma) as described in the manufacturer's manual. Reactions were run in a PCR System 7000HT (Applied Biosystems). Data were analyzed using SDS 2.0 software (Applied Biosystems) and then exported to an Excel file. Data was normalized as described previously [25]. The nucleotide sequences of the primers were as follows: forward 5'-AACAATCACTGCTGACACTT-3' and reverse 5'-AGAGTTGCTCTCTGGACAGT-3' for EPO; forward 5'-TATGAGGGGTCTCTGACTACA-3' and reverse 5'-TTCTCATCTGCACAAGGAAC-3' for CA9; forward 5'-GATTATGATCACAGGGTGCT-3' and reverse 5'-ATCCACTGGCAGTCTATGTC-3' for LOX; forward 5'-TTCGTATTGCGCCGCTAGA-3' and reverse 5'-CTTCGCTCTGGTCCGTCTT-3' for 18S ribosomal RNA.
Reporter assay
Luciferase reporter plasmid containing HRE of the human EPO gene was a gift from Dr. Eric Huang (University of Utah, Utah). Constructs of HIF-1α-5'-UTR (1~284) cloned into pGL3 (Tk/HIF-1α-5'-UTR-Luciferase) and between the GFP gene and the luciferase gene (CMV/GFP-5'UTR-Luciferase) were described in the previous report [26]. Cells were transfected with 1 µg of reporter plasmid and 0.4 µg of β-gal DNA per well of a 12-well plate using Lipofectamine 2000 (Invitrogen) and then lysed to determine luciferase and β-gall activities according to the manufacturer's manual for the Luciferase assay kit from Promega (Madison, WI). Luciferase activities were measured using a Lumat LB9507 luminometer (Berthold), and initial activities were divided by β-gal activities to normalize transfection efficiencies.
RESULTS
KN-62 inhibits the hypoxic induction of HIF-1α protein in hepatocellular carcinoma
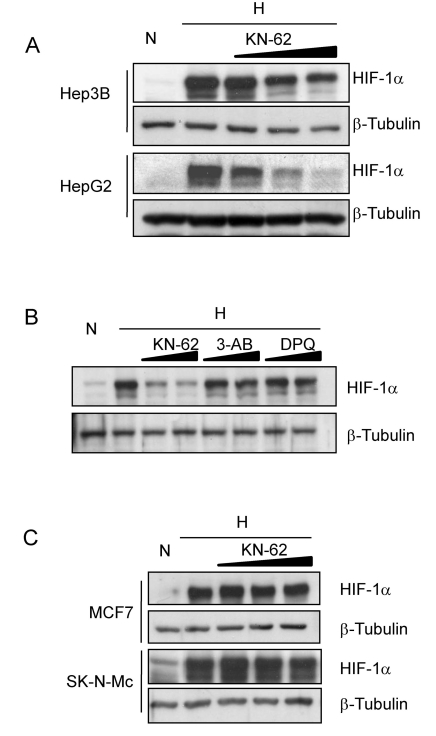
To see the effect of KN-62 CaMKII inhibitor on HIF-1α expression in hepatoma cells, we treated Hep3B and HepG2 cells with KN-62 in hypoxia and then detected HIF-1α protein by Western blotting. KN-62 substantially reduced HIF-1α levels in a dose-dependent manner, but PARP-1 inhibitors, 3-AB and DPQ did not (Fig. 1A and 1B). In contrast to hepatoma cells, MCF7 (breast cancer) and SK-N-MC (neuroblastoma) cells did not respond to KN-62, even at high concentrations (Fig. 1C), which suggests that HIF-1α suppression by KN-62 is cancer type-dependent.
Fig. 1.
HIF-1α protein level is regulated by CamKII inhibitor KN-62 in Hepatoma cell lines. (A) Hep3B and HepG2 cells were pre-treated with 0, 1, 2, or 5 µM KN-62 and incubated with 1% O2 hypoxia for 8 hr. Western blot analysis was done with anti-HIF-1α and anti-β-Tubulin antibodies. (B) Western blot analysis of HIF-1α and β-Tubulin in Hep3B cell line before (N) and after (H) 8 hr incubation in 1% O2 in the presence of 2 µM and 5 µM KN-62, 100 µM and 500 µM 3-AB, or 10 µM and 30 µM DPQ. (C) MCF7 and SK-N-MC cells were incubated in 1% O2 for 8 hr in the presence of 0, 5, 10, or 15 µM KN-62 and western blot analysis was performed with anti-HIF-1α and anti-β-Tubulin antibodies.
