Abstract
Lipid raft microdomains are enriched in sphingomyelin and cholesterol and function as platforms for signal transduction and as the site of budding of several enveloped viruses, including influenza virus. The influenza virus hemagglutinin (HA) glycoprotein, which mediates both viral-cell attachment and membrane fusion, associates intrinsically with lipid rafts. Residues in the HA transmembrane (TM) domain are important for raft association as sequence substitutions in the HA TM domain ablate HA association with rafts (nonraft HA). Cells expressing either WT or nonraft HA cause complete fusion (lipid mixing and content mixing) over widely varying HA expression levels. However, the number of fusion events measured for nonraft HA mutant protein at all HA surface densities was reduced to ≈55% of the events for WT HA protein. Mutant influenza viruses were generated that contain the nonraft HA TM domain alterations. Electron microscopy experiments showed that WT HA was distributed at the cell surface in clusters of 200–280 nm in diameter, whereas nonraft HA was distributed mostly randomly at the plasma membrane. Nonraft HA virus showed reduced budding, contained reduced amounts of HA protein, was greatly reduced in infectivity, and exhibited decreased virus–membrane fusion activity. Cholesterol depletion of virus did not affect the ability of virions to cause either virus–cell lipid mixing or virus-mediated hemolysis, a surrogate for content mixing. Taken together, the data suggest that HA clusters in rafts to provide a sufficient concentration of HA in budding virus to mediate efficient virus–cell fusion.
The lipids of the plasma membrane are thought to show lateral organization resulting from preferential packaging of sphingolipids and cholesterol into moving platforms, or rafts, in which specific membrane proteins become incorporated (1, 2). Although individual raft domains are small (≈50 nm in diameter) (3, 4), they are dynamic structures that can undergo changes in size and protein/lipid composition in response to intracellular or extracellular triggering processes (5). Generally, rafts are assumed to function as selective concentration devices for proteins and protein complexes and to provide platforms for signal transduction (reviewed in refs. 1, 2, 5, and 6). In addition, it has been shown that several enveloped viruses, including influenza virus, Ebola virus, and HIV-1, assemble at rafts and bud from these rafts (refs. 7–12 and reviewed in ref. 2). For influenza virus it has been shown that purified virus has a lipid composition more similar to that of a raft (high sphingomyelin and cholesterol content) than the overall plasma membrane (9, 10).
Raft microdomains (also called detergent-insoluble glycolipid complexes, DIGs) can be isolated biochemically because of their insolubility in nonionic detergent at low temperature [0.25–1.0% Triton X-100 (TX-100) at 4°C] (13). DIGs float on sucrose gradients, and thus proteins in DIGS can be separated from other detergent-insoluble material, e.g., proteins associated with the cytoskeleton (13, 14).
Influenza virus hemagglutinin (HA) glycoprotein associates intrinsically with lipid rafts, and it has been found that residues in the HA transmembrane (TM) domain are important for raft association. Expression of HA-containing mutations in the TM domain and in the HA cytoplasmic tail show reduced association with detergent-insoluble glycolipid complexes (10, 15, 16). It is not known whether the HA TM domain changes cause a direct alteration of HA with specific lipids or whether an association of HA with other proteins such as VIP17/MAL and annexin XIIIb is changed (17–19).
The catalog of enveloped viruses budding from rafts is growing (2). However, even for the best-studied example, influenza virus, evidence has been lacking to explain why influenza virus buds from rafts. Here, we provide evidence that suggests that HA clusters in rafts to provide a sufficient concentration of HA in budding virus to mediate efficient virus–cell fusion.
Materials and Methods
Cells, Viruses, and Plaque Assays. 293T cells, Madin-Darby canine kidney (MDCK) cells, MDCK-HA cells, HeLa-T4 cells, BHK cells, and Vero cells were maintained as described (20). Blasticidin-resistant MDCK cells stably expressing the influenza HA (A/Udorn/72) or mutant HA proteins were generated by standard methods (21). WT and mutant influenza A (A/Udorn/72) viruses were generated from cDNAs as described (22, 23). Influenza virus and vesicular stomatitis virus (VSV) stocks were grown as described, and virus infections and plaque assays were performed as described (20).
Plasmids, Alanine Scanning Mutagenesis, and cDNA Expression. Eight pHH21 plasmids and four pcDNA3.1 plasmids to generate influenza virus have been described (22). Eukaryotic expression plasmid pCAGGS was used to express the HA, neuraminidase (NA), M1, and M2 proteins of influenza virus and VSV G protein. Site-specific mutagenesis and DNA sequencing were performed as described (22).
TX-100 Extraction and Flotation Centrifugation. Virus-infected or transfected cells were metabolically labeled by using pulse–label–chase protocols as described (10). Cells were Dounce-homogenized in Dounce buffer (25 mM NaCl/25 mM Hepes, pH 7.3) and extracted with 0.25% (wt/vol) TX-100 in NTE (10 mM Tris, pH 7.4/100 mM NaCl/1 mM EDTA) on ice for 30 min. For separation of the soluble and insoluble fractions the cells were centrifuged at 14,000 × g for 30 min at 4°C. For flotation gradient analysis, 0.5 ml of the TX-100-extracted cell lysate was mixed with 1.5 ml of 65% (wt/vol) sucrose–NTE, layered on the bottom of a SW60 centrifuge tube, and overlaid with 1.5 ml of 30% sucrose–NTE and 0.7 ml of 5% sucrose–NTE. The gradients were subjected to centrifugation in a Beckman SW60Ti rotor at 210,000 × g at 4°C for 18 h. Samples of 0.7 ml were fractionated from the top.
Antibodies, Immunoprecipitation, and SDS/PAGE. Antisera used were as follows: for HA, an HA-specific mAb (C45/3); for M2, mAb 14C2; for HA, NA, M1, and NP proteins, goat serum raised to purified influenza A/Udorn/72 virus; for VSV-G protein, goat anti-VSV serum; and for transferrin receptor, mAb H68.4, (Zymed). Cell lysates or sucrose gradient fractions were solubilized in RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% TX-100/1% sodium deoxycholate/0.1% SDS), and immunoprecipitation and SDS/PAGE were performed as described (20). Quantification of radioactivity was performed with a Fuji BioImager 1000 (Fuji Medical Systems, Stanford, CT).
