Abstract
Context
Reliable methods to screen newborns for congenital cytomegalovirus (CMV) infection are needed for identification of infants at increased risk for hearing loss. Since dried blood spots (DBS) are routinely collected for metabolic screening from all newborns in the United States, there has been interest in using DBS polymerase chain reaction (PCR)-based methods for newborn CMV screening.
Objective
To determine the diagnostic accuracy of DBS real-time PCR assays for newborn CMV screening
Design, Setting, and Participants
Between March 2007 and May 2008, infants born at seven medical centers in the U.S. were enrolled in the CMV and Hearing Multicenter Screening (CHIMES) study. Newborn saliva specimens were tested for the detection of early antigen fluorescent foci (DEAFF). Results of saliva DEAFF were compared with a single-primer (from 03/07 to 12/07) and a two-primer (from 01/08 to 05/08) DBS real-time PCR. Infants positive by screening DEAFF or PCR were enrolled in follow-up to confirm congenital infection by the reference standard method, DEAFF on saliva or urine.
Main Outcome Measures
Sensitivity, specificity and positive and negative likelihood ratios (LRs) of single-primer and two-primer DBS real-time PCR assays for identifying infants with confirmed congenital CMV infection.
Results
Congenital CMV infection was confirmed in 92 of 20,448 (0.45%; 95% CI, 0.36–0.55) infants. Ninety-one of 92 infants were saliva DEAFF positive on screening. Of the 11,422 infants screened using the single-primer DBS PCR, 17 of 60 (28%) infants were positive with this assay, whereas, among the 9,026 infants screened using the two-primer DBS PCR, 11 of 32 (34%) infants were positive. The single-primer DBS PCR identified congenital CMV infection with a sensitivity of 28.3% (95% CI, 17.4–41.4%), specificity, 99.9% (95% CI, 99.9–100%), positive LR, 803.7 (95% CI, 278.7–2317.9), and negative LR, 0.7 (95% CI, 0.6–0.8). The positive and negative predictive values of the single-primer DBS PCR were 80.9% (95% CI, 58.1–94.5%) and 99.6% (95% CI, 99.5–99.7%), respectively. The two-primer DBS PCR assay identified infants with congenital CMV infection with a sensitivity of 34.4% (95% CI, 18.6–53.2%), specificity, 99.9% (95% CI, 99.9–100%), positive LR, 3088.9 (95% CI, 410.8–23226.7), and negative LR, 0.7 (95% CI, 0.5–0.8). The positive and negative predictive values of the two-primer DBS PCR were 91.7% (95% CI, 61.5–99.8%) and 99.8% (95% CI, 99.6–99.9%), respectively.
Conclusions
Among newborns, CMV testing with DBS real-time PCR compared with saliva rapid culture had low sensitivity, limiting its value as a screening test.
Cytomegalovirus (CMV) is an important cause of congenital infection and a leading cause of sensorineural hearing loss (SNHL) in children.1–5 Of the estimated 20,000 to 40,000 infants born each year with congenital CMV infection in the U.S., most (90% to 95%) have no detectable clinical abnormalities at birth and, thus, will not be identified by routine clinical examination.2, 6, 7 Furthermore, SNHL occurs in approximately 10–15% of infants with clinically inapparent congenital CMV infection and the majority of children with CMV-related SNHL will have late onset and/or progressive losses.1, 8, 9 Therefore, both routine physical examination and newborn hearing screening will miss many children who develop SNHL secondary to congenital CMV infection. To identify these at-risk infants early in life, rapid, reliable, and relatively inexpensive methods to screen newborns for congenital CMV infection are needed (NIDCD Workshop on Congenital Cytomegalovirus Infection and Hearing Loss: http://www.nidcd.nih.gov/funding/programs/hb/cmvwrkshop.htm). Identification of children at increased risk for CMV-associated SNHL early in life will allow targeted monitoring of these children in order to intervene at critical stages of acquisition of speech and language skills.10
Although traditional virus isolation from saliva or urine samples in tissue culture is considered the standard method for identification of infants with congenital CMV infection, it is not amenable to mass screening (even when modified to produce rapid results) because it is labor- and resource-intensive and requires tissue culture facilities. Real-time polymerase chain reaction (PCR) technology, in contrast, is well-suited for mass screening. A variety of newborn specimens including saliva, urine and dried blood spots (DBS) can be tested with PCR-based methods for the diagnosis of congenital CMV infection.11–18 Since DBS are collected routinely for newborn metabolic screening from all infants born in the U.S., there has been considerable interest in utilizing PCR assays for the detection of CMV in newborn DBS samples. The advantages of DBS PCR for newborn CMV screening include: 1) the specimens are already routinely collected for metabolic screening in all states; 2) PCR can detect viral DNA in DBS samples from CMV-infected infants; 3) PCR requires no tissue culture facilities; and 4) PCR is amenable to automation, so large numbers of specimens may be screened at relatively low cost. Despite the benefits of DBS PCR-based methods for screening infants for congenital CMV infection, the sensitivity and specificity of these assays for universal newborn screening have not been determined. Most reports have studied selected infant populations and none has prospectively compared the results of a DBS PCR assay to a standard (i.e., tissue culture) method for identifying CMV infection in an unselected newborn population.12–15, 19–21 This study examined the diagnostic accuracy of real-time PCR analysis of DBS as an approach for mass screening of newborns for congenital CMV infection.