KN-62 attenuates the hypoxic activation of HIF-1 in Hep3B cells
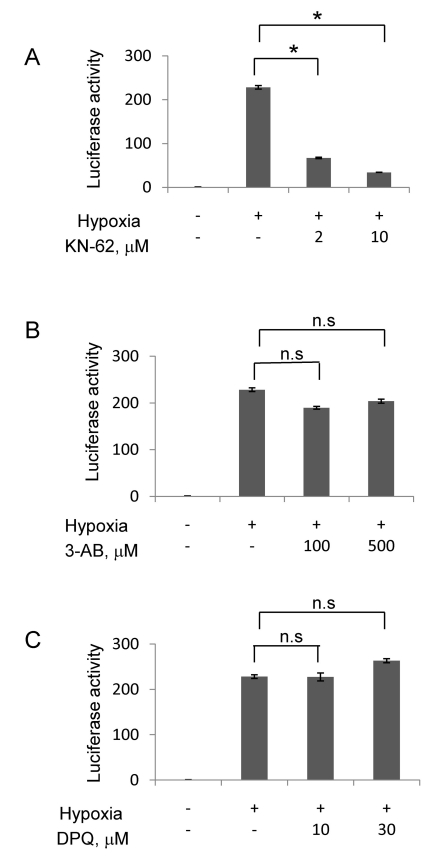
To examine whether KN-62 functionally inhibits HIF-1, we analyzed the Epo-reporter activity, which reflects on HIF-1-driven gene expression, in Hep3B cells. The Epo-reporter activity markedly increased by about 200-fold in hypoxia versus in normoxia, and this activation was significantly attenuated by KN-62 (Fig. 2A). In contrast, PARP inhibitors like 3-AB and DPQ failed to inhibit the hypoxic activation of the Epo-reporter even though they were administered at much higher concentrations than KN-62 (Fig. 2B and 2C).
Fig. 2.
KN-62 repressed HIF-1α activity in Hep3B cell line. Hep3B cells were transiently transfected with an Epo-Luc construct. The cells were treated with increasing concentrations of KN-62 (A), 3AB (B) and DPQ (C) in 21% (Normoxia) or 1% (Hypoxia) O2 for 16 h. Luciferase activity is shown as the fold change from the value with 21% O2 and no drug. *Denotes p<0.01 between indicated groups. n.s., not significant.
KN-62 blunts the HIF-1-driven transcription during hypoxia in Hep3B cells
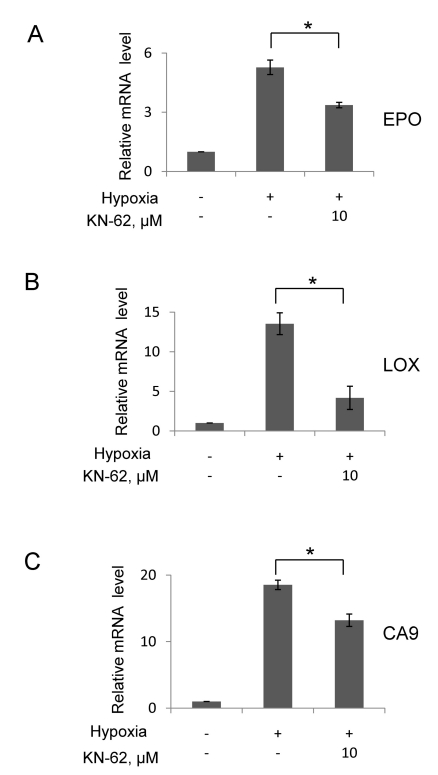
To further examine the effect of KN-62 on HIF-1 activity, we analyzed mRNAs of HIF-1 target genes using quantitative RT-PCR. The mRNAs of erythropoietin (Epo), carbonic anhydrase 9 (CA9) and lysyl oxidase (LOX) were induced after hypoxic incubation, and KN-62 significantly reduced the mRNA levels (Fig. 3). These results robustly indicate that KN-62 deregulates hepatoma cell response to hypoxia by inhibiting the HIF-mediated signaling.