Analysis of HA Cell Surface Expression. HA cell surface expression was analyzed by using HA-specific C45/3 as the primary antibody and FITC-conjugated donkey anti-goat IgG (Jackson ImmunoResearch) as the secondary antibody. The cell surface fluorescence of 10,000 cells was analyzed with a FACSCalibur flow cytometer (Becton Dickinson).
Analysis of Cell–Cell Fusion. In vivo cell–cell fusion assays were performed essentially as described (24). BHK cells transiently expressing WT and mutant HA proteins were bound with human RBCs (1.0 ml of 0.05% hematocrit) that were dual-labeled with the lipid probe octadecyl rhodamine (R18) (Molecular Probes) and the aqueous probe carboxyfluorescein (CF) (Molecular Probes). Fusion was triggered by low pH (pH 4.8) treatment (25). Dye transfer and fluorescence dequenching was observed by scanning confocal microscopy (Zeiss LSM 410). Fusion was quantified by counting positive events of R18 or CF dye transfer from the RBCs to the BHK cells and averaging the fusion events from five to seven fields.
Purification of Virions and Immunoblotting. Influenza viruses were purified on sucrose density gradients as described (20). Immunoblotting of virions and infected cell polypeptides was performed as described (10). The membranes were incubated with goat anti-Udorn serum in blocking buffer (Li-Cor, Lincoln, NE) followed by incubation with Alexa Fluor 680-conjugated donkey anti-goat secondary antibody (Molecular Probes). Polypeptides were detected and quantified with an Odyssey Infrared Imaging system (Li-Cor).
Electron Microscopy. Negative staining of purified virions was done as described (26). Analysis of the distribution of HA at the plasma membrane was done by pre-embedding immunogold labeling followed by examination of thin sections as described (27). Quantification of gold-labeled HA on the cell surface was performed as described (28). A JEOL JEM-100CX II electron microscope was used to visualize specimens.
Fluorescent Dequenching Fusion Assay. Virus–cell fusion assays between sealed RBC ghosts and purified viruses labeled with R18 at a self-quenching concentration were performed as described (24). Fusion between the R18-labeled virus and the ghosts was triggered by lowering the pH from 7.0 to 5.0 by adding citric acid into the virus–ghost suspension. Fluorescence dequenching was monitored continuously with a fluorescence spectrometer (series 2; SLM–Aminco-Bowman, Urbana, IL) as described (24). To inactivate the HA proteins, the R18-labeled virus was incubated in PBS (pH 5.0) for 15 min at 37°C before incubation with the RBC ghosts.
Results
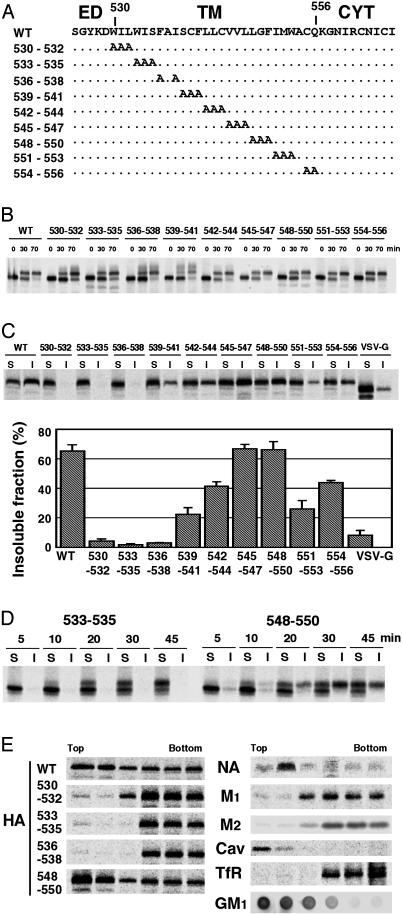
Characterization of HA Protein TM Domain Mutants. Scanning mutagenesis analysis showed previously that residues in the TM domain of HA protein derived from the H2 subtype HA (influenza A/Japan/305 virus) specify raft localization (15, 16). To analyze HA raft association in the context of an influenza virus infection by using a well characterized reverse genetics system (22), we used the H3 subtype HA of inf luenza A/Udorn/72 virus in the present study. Because the TM domain of H2 and H3 HAs have very different amino acid sequences, intracellular transport properties and raft association of H3 HA were examined. Scanning alanine mutagenesis of the HA TM domain was performed (Fig. 1A), and the substitution mutants were expressed constitutively in MDCK cells. Pulse–label and pulse–chase analysis indicated the carbohydrate chains of the mutant HA proteins were modified to the Golgi mature form with kinetics similar to WT HA (Fig. 1B), but some of the HA mutants (530–532, 533–535, 536–538, and 539–541) did show heterogeneously migrating HA bands indicative of additional carbohydrate modifications. Analysis of cell surface expression levels of MDCK cells and 293T cells transiently expressing HAs showed that all HA mutants were expressed at levels equivalent to or greater than WT HA (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Analysis of the solubility of mutant HAs in 0.25% TX-100 at 4°C showed that WT HA was ≈70% insoluble, whereas the control protein VSV G protein was >90% soluble, which is in agreement with previous results (10, 13) (Fig. 1C). Three HA mutants (530–532, 533–535, and 536–538) were solubilized almost completely by TX-100 (Fig. 1C), suggesting these three mutant HA proteins were not associated with rafts (nonraft). To confirm that HA raft association is a dynamic process (29), a pulse–label–chase analysis was coupled with TX-100 extraction and preparation of soluble and insoluble fractions. It was observed for raft-associated HA mutant 548–550 that TX-100 insolubility occurred at a time concomitant with acquisition of Golgi mature carbohydrate chains (Fig. 1D). Mutant HA 533–535 acquired Golgi mature carbohydrate chains but not TX-100 insolubility, confirming that segregation of HA into rafts occurs within Golgi cisternae.