METHODS
Study population
Between March 2007 and May 2008, infants born at seven medical centers in different geographic regions of the U.S. were enrolled prospectively in the National Institute on Deafness and Other Communication Disorders (NIDCD) CMV and Hearing Multicenter Screening (CHIMES) study. The study hospitals where the infants were born included: University of Alabama Hospital, Birmingham, AL; The University of Mississippi Medical Center, Jackson, MS; Carolinas Medical Center, Charlotte, NC; Saint Peter’s University Hospital, New Brunswick, NJ; Good Samaritan Hospital, Cincinnati, OH; Magee Women’s Hospital, Pittsburgh, PA; and Parkland Memorial Hospital, Dallas, TX. Mothers were approached postpartum to obtain informed consent for their newborn’s enrollment in the study. Upon enrollment, saliva specimens were collected from infants along with additional blood spot obtained at the time of newborn metabolic screening. The DBS sample for the study was collected only after the completion of metabolic screening and infants were not subjected to additional heel sticks for the CHIMES study. Infants with positive saliva or DBS screening samples were enrolled in the follow-up component of the study to confirm congenital CMV infection. The follow-up is ongoing with infants being evaluated for hearing outcome during the first 4 years of life. Data on race and ethnicity for all participating infants as self reported by their parents were collected since the prevalence of congenital CMV infection has been shown to vary according to racial and ethnic composition of the delivery population.22, 23 Written informed consent was obtained from a parent or guardian for each infant enrolled in the CHIMES study and Institutional Review Board approval was obtained at each site.
Specimen Collection
Saliva specimens were collected from the enrolled newborns at a mean age of 0.9 ± 0.6 days and before nursery discharge. DBS specimens were collected at the time of newborn metabolic screening and the mean age at collection of DBS samples was 1.9 ± 1.8 days. The saliva samples were collected by swabbing inside the baby’s mouth using a sterile polyester fiber-tipped applicator (PurFybr Inc., Munster, IN) and placed in 1.0 mL of transport medium containing sucrose phosphate.24 The samples were stored at 4°C and transported to the Central Laboratory at the University of Alabama at Birmingham on ice within a week of collection. A temperature monitoring device was included in shipments to monitor for temperature variation during transport (TL20, 3M, St. Paul, MN). At the time of routine newborn metabolic screening, additional blood spots were collected on a separate filter paper (Whatman 903®, Florham Park, NJ) for the CHIMES study infants, placed in individual envelopes and stored in plastic re-sealable bags containing desiccant. DBS specimens were maintained at room temperature and shipped to the Central Laboratory once weekly.
Detection of CMV in saliva specimens
Mean interval between the collection of initial saliva samples and testing was 7.4 ± 4.0 days. The presence of CMV in saliva specimens was detected by a rapid culture method for the detection of early antigen fluorescent foci (DEAFF) using a monoclonal antibody against the CMV major immediate early antigen (IE-1) in duplicate wells of a 96-well microtiter plate.24, 25 Each run included two positive control wells inoculated with the AD169 strain of CMV at a titer producing approximately 100 infectious foci per well. A sample was considered positive if at least one focus of distinct nuclear fluorescence was detected in at least one well. This assay has been used to screen newborns for the diagnosis of congenital CMV infection at the Central Laboratory.24, 25 Individuals ascertaining the results of the DEAFF assay were blinded to the results of the DBS PCR assay and vice versa.
DNA extraction from DBS samples
From each DBS, two 3 mm discs were punched using the BSD600 automated filter paper puncher (BSD Robotics, Acacia Ridge, Queensland, Australia) into 1.5 mL sample tubes. The punched filter paper discs were processed to extract DNA using the Qiagen M48 robotic system with MagAttract technology according to the manufacturer’s instructions (Qiagen, Inc., Valencia, CA). The extracted DNA samples were stored at −20°C. A blank filter card was punched and included in each extraction run to serve as a negative control for DNA extraction and to monitor for cross contamination. In addition, a filter paper spotted with 10,000 copies of AD169 strain of CMV was punched and included in the extraction run to serve as a positive control and to monitor for consistency and reliability of the extraction protocol.