Fig. 3.
The mRNA level of HIF-1α downstream gene was decreased in the presence of KN-62 under hypoxia. Quantitative real-time measurement of Epo (A), LOX (B) and CA9 (C) mRNA level was done using total RNA from Hep3B cells treated with 16-hr hypoxia with or without KN-62. 18S signal was used for normalization of signals and values were shown as relative value to normoxia sample. *Denotes p<0.05 between indicated groups.
KN-62 demotes the synthesis, rather than the stability, of HIF-1α protein
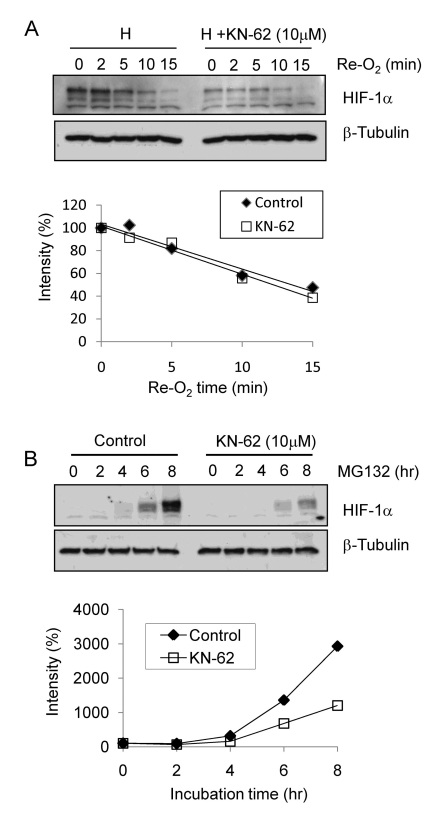
To determine the mechanism by which HIF-1α is suppressed by KN-62, we first checked the stability of HIF-1α protein. The degradation of HIF-1α protein was induced by supplying 20% oxygen to Hep3B cells subjected to 8-h hypoxia. Although substantial differences in HIF-1α levels during hypoxia were found between DMSO- and KN-62-treated cells, there were no apparent differences in the degradation rates of HIF-1α after reoxygenation (Fig. 4A, upper panel). To estimates the degradation rates of HIF-1α, the intensities of the protein bands were analyzed using the ImageJ 1.36b program and the first-order kinetics were plotted. No significant changes in HIF-1α degradation were observed between the two groups (Fig. 4A, lower panel). Next, we analyzed the de novo synthesis of HIF-1α protein in the presence of a proteasome inhibitor, MG132, to force the accumulation of synthesized HIF-1α. HIF-1α protein gradually accumulated after MG132 treatment in both groups. However, the accumulation of HIF-1α protein was noticeably retarded in KN-62-treated cells (Fig. 4B, upper panel). ImageJ analysis confirmed that KN-62 inhibited HIF-1α protein synthesis at the translational level (Fig. 4B, lower panel).
Fig. 4.
The protein level of HIF-1α was regulated by synthesis rate. (A) Top: Hep3B cells were treated with 1% O2 for 8 hr with or without KN-62 and re-oxygenated for the indicated time. Western blot signals of HIF-1α and β-tubulin are detected. Bottom: The HIF-1α protein band densities were quantified using the ImageJ program and plotted as a function of time. (B) Top: Western blot signals of HIF-1α and β-tubulin are detected with Hep3B cell extract treated with MG132 with or without KN-62 for the indicated time. Bottom: The protein band densities were quantified using ImageJ and plotted as a function of time.