Fig. 1.
Alanine-scanning mutagenesis and characterization of the mutant HA proteins. (A) Schematic diagram showing the amino acid substitutions made in the influenza virus HA TM domain. ED, ectodomain; CYT, cytoplasmic tail. (B) Carbohydrate maturation of WT and mutant HA proteins in MDCK cells stably expressing WT or mutant HAs. (C) Solubility of HA in 0.25% TX-100 at 4°C in BHK cells transiently expressing WT and mutant HAs. S, soluble fractions; I, insoluble fractions. (D) Correlation between HA carbohydrate maturation and TX-100 insolubility in BHK cells transiently expressing HAs. S, soluble fractions; I, insoluble fractions. (E) Flotation sucrose density gradient of 0.25% TX-100-lysed HeLa-T4 cells transiently expressing WT and mutant HAs, NA, M1, and M2 proteins. Cav, caveolin; TfR, transferrin receptor; GM1, GM1 ganglioside. GM1 was detected by a dot blot analysis using a peroxidase-conjugated cholera toxin B subunit.
To conduct a more rigorous analysis for raft association, a flotation sucrose density analysis was performed on TX-100-solubilized cells, and polypeptides were immunoprecipitated. The raft markers, ganglioside GM1, and caveolin (30) together with WT HA and influenza virus NA integral membrane protein were found to float in light density fractions (Fig. 1E), as found (10). In contrast, the known nonraft-associated cellular protein transferrin receptor, the influenza virus matrix protein (M1), and the influenza virus M2 ion channel protein did not float and were found in the loading fraction at the bottom of the gradient. Under these conditions the three HA mutants 530–532, 533–535, and 536–538 were found in the bottom fractions, indicating that they are nonraft proteins.
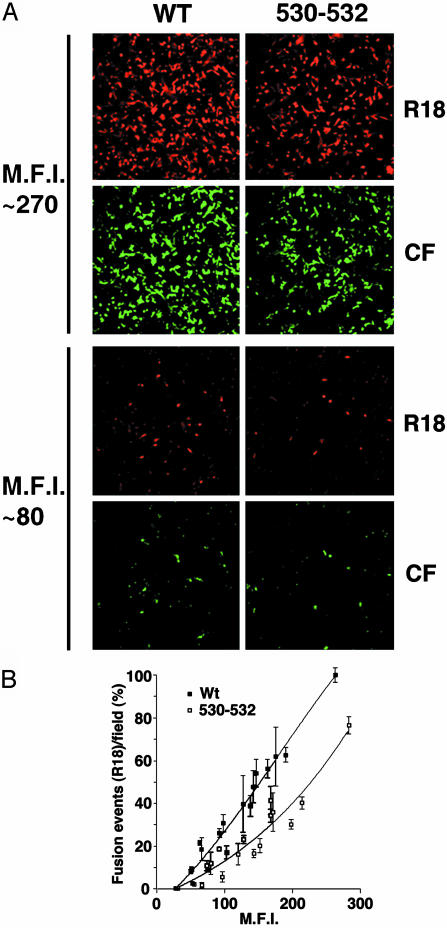
Higher Cell Surface Densities of Nonraft HA Are Required to Cause Equivalent Levels of Complete Fusion as WT HA. HA protein has two known biological activities: sialic acid receptor binding (hemadsorption) and virus–cell or cell–cell fusion (reviewed in ref. 31). Hemadsorption activity of WT and nonraft mutant HA was examined at varying cell surface densities of HA, and no difference between WT and nonraft HA was observed (data not shown). To examine fusion activity of WT and nonraft mutant HA, lipid mixing and aqueous content mixing assays were performed by using BHK effector cells expressing different surface densities of HA and dual-labeled human erythrocyte (RBC) target cells (R18 for lipid mixing and CF for content mixing) (Fig. 2A). The number of fusion events between dual-labeled R18/CF-labeled RBCs and the HA protein-expressing BHK cells increased with increasing cell surface expression of HA protein (Fig. 2B). At all HA expression levels, the cells showed both lipid mixing and content mixing for either WT or nonraft HA 530–532-expressing cells. Unrestricted hemifusion (R18 dye spread) in the complete absence of content mixing (no CF dye spread) was not observed even at low HA expression levels. However, the number of the fusion events measured for nonraft HA 530–532 protein was reduced compared to WT HA protein (Fig. 2B). Thus, the data suggest that although the nonraft HA 530–532 protein is competent for inducing complete fusion, association of HA with rafts promotes fusion activity, perhaps because rafts concentrate sufficient HA molecules for efficient fusion activity.
Fig. 2.
Nonraft HA has reduced cell–cell fusion capacity as compared with WT HA at equivalent overall surface density of HA. (A) HA fusion activity of WT and nonraft mutant HA 530–532 was measured by binding RBCs dual-labeled with lipid probe octadecyl rhodamine (R18) and aqueous cytoplasmic probe CF to BHK cells transiently expressing HAs and triggering fusion by low pH treatment (pH 4.8 for 1 min). MFI, mean fluorescent intensity. (B) Quantification of fusion events for BHK cells expressing different cell surface densities of HAs as measured by flow cytometry. Average fusion events and standard deviations are from three to five microscopic fields per experiment. MFI, mean fluorescent intensity.