Real-Time Polymerase Chain Reaction
Mean interval between DBS specimen collection and PCR analysis was 14.6 ± 9.6 days. The detection of CMV DNA was performed using the ABI 7500 Real-time PCR System (Applied Biosystems Inc., Foster City, CA) and ABsolute™ QPCR Low ROX Mix (ABgene USA, Rockford, IL). The reaction mixture contained primers at a concentration of 900 nM and the probe at 250 nM. Each sample was run in duplicate and the 25 µL of reaction mixture contained 20 µL of master mix and 5 µL of test sample. To generate standard curves, each plate contained plasmid standards incorporating the target sequences in 10-fold dilutions ranging between 100,000 and 10 genomic equivalents (ge) per reaction. The real-time PCR amplification conditions have been described.26, 27 During the first ten months of the study, the real-time PCR assay included primers to detect the highly conserved AD-1 region of the major envelope glycoprotein B (gB).26–28 During the final five months of the study, the PCR method was modified to include a second primer set from the highly conserved immediate early 2 (IE-2) exon 5 region (forward primer GAG CCC GAC TTT ACC ATC CA, reverse primer CAG CCG GCG GTA TCG A and probe VIC - ACC GCA ACA AGA TT – MGBNFQ) in the PCR reaction in an effort to improve the sensitivity of the assay (GeneBank accession numbers, GU179001, AY446871, AY446870, FJ616285, AY446868). The real-time PCR was repeated on all samples with a positive signal in either well and a sample was considered positive if one or more ge per reaction were detected on both PCR runs. In addition, real-time PCR was repeated on DBS samples from infants with a positive saliva DEAFF assay that were negative on the first PCR run. The detection limit of our real-time PCR assay as determined by the sensitivity titration analysis was 250 ge/mL for the single-primer assay and 50 ge/mL for the two-primer assay (eAppendix).
Efficiency of DNA extraction DBS PCR performance characteristics
To determine whether the sensitivity of DBS real-time PCR for CMV DNA detection was influenced by the extraction method, detection of CMV DNA by the two-primer real-time PCR protocol was compared between a commercial column-extraction method (Qiagen, Inc., Valencia, CA) and the robot-extraction protocol used in this study (eAppendix). In addition, the amount of genomic DNA as determined by real-time PCR amplification of RNase P (TaqMan® RNase P control reagents kit, Applied Biosystems Inc., Foster City, CA) in 185 randomly selected DBS specimens from CMV-negative infants was compared between the robot- and column-extraction methods (eAppendix). A comparison of the two-primer real-time PCR assay and a previously described nested PCR protocol was undertaken to assess whether our real-time PCR method would be as or more sensitive for detecting CMV DNA than a standard nested PCR method (eAppendix). 12
Confirmation of Screening Results
To account for the possibility that saliva DEAFF assay may be less than 100% sensitive in identifying CMV-infected newborns, infants with positive saliva or DBS screening samples were enrolled in the follow-up component of the CHIMES study to confirm congenital CMV infection.24 The urine and repeat saliva obtained from these infants at the enrollment visit for the follow-up study were tested for CMV using the DEAFF assay described above. The DEAFF assay on the follow-up saliva or urine sample was considered the reference standard for this study and therefore, a confirmed congenital CMV infection was defined as identification of CMV in either saliva or urine obtained at enrollment into the follow-up study. Infants were considered to be uninfected if both the saliva and the urine samples were negative by the DEAFF method. Newborns who were negative for CMV by both screening assays (saliva DEAFF and DBS PCR) were enrolled in follow-up and not retested with the reference standard assay.
Data Analysis
Only infants who enrolled in the follow-up component of the study for confirmation of congenital CMV infection status were included in determining the diagnostic ability of our DBS real-time PCR assays. Sensitivity, specificity and predictive values for both the single-primer and the two-primer DBS real-time PCR assays were calculated using standard methods for proportions and exact 95% confidence limits. The positive predictive value was the ratio of true positives to all positive DBS PCR results and the negative predictive value was the ratio of true negatives to all negative DBS test results. Likelihood ratios (LRs) were calculated for summarizing the diagnostic accuracy of the DBS PCR assays. Positive LR was sensitivity/(1-specificity) and the negative LR was (1-sensitivity)/specificity. Confidence intervals for LRs were determined using the method described by Simel and colleagues.29 Statistical differences between nested and real-time PCR methods were calculated using the Chi-square test. All statistical analyses were performed using SAS software (SAS Institute, Inc., Cary, NC).
RESULTS
Study Population and Specimens