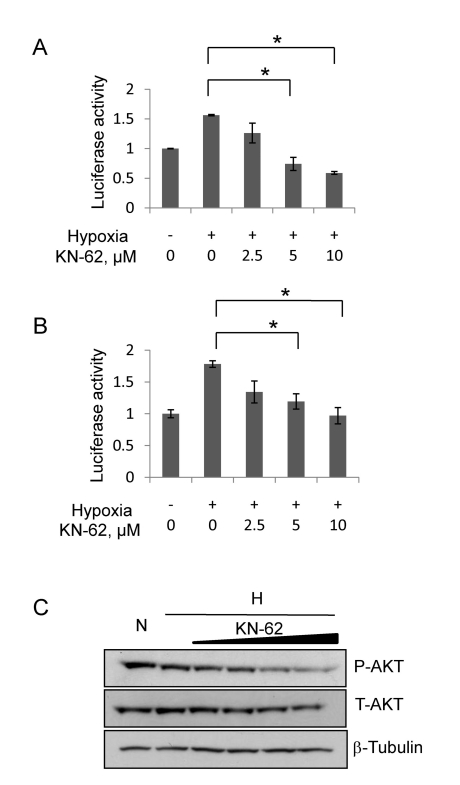
KN-62 repressed the 5'-UTR-driven translation of HIF-1α by inactivating AKT
HIF-1α protein is synthesized via 5' cap-dependent translation and IRES-dependent translation [9,27]. To determine which translational pathway is regulated by KN-62, we used two reporter plasmids. One is Tk/HIF-1α/5'-UTR-luciferase reporter designed for cap-dependent translation and the other is CMV/GFP-5'UTR-Luciferase reporter for IRES-dependent translation. Hep3B cells were transfected with either Tk/HIF-1α/5'-UTR-luciferase vector (Fig. 5A) or CMV/GFP-5'UTR-Luciferase vector (Fig. 5B), and incubated in hypoxic conditions with or without KN-62. As shown in Fig. 5, Tk/HIF-1α/5'-UTR reporter activity (Fig. 5A) and CMV/GFP-5'UTR-Luc reporter (Fig. 5B) were both increased after 16-hr hypoxia, but significantly attenuated by KN-62 in a dose-dependent manner. Given that the AKT signaling determines the HIF-1α/5'-UTR-driven translation [28], we measured the cellular levels of phospho Ser473-AKT (active form of AKT), and found that KN-62 noticeably inhibits the AKT phosphorylation (Fig. 5C). These results suggest that KN-62 inhibits the AKT signaling and by doing so blocks the de novo synthesis of HIF-1α in hepatoma cells.
Fig. 5.
HIF-1α protein translation was decreased by KN-62 treatment via inhibition of Akt signaling. (A) The reporter activity of Tk/HIF-1α-5'-UTR Luciferase in Hep3B cells treated with Normoxia, Hypoxia or Hypoxia with KN-62 was analyzed. Values are shown as the relative value to normoxia sample. (B) The reporter activity of CMV/GFP-5'UTR-Luciferase in Hep3B cells treated with Normoxia, Hypoxia or Hypoxia with KN-62 was analyzed. Values are shown as relative value to normoxia sample. (C) KN-62 inhibits phosphorylation of AKT. After 8-hr incubation under hypoxic conditions with 0, 1, 2, 5, or 10 µM KN-62, Hep3B cells were lysed and subjected to Western blotting with anti-phospho S473 Akt antibody, anti-total Akt antibody and anti-β tubulin antibody. *Denotes p<0.05 between indicated groups.
DISCUSSION
In the present study, we found that a CaMKII inhibitor, KN-62, attenuates the hypoxic gene regulation in hepatoma cells. As the mode-of-action of KN-62, we propose that KN-62 inhibits AKT signaling, which in turn demotes the translation of HIF-1α, and this results in deregulation of HIF-1 activity and the hypoxic induction of HIF-1 target genes. These results suggest that KN-62 could be a candidate to be developed as HIF-1-targeting anti-hepatoma agent.