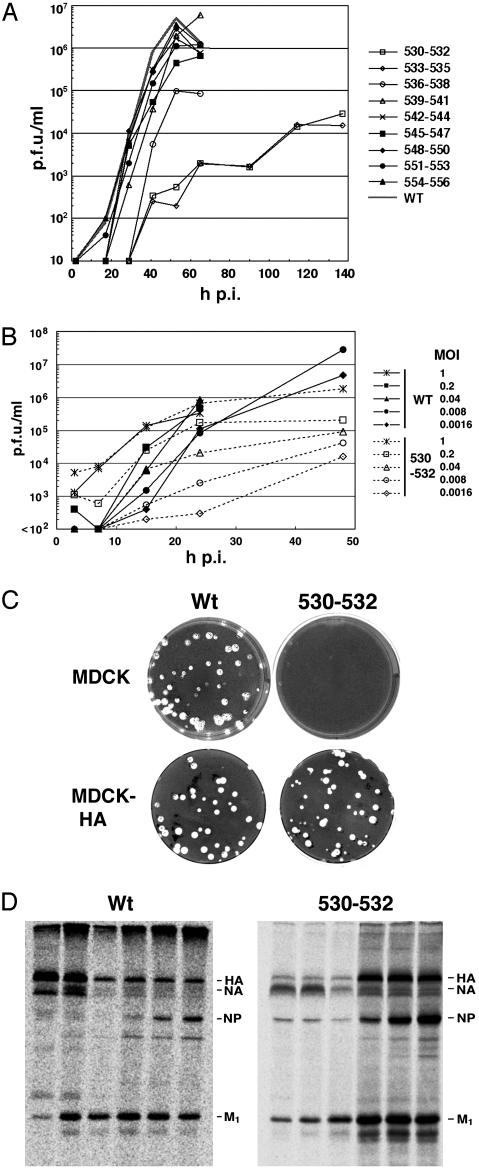
Raft Association of the HA Protein Is Essential for Efficient Virus Replication. To examine the effect of HA mutants that do not associate with lipid rafts on virus replication, the HA alanine-scanning TM domain mutations (Fig. 1 A) were introduced into influenza virus (A/Udorn/72) by using reverse genetics procedures (22, 23). Nonraft HA mutant viruses were recovered on a time scale significantly slower than that for WT virus or other mutant viruses where HA remained associated with rafts (Table 2, which is published as supporting information on the PNAS web site). The complete nucleotide sequence of the HA gene of all recovered mutant viruses was determined and validated.
Growth curves of the recovered viruses that contain HA TM domain alterations were examined after infection of MDCK cells at low multiplicity of infection (moi) [0.001 plaque-forming units (pfu) per cell] (Fig. 3A). The nonraft HA mutants 530–532 and 533–535 had a slower growth rate and a titer ≈3 logs lower than WT virus. Nonraft HA mutant 536–538 had a somewhat slower growth rate and a titer 1.5 log lower than WT virus. Mutations in the HA TM domain, which did not affect raft association, showed growth rates and titers very similar to WT virus. The growth defect of nonraft HA virus (e.g., mutant 530–532, Fig. 3B) was more obvious at low moi, when the virus has to go through multiple cycles of infection. As described below, this finding is likely caused by the combination of two defects: a reduction in budding, combined with a reduction in fusion.
Fig. 3.
Characterization of influenza viruses containing HA TM alanine substitutions: replication kinetics, trans complementation, and flotation gradient analysis. WT influenza virus and nine HA TM domain mutant viruses were recovered by using reverse genetics methods from cloned DNAs. (A) Growth curve of influenza viruses by using a moi of 0.001 pfu per cell. Viral titers were determined by plaque assay on MDCK-HA cells. h p.i., hours postinfection. (B) Effect of different moi on nonraft HA 530–532 virus growth rates. Viruses were grown in MDCK cells at the moi shown and plaqued on MDCK-HA cells. (C) Viral plaques of WT virus and HA 530–532 virus on MDCK and MDCK-HA cells. (D) TX-100 solubilization and flotation analysis of HeLa-T4 cells infected with WT or HA 530–532 viruses.
The growth curve plaque assays were performed on an HA-complementing cell line in which HA was expressed constitutively (MDCK-HA) (Fig. 3C). The normal size of plaques on MDCK-HA cells of mutant HA 530–532 virus confirms that the HA 530–532 mutation affects only the HA protein and makes it unlikely there is an additional effect on a property of the genome HA RNA segment, such as RNA packaging (32). For further characterization of a virus containing a nonraft HA TM domain mutation, we selected the HA 530–532 virus.
To examine the interactions of viral proteins with lipid rafts in virus-infected cells, HeLa-T4 cells were infected with WT and HA 530–532 virus, and TX-100 lysates were subjected to flotation analysis and polypeptide immunoprecipitation (Fig. 3D). As we found (10), in WT virus-infected cells both glycoproteins HA and NA floated to the top fractions. In contrast, for the HA 530–532 virus-infected cells, the NA protein floated to the top fractions, whereas the HA protein remained in the soluble bottom fractions. These data indicate that the nonraft HA protein can be segregated from the raft-associated NA protein in virus-infected cells.
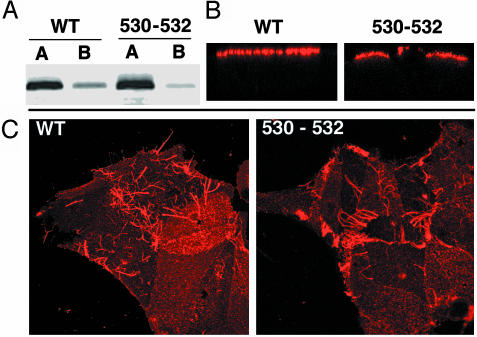
Both HA and NA proteins are targeted to the apical cell surface of polarized epithelial cells, and budding of influenza virus occurs from apical surfaces (reviewed in ref. 33). To ensure that the nonraft HA 530–532 mutation had not caused an alteration to the distribution of HA in polarized cells, we examined virus-infected polarized MDCK cells both by surface biotinylation of HA at apical and basolateral surfaces and by indirect immunofluorescence microscopy and Z-scans of cells. The WT HA and HA 530–532 distribution patterns were indistinguishable and predominantly at the apical surface (Fig. 4 A and B). Filamentous influenza virus particle formation is characteristic for field-isolated strains, including A/Udorn/72, and can be readily visualized by immunofluorescence microscopy (34). Examination of cells infected with WT and nonraft HA 530–532 virus showed indistinguishable patterns for the formation of filamentous virus (Fig. 4C). Thus, in the context of an influenza virus infection nonraft mutant HA was targeted to apical surfaces. Furthermore, nonraft HA did not affect the formation of filamentous influenza A/Udorn/72 virus.
Fig. 4.