Ca2+ signaling, which is one of the most prevalent and essential signaling pathways, is involved in diverse cellular processes including apoptosis, ion channel regulation, cell cycling, and cellular responses to stress [29,30]. As HIF-1α is induced Ca2+-dependently during intermittent hypoxia, Ca2+ signaling is thought to participate in HIF-1α regulation. Indeed, HIF-1α has been found to be regulated by p300 coactivator, phospholipase C-γ, and protein kinase C, all of which are activated by Ca2+ signaling. Moreover, overexpression of constitutively active CaMKII enhances HIF-1α activity and the CaMKII inhibitor KN-93 represses it in PC12 cells [14,31,32]. The involvement of CaMKII in HIF-1α activation was also demonstrated in macrophages; that is, CaMKII inhibitors SMP-114 and KN-93 down-regulated HIF-1α and VEGF in THP-1 monocytic cells [33-35]. However, the role of CaMKII in HIF-1α regulation remains controversial because a specific CaMKII inhibitor SMP-114 had no effect on HIF-1α and VEGF expressions in rheumatoid synovial fibroblasts [34]. Therefore, the CaMKII regulation of HIF-1α is regarded as a cell type-dependent event. In this study, the CamKII inhibitor KN-62 down-regulated HIF-1α in hepatoma cells, but this KN-62 effect was not reproduced in other cancer cells. These results also support the cell type-dependent action of CamKII against HIF-1α.
The role of PARP in HIF-1α regulation is also controversial. Several studies using PARP-/- MEF and PARP inhibitors showed that active PARP is required for HIF-1α expression and activation [20-22]. In other studies, however, PARP did not affect HIF-1α expression but functioned as a coactivator of HIF-1α [23]. We also tested the effects of PARP-1 inhibitors 3-AB and DPQ on HIF-1α expression and activation, but found no effects of the inhibitors even at high concentrations. The mRNA levels of HIF-1α downstream genes like EPO, LOX and CA9 were slightly reduced in Hep3B cells by DPQ, but the DPQ effects were much lower than those of KN-62 (data not show). Based on these results, PARP is unlikely to participate in HIF-1α regulation, and thus it is doubtful that PARP inhibitors will be developed as HIF-1-targeting anticancer agents at least for hepatoma.
Malignant hepatoma (also called hepatocellular carcinoma) is one of the most common cancers in East Asian countries including Korea, and ranks as the third leading cause of cancer mortality worldwide [36,37]. In the case of advanced hepatoma, systemic chemotherapy used to be prescribed to increase patient survival, but conventional anticancer agents seldom improve the clinical outcome. We have therefore made many efforts to develop various classes of anticancer agents. Recent advances in the understanding of cancer progression have provided the scientific background to develop new molecular-targeted therapies [38,39]. Moreover, a growing body of evidence indicates that HIF-1α is a valuable target for cancer therapy. Considering that hepatoma is a hyper-vascular tumor with high levels of HIF-1α, anti-HIF-1 agents are expected to be available for molecular-targeted hepatoma therapy. It is thus worthwhile to search for new anti-hepatoma agents based on anti-HIF-1 action. Here we propose KN-62 as a potential anti-HIF-1 agent effective against hepatoma. Further studies of its anti-hepatoma activity remain to be conducted.