Nonraft HA 530–532 is targeted to the apical surface of polarized cells. (A) MDCK cells were grown on 24-mm diameter, 0.4-μm pore size Transwell polycarbonate filters (Costar) and infected with WT or nonraft HA 530–532 virus from the apical cell surface at a moi of 3.0 pfu per cell. Monolayers were biotinylated at 5 h p.i. as described (10) and immunoprecipitated with HA-specific mAb C45/3 (26). Polypeptides were separated by SDS/PAGE and blotted to poly(vinylidene difluoride) membranes, and the surface-biotinylated proteins were detected by using Alexa 680-conjugated streptavidin and binding quantified with an Odyssey Infrared Imaging system (Li-Cor). (B) Localization of HA on polarized virus-infected MDCK cells. Cells were infected as above and stained with mAb C45/3 followed by incubation with Texas red-conjugated donkey anti-goat secondary antibody (Jackson ImmunoResearch). Fluorescence was observed with a Zeiss LSM 410 confocal microscope. Z-scans are shown. (C) Staining of virus-infected MDCK cells with goat anti-Udorn serum to examine whether production of filamentous virus production (a characteristic of field-isolated influenza A viruses including the A/Udorn/72 strain) depends on the interaction with rafts.
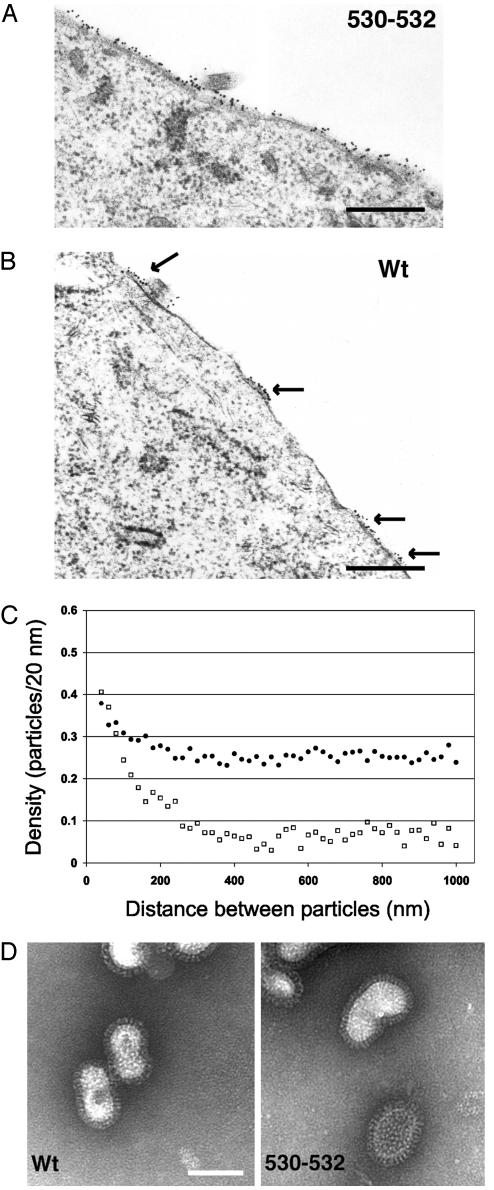
WT HA Is in Clusters at the Cell Surface; Nonraft HA Is Distributed More Uniformly and Is Budding Defective. It has been known for >30 years that in influenza virus-infected cells HA is distributed at the cell surface in clusters (e.g., ref. 35) in regions of the membrane that are not actively involved in budding. Moreover, it is generally thought that the locations of HA clusters become budding sites. The distribution of nonraft HA 530–532 at the surface of virus-infected cells was compared with that of WT HA. HA was stained by pre-embedding indirect immunogold labeling, and thin sections were examined in the electron microscope (Fig. 5 A and B). The distribution of gold particles in random regions of the plasma membrane not actively involved in budding (3,000 nm total membrane length measured from ≈30 cell profiles) were quantified by using the method of Brown and Lyles (28). Further information on the method of quantification is in Supporting Text, which is published as supporting information on the PNAS web site. As shown in Fig. 5A, for HA 530–532 virus-infected cells, 15-nm gold particles were observed in a wide distribution across the plasma membrane. In contrast, for WT HA virus-infected cells, HA was observed in clusters (arrows) with considerable regions of the cell surface devoid of HA staining. The y intercept of the plateau line represents the average concentration of label on the membrane, including both “raft associated” and “inter-raft” labeling. The x intercept of the ascending region represents the average size of the microdomain. For WT HA the majority of gold particles were localized to microdomains that have a diameter of 200–280 nm (Fig. 5C). For nonraft HA 530–532 there was no clear size of clusters, suggesting that little HA was found in microdomains, with the caveat that the microdomains could be extremely variable in size with a high degree of label found between the microdomains. We also observed that there was less overall density of gold particles for WT HA than nonraft HA 530–532, most likely reflecting a loss of WT HA caused by efficient budding (mutant HA 530–532 virus is budding impaired; see below). However, the lower overall density of WT HA may reflect less efficient gold labeling of HA because of tight packing in rafts. The simplest interpretation of these data is that WT HA is localized in clusters that most likely represent amalgams of rafts (barges of rafts, ref. 8), whereas nonraft HA 530–532 does not concentrate in clusters.
Fig. 5.
Distribution of HA at the plasma membrane: WT HA but not nonraft HA is clustered in microdomains. (A and B) MDCK cells were infected with nonraft HA virus (A) and WT virus (B) at a moi of 5 pfu per cell, and at 4.5 h p.i. cells were incubated with mAb C78/1 specific for Udorn HA protein followed by a secondary antibody conjugated to 15-nm gold particles and processed for sectioning, staining, and electron microscopy. Cell profiles were randomly selected and photographed at ×36,000. (Bar = 0.5 μm.) (C) The plasma membrane length and the gold particle distribution of ≈30 random cell profiles were measured by using the method of Brown and Lyles (28). •, Nonraft HA virus. □, WT virus. (D) Negatively stained purified influenza virions. Images are at ×89,360. (Bar = 100 nm.)
Virus budding was analyzed by quantifying on electron micrographs the number of virions budding from ≈500 μm of plasma membrane by using thin sections of WT virus and nonraft HA 530–532 virus-infected MDCK cells. It was found that for WT virus there were 1.79 ± 0.35 virions per μm, whereas for nonraft HA 530–532 virus there were 0.57 ± 0.09 virions per μm, a 3-fold drop in budding particles. Examination of the purified virions released from MDCK cells by negative staining and electron microscopy indicated the particles were indistinguishable (Fig. 5D). Thus, taken together these data suggest that although raft association of the HA protein is not necessary to form a morphologically normal virus, raft association does facilitate efficient virus budding.
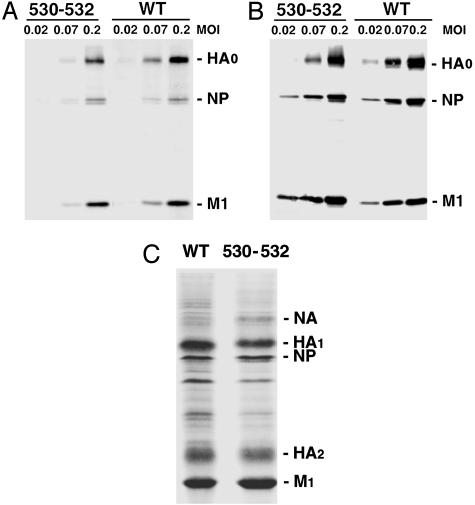
Raft Association Promotes Efficient Incorporation of HA into Virions. To compare the protein composition of WT and nonraft HA virions, MDCK cells were infected with the viruses at different moi, and particles released into the media were purified. The amount of each viral protein incorporated into virus particles as compared with the amount of each viral protein synthesized in infected cells was determined by immunoblotting and quantification. Comparable amounts of WT and nonraft HA virus proteins were detected in infected cells (Fig. 6B). However, the amount of the HA protein in the nonraft HA virus was less (≈60%) than that for the WT virus when normalized to the amounts of the M1 protein from purified WT and nonraft HA 530–532 (Fig. 6A). To confirm this observation, the polypeptide composition of purified WT and the nonraft HA viruses was compared after Coomassie brilliant blue staining and imaging (Fig. 6C). When the amounts of the M1 protein from purified WT and nonraft HA 530–532 were normalized, it was found that the amount of the HA protein in the nonraft HA 530–532 virus was ≈55% of that found in WT virus. It was also observed that nonraft HA virions contained an increased amount of NA protein (Fig. 6C). It is possible that the decreased amount (≈55%) of HA protein may be compensated for by increased incorporation of NA protein spike. As HA and NA are hard to distinguish in virus particles by electron microscopy, it is difficult to observe a difference in density of the glycoprotein spikes between the WT and the nonraft HA viruses (Fig. 5D). Taken together the data suggest that raft association of HA protein promotes its efficient incorporation virions at the raft-lipid budding sites.
Fig. 6.
Nonraft HA virions have decreased HA protein composition. (A and B) Protein composition of WT and nonraft HA virus. MDCK cells were infected with WT and nonraft mutant (530–532) viruses at varying moi indicated. At 18 h p.i., the culture media (A) and the cells (B) were harvested. The polypeptide composition of both cell lysates and purified virions was analyzed by immunoblotting using an anti-influenza virus specific serum. The ratio of HA, NP, and M1 proteins in virions as compared with cells was quantified. (C) Coomassie brilliant blue-stained polypeptides of purified WT and mutant (530–532) viruses grown in MDCK cells are shown.
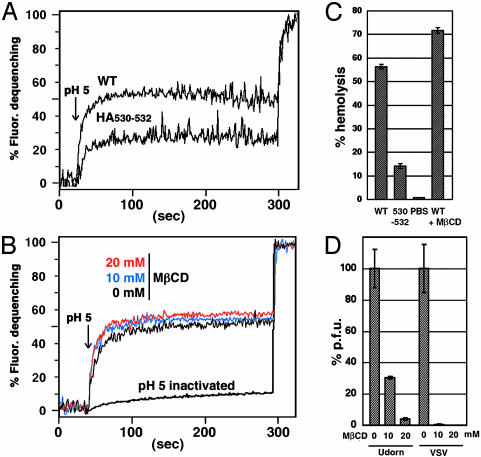
Nonraft HA Virus Shows Reduced Fusion Activity. The data presented above suggest that influenza virus HA is associated with rafts to concentrate HA into virus particles during budding. However, we also thought it possible that the nonraft HA in the context of a virus infection may be defective in biological activity. This notion seemed possible based on the observation of the large drop in infectivity (3 logs) of nonraft HA 530–532 virus when grown at low multiplicity (Fig. 3A), compared with the observation that the amount of HA protein in purified nonraft HA 530–532 virus when grown at high multiplicity was reduced only ≈40%. Furthermore, we have shown that nonraft HA 530–532 has reduced fusion activity for an equivalent surface density of HA as compared with WT HA (Fig. 2). As hemadsorption was not affected (data not shown), fusion activity of purified virions was tested by using a fluorescence dequenching fusion assay. For an equivalent amount of purified virus (measured by protein concentration), the nonraft HA virus exhibited a decrease in both the initial rate and the final extent of virus fusion to RBC ghosts as compared with WT virus (Fig. 7A).
Fig. 7.