ABBREVIATIONS
- HIF-1α
hypoxia inducible factor-1α
- CamKII
calcium/calmodulin-dependent protein kinase II
- PARP
poly (ADP-ribose) polymerase
- pVHL
von Hippel-Lindau protein
- PHD
prolyl hydroxylase
- Epo
erythropoietin
- LOX
lysyl oxidase
References
- 1.Hockel M, Vaupel P. Tumor Hypoxia: Definitions and Current Clinical, Biologic, and Molecular Aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G, Wang G. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O-2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berra E, Ginouvès A, Pouysségur J. The hypoxia-inducible-factor hydroxylases bring fresh air into hypoxia signalling. EMBO Reports. 2006;7:41–45. doi: 10.1038/sj.embor.7400598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugh C, O'Rourke J, Nagao M, Gleadle J, Ratcliffe P. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J Biol Chem. 1997;272:11205. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 7.Arany Z, Huang L, Eckner R, Bhattacharya S, Jiang C, Goldberg M, Bunn H, Livingston D. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza G, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 9.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of Hypoxia-Inducible Factor 1alpha Expression and Function by the Mammalian Target of Rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducibella T, Schultz RM, Ozil JP. Role of calcium signals in early development. Semin Cell Dev Biol. 2006;17:324–332. doi: 10.1016/j.semcdb.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Zayzafoon M. Calcium/calmodulin signaling controls osteoblast growth and differentiation. J Cell Biochem. 2006;97:56–70. doi: 10.1002/jcb.20675. [DOI] [PubMed] [Google Scholar]
- 12.Minami H, Inoue S, Hidaka H. The Effect of KN-62, Ca2+/Calmodulin Dependent Protein Kinase II Inhibitor on Cell Cycle. Biochem Biophys Res Commun. 1994;199:241–248. doi: 10.1006/bbrc.1994.1220. [DOI] [PubMed] [Google Scholar]
- 13.Tombes R, Grant S, Westin E, Krystal G. G1 cell cycle arrest and apoptosis are induced in NIH 3T3 cells by KN-93, an inhibitor of CaMK-II (the multifunctional Ca2+/CaM kinase) Cell Growth Differ. 1995;6:1063–1070. [PubMed] [Google Scholar]
- 14.Hui AS, Bauer AL, Striet JB, Schnell PO, Czyzyk-Krzeska MF. Calcium signaling stimulates translation of HIF-1 alpha during hypoxia. FASEB J. 2006;20:466–475. doi: 10.1096/fj.05-5086com. [DOI] [PubMed] [Google Scholar]
- 15.Youn H-D, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. Activation of the β myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J Cell Physiol. 2007;211:138–148. doi: 10.1002/jcp.20916. [DOI] [PubMed] [Google Scholar]
- 17.Rhun YL, Kirkland JB, Shah GM. Cellular Responses to DNA Damage in the Absence of Poly (ADP-ribose) Polymerase. Biochem Biophys Res Commun. 1998;245:1–10. doi: 10.1006/bbrc.1998.8257. [DOI] [PubMed] [Google Scholar]
- 18.Ermak G, Davies KJ. Calcium and oxidative stress: from cell signaling to cell death. Mol Immunol. 2002;38:713–721. doi: 10.1016/s0161-5890(01)00108-0. [DOI] [PubMed] [Google Scholar]
- 19.Virag L, Scott GS, Antal-Szalmas P, O'Connor M, Ohshima H, Szabo C. Requirement of intracellular calcium mobilization for peroxynitrite-induced poly (ADP-ribose) synthetase activation and cytotoxicity. Mol Pharmacol. 1999;56:824–833. [PubMed] [Google Scholar]
- 20.Martin-Oliva D, Aguilar-Quesada R, O'Valle F, Muñoz-Gámez JA, Martínez-Romero R, García del Moral R, Ruiz de Almodóvar JM, Villuendas R, Piris MA, Oliver FJ. Inhibition of poly (ADP-Ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Research. 2006;66:5744–5756. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Romero R, Martínez-Lara E, Aguilar-Quesada R, Peralta A, Oliver FJ, Siles E. PARP-1 modulates deferoxamine-induced HIF-1alpha accumulation through the regulation of nitric oxide and oxidative stress. J Cell Biochem. 2008;104:2248–2260. doi: 10.1002/jcb.21781. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Romero R, Cañuelo A, Martínez-Lara E, Oliver FJ, Cárdenas S, Siles E. Poly (ADP-ribose) polymerase-1 modulation of in vivo response of brain hypoxia-inducible factor-1 to hypoxia/reoxygenation is mediated by nitric oxide and factor inhibiting HIF. J Neurochem. 2009;111:150–159. doi: 10.1111/j.1471-4159.2009.06307.x. [DOI] [PubMed] [Google Scholar]