Nonraft HA virus has reduced fusion activity and does not require cholesterol for fusion. (A) Virus–RBC ghost fusion was assayed by fluorescence dequenching. Purified WT and nonraft 530–532 virions were labeled with R18 and bound to sealed human RBC ghosts. Fusion between the R18-labeled virus and the RBC ghosts was triggered by lowering the pH from 7.0 to 5.0 (arrow). After 300 s of fluorescence recording TX-100 was added to the cuvette to obtain the maximum dequenching. (B) Fusion activity of MβCD-treated virus. R18-labeled WT virus was incubated in PBS (pH 7.0) for 15 min at 37°C in the absence or presence of 10 mM or 20 mM MβCD, and virions were repurified through a sucrose cushion. Fusion kinetics between the viruses and ghosts were analyzed as described above. To inactivate HA protein the R18-labeled virus was incubated in PBS (pH 5.0) for 15 min at 37°C. (C) Hemolysis assay as a surrogate for membrane fusion content mixing. WT and nonraft 530–532 virus (3.3 μg protein) or PBS were incubated with human RBCs for 45 min at 4°C. The RBCs were then incubated in a low pH buffer (PBS pH 5.0) for 1 min at room temperature. After replacing the low pH buffer with a neutral pH PBS, the RBCs were incubated for 5 min at 37°C. The amount of released hemoglobin was determined spectrophotometrically at 540 nm. The hemolysis capacity of WT virus treated with 20 mM MβCD (WT + MβCD) was also determined. RBCs lysed in 1% TX-100 was set at 100% hemolysis. (D) Reduction of plaque-forming capacity by MβCD. Influenza virus and VSV were incubated in PBS (pH 7.0) for 15 min at 37°C in the absence or presence of 10 mM or 20 mM MβCD. MβCD was removed by pelleting the virus through a 30% sucrose cushion, and infectivity of the pelleted virus was determined by plaque assay. Pfu of the MβCD-untreated viruses are shown as 100%.
Although a lower density of HA in nonraft HA virus explains its lowered virus to cell fusion activity, it is also possible that HA protein is stabilized or clustered via raft lipids for efficient virus to cell fusion. A requirement for cholesterol in HA-mediated fusion using the H1 and H2 subtype HAs has been tested but not found (36–38). However, to test a possible requirement for cholesterol in the context of a viral infection with the H3 subtype HA, WT A/Udorn/72 virus was depleted of cholesterol by treating with methyl-β-cyclodextrin (MβCD) (10 mM MβCD removed >75% of the cholesterol in 4.0 μg virus as determined with the Molecular Probes assay), the virus was repurified, and virus to cell fusion activity was analyzed. MβCD-treated virus showed similar R18 fluorescence dequenching kinetics to untreated virus (Fig. 7B), indicating lipid mixing occurs normally.
Nonetheless, MβCD-treated virus might be able to permit lipid mixing (hemifusion) but fail to cause aqueous content mixing. However, this is a difficult point to prove because a simple dye transfer assay for influenza virus–cell fusion has not been developed. Moreover, we cannot interpret meaningfully the fusion activity of virus grown in cells treated with simvastatin, an inhibitor of cholesterol synthesis, because such virus would be the equivalent of nonraft mutant HA virus. Furthermore, cholesterol is required for surface transport of HA (39). Thus, as a surrogate assay for content mixing, we measured the ability of 3.3 μg each of WT and nonraft HA mutant virus to cause hemolysis of RBCs. The nonraft HA virus was found to cause hemolysis less efficiently than the WT virus; whereas MβCD-treated WT virus caused hemolysis as efficiently as the non-MβCD-treated WT virus (Fig. 7C). Even though MβCD treatment of influenza virus did not affect HA-mediated lipid mixing or hemolysis, MβCD treatment drastically reduces infectivity of both influenza virus (>95% at 20 mM MβCD) and VSV (>3 logs at 20 mM MβCD) (Fig. 7D) by an unknown mechanism, possibly related to the finding that MβCD treatment of HIV-1 causes perforation of the viral membrane with loss of viral genomes (40).
Perspectives
Although the association of HA with rafts is established (10, 14–16, 29) and it is known that influenza virus buds from rafts (10, 16), we have shown here that the lipid raft is used as a platform to concentrate sufficient HA into virions for efficient virus fusion. When HA cannot associate with rafts, virus infectivity decreases ≈3 logs. HA is a trimer, and there has been speculation as to the number of trimeric molecules that are required for membrane fusion (reviewed in ref. 31). In a detailed study in which fusion kinetics of cells expressing different surface densities of HA was measured it was calculated that minimally three to four trimers were required for fusion (41). Other data also suggest that the fusion site is ringed by multiple copies of the HA trimer that act synergistically to provide the conformational energy needed to drive the fusion reaction (42, 43). An extrapolation of our work corroborates the notion that multiple HA trimers are concentrated to mediate membrane fusion. However, our experiments cannot formally eliminate a requirement for cholesterol in the virus–cell fusion process, particularly the final aqueous content mixing stage, because of the lack of an assay for direct virus–cell content mixing. Another possible related model for HA concentrating in rafts is that the TM domains of trimeric HA promote association with other HA trimers. These larger arrays of HA trimers might have affinity for raft lipids and increased fusion activity. A lack of raft association of mutant HAs could be a consequence of a failure to form a higher-order structure independent of lipid rafts.
The quantitative electron microscopy approach (28) to determine the distribution of WT HA and nonraft mutant HA on the cell surface is a powerful method for examining virus assembly. The sites of clusters of HA, ranging from 200 to 280 nm, are consistent with these sites evolving to become budding sites, and the size of these HA clusters adds credence to the notion of individual rafts condensing to become barges of rafts (8). Reduced budding of nonraft HA virus is consistent with concentration of HA at budding sites and the formation of contacts between the HA and NA cytoplasmic tails and the viral M1 protein. Although there is some redundancy in the presumed M1 protein to HA or NA cytoplasmic tail interactions (26), a reduced density of nonraft HA would be anticipated to cause a deficiency in viral assembly.
The influenza virus NA protein is also targeted to lipid rafts and alterations to the NA TM domain ablate raft association (reviewed in ref. 33). We anticipate that NA is concentrated in rafts for its normal incorporation into virions. In the future it will be of interest to construct a double mutant virus that is nonraft HA and nonraft NA and to examine its efficiency of budding. It seems most likely that budding would be impaired as neither HA nor NA would be concentrated and the M1 protein would have more difficulty coalescing with the HA and NA cytoplasmic tails.