- 23.Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, Keller S, Gassmann M, Hottiger MO. Poly (ADP-Ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Molecular Cancer Research. 2008;6:282–290. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- 24.Chun YS, Choi E, Yeo EJ, Lee JH, Kim MS, Park JW. A new HIF-1 alpha variant induced by zinc ion suppresses HIF-1-mediated hypoxic responses. J Cell Sci. 2001;114:4051–4061. doi: 10.1242/jcs.114.22.4051. [DOI] [PubMed] [Google Scholar]
- 25.Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin HW, Cho CH, Kim TY, Park JW. Sunitinib deregulates tumor adaptation to hypoxia by inhibiting HIF-1alpha synthesis in HT-29 colon cancer cells. Biochem Biophys Res Commun. 2010;398:205–211. doi: 10.1016/j.bbrc.2010.06.060. [DOI] [PubMed] [Google Scholar]
- 27.Lang KJD, Kappel A, Goodall GJ. Hypoxia-inducible Factor-1alpha mRNA Contains an Internal Ribosome Entry Site That Allows Efficient Translation during Normoxia and Hypoxia. Mol Biol Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arsham AM, Plas DR, Thompson CB, Simon MC. Phosphatidylinositol 3-kinase/Akt signaling is neither required for hypoxic stabilization of HIF-1 alpha nor sufficient for HIF-1-dependent target gene transcription. J Biol Chem. 2002;277:15162–15170. doi: 10.1074/jbc.M111162200. [DOI] [PubMed] [Google Scholar]
- 29.Bootman MD, Collins TJ, Peppiatt CM, Prothero LS, MacKenzie L, De Smet P, Travers M, Tovey SC, Seo JT, Berridge MJ, Ciccolini F, Lipp P. Calcium signalling--an overview. Semin Cell Dev Biol. 2001;12:3–10. doi: 10.1006/scdb.2000.0211. [DOI] [PubMed] [Google Scholar]
- 30.Howe CJ, LaHair MM, McCubrey JA, Franklin RA. Redox Regulation of the Calcium/Calmodulin-dependent Protein Kinases. J Biol Chem. 2004;279:44573–44581. doi: 10.1074/jbc.M404175200. [DOI] [PubMed] [Google Scholar]
- 31.Yuan G, Nanduri J, Bhasker C, Semenza G, Prabhakar N. Ca2+/calmodulin kinase-dependent activation of hypoxia inducible factor 1 transcriptional activity in cells subjected to intermittent hypoxia. J Biol Chem. 2005;280:4321–4328. doi: 10.1074/jbc.M407706200. [DOI] [PubMed] [Google Scholar]
- 32.Yuan G, Nanduri J, Khan S, Semenza GL, Prabhakar NR. Induction of HIF-1alpha expression by intermittent hypoxia: Involvement of NADPH oxidase, Ca2+ signaling, prolyl hydroxylases, and mTOR. J Cell Physiol. 2008;217:674–685. doi: 10.1002/jcp.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westra J, Brouwer E, Bos R, Posthumus MD, Meer BD-VD, Kallenberg CGM, Limburg PC. Regulation of Cytokine-Induced HIF-1alpha; Expression in Rheumatoid Synovial Fibroblasts. Ann N Y Acad Sci. 2007;1108:340–348. doi: 10.1196/annals.1422.035. [DOI] [PubMed] [Google Scholar]
- 34.Westra J, Brouwer E, Bouwman E, Meer BD-vd, Posthumus MD, Leeuwen MAv, Limburg PC, Ueda Y, Kallenberg CGM. Role for CaMKII Inhibition in Rheumatoid Arthritis. Ann N Y Acad Sci. 2009;1173:706–711. doi: 10.1111/j.1749-6632.2009.04736.x. [DOI] [PubMed] [Google Scholar]
- 35.Westra J, Brouwer E, van Roosmalen IA, Doornbos-van der Meer B, van Leeuwen MA, Posthumus MD, Kallenberg CG. Expression and regulation of HIF-1alpha in macrophages under inflammatory conditions; significant reduction of VEGF by CaMKII inhibitor. BMC Musculoskelet Disord. 2010;11:61. doi: 10.1186/1471-2474-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: Worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Jee SH, Ohrr H, Sull JW, Samet JM. Cigarette Smoking, Alcohol Drinking, Hepatitis B, and Risk for Hepatocellular Carcinoma in Korea. J Natl Cancer Inst. 2004;96:1851–1856. doi: 10.1093/jnci/djh334. [DOI] [PubMed] [Google Scholar]
- 38.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 39.Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H, Haga N. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Science. 2003;94:15–21. doi: 10.1111/j.1349-7006.2003.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]