Supplementary Material
Acknowledgments
We thank Dr. Anthony Schmitt for his help with the raft biochemical fractionation, Dr. Koji Sakai for providing pKS336 plasmid used for establishing the MDCK-HA cell lines, and all members of the Lamb laboratory and Drs. Richard W. Compans, Robert W. Doms, Douglas S. Lyles, John K. Rose, Fred S. Cohen, and Kai Simons for helpful suggestions. This research was supported in part by National Institute of Allergy and Infections Diseases Grants R37 AI-20201 and R01 AI-23173. M.T. and C.J.R. are Associates of the Howard Hughes Medical Institute, and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: TX-100, Triton X-100; HA, hemagglutinin; TM, transmembrane; MDCK, Madin-Darby canine kidney; VSV, vesicular stomatitis virus; CF, carboxyfluorescein; NA, neuraminidase; moi, multiplicity of infection; pfu, plaque-forming units; MβCD, methyl-β-cyclodextrin; h p.i., hours postinfection.
See accompanying Biography on page 14607.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
References
- 1.Simons, K. & Ikonen, E. (1997) Nature 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 2.Suomalainen, M. (2002) Traffic 3, 705–709. [DOI] [PubMed] [Google Scholar]
- 3.Pralle, A., Keller, P., Florin, E.-L., Simons, K. & Horber, J. (2000) J. Cell Biol. 148, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varma, R. & Mayor, S. (1998) Nature 394, 798–801. [DOI] [PubMed] [Google Scholar]
- 5.Simons, K. & Toomre, D. (2000) Nat. Rev. Mol. Cell. Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 6.Cary, L. A. & Cooper, J. A. (2000) Nature 404, 945–947. [DOI] [PubMed] [Google Scholar]
- 7.Ono, A. & Freed, E. O. (2001) Proc. Natl. Acad. Sci. USA 98, 13925–13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindwasser, O. W. & Resh, M. D. (2001) J. Virol. 75, 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheiffele, P., Rietveld, A., Wilk, T. & Simons, K. (1999) J. Biol. Chem. 274, 2038–2044. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, J., Pekosz, A. & Lamb, R. A. (2000) J. Virol. 74, 4634–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali, A., Avalos, R. T., Ponimaskin, E. & Nayak, D. P. (2000) J. Virol. 74, 8709–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen, D. H. & Hildreth, J. E. K. (2000) J. Virol. 74, 3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, D. A. & Rose, J. K. (1992) Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- 14.Fiedler, K., Kobayashi, T., Kurzchalia, T. V. & Simons, K. (1993) Biochemistry 32, 6365–6373. [DOI] [PubMed] [Google Scholar]
- 15.Lin, S., Naim, H. Y., Rodriguez, A. C. & Roth, M. G. (1998) J. Cell Biol. 142, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheiffele, P., Roth, M. G. & Simons, K. (1997) EMBO J. 16, 5501–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong, K. H., Zacchetti, D., Schneeberger, E. E. & Simons, K. (1999) Proc. Natl. Acad. Sci. USA 96, 6241–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiedler, K., Lafont, F., Parton, R. G. & Simons, K. (1995) J. Cell Biol. 128, 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puertollano, R., Martin-Belmonte, F., Millan, J., del Carmen de Marco, M., Albar, J. P., Kremer, L. & Alonso, M. A. (1999) J. Cell Biol. 145, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paterson, R. G. & Lamb, R. A. (1993) in Molecular Virology: A Practical Approach, eds. Davidson, A. & Elliott, R. M. (IRL, Oxford), pp. 35–73.
- 21.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1994) Current Protocols in Molecular Biology (Wiley, New York), Vols. 1–3.
- 22.Takeda, M., Pekosz, A., Shuck, K., Pinto, L. H. & Lamb, R. A. (2002) J. Virol. 76, 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, G., Watanabe, T., Ito, H., Watanabe, S., Goto, H., Gao, P., Hughes, M., Perez, D. R., Donis, R., Hoffmann, E., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9345–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterson, R. G., Russell, C. J. & Lamb, R. A. (2000) Virology 270, 17–30. [DOI] [PubMed] [Google Scholar]
- 25.Jin, H., Subbarao, K., Bagai, S., Leser, G. P., Murphy, B. R. & Lamb, R. A. (1996) J. Virol. 70, 1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, H., Leser, G. P., Zhang, J. & Lamb, R. A. (1997) EMBO J. 16, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leser, G. P., Ector, K. J. & Lamb, R. A. (1996) Mol. Biol. Cell 7, 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, E. L. & Lyles, D. S. (2003) Virology 310, 343–358. [DOI] [PubMed] [Google Scholar]
- 29.Skibbens, J. E., Roth, M. G. & Matlin, K. S. (1989) J. Cell Biol. 108, 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harder, T., Scheiffele, P., Verkade, P. & Simons, K. (1998) J. Cell Biol. 141, 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- 32.Fujii, Y., Goto, H., Watanabe, T., Yoshida, T. & Kawaoka, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 2002–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayak, D. P. & Barman, S. (2002) Adv. Virus Res. 58, 1–28. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, P. C., Lamb, R. A. & Compans, R. W. (1998) Virology 240, 127–137. [DOI] [PubMed] [Google Scholar]
- 35.Compans, R. W. & Dimmock, N. J. (1969) Virology 39, 499–515. [DOI] [PubMed] [Google Scholar]
- 36.White, J., Kartenbeck, J. & Helenius, A. (1982) EMBO J. 1, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn, A., Gibbons, D. L. & Kielian, M. (2002) J. Virol. 76, 3267–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Razinkov, V. I. & Cohen, F. S. (2000) Biochemistry 39, 13462–12468. [DOI] [PubMed] [Google Scholar]
- 39.Keller, P. & Simons, K. (1998) J. Cell Biol. 140, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham, D. R., Chertova, E., Hilburn, J. M., Arthur, L. O. & Hildreth, J. E. (2003) J. Virol. 77, 8237–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danieli, T., Pelletier, S. L., Henis, Y. I. & White, J. M. (1996) J. Cell Biol. 133, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blumenthal, R., Sarkar, D. P., Durell, S., Howard, D. E. & Morris, S. J. (1996) J. Cell Biol. 135, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markovic, I., Leikina, E., Zhukovsky, M., Zimmerberg, J. & Chernmordik, L. V. (2001) J. Cell Biol. 155, 833–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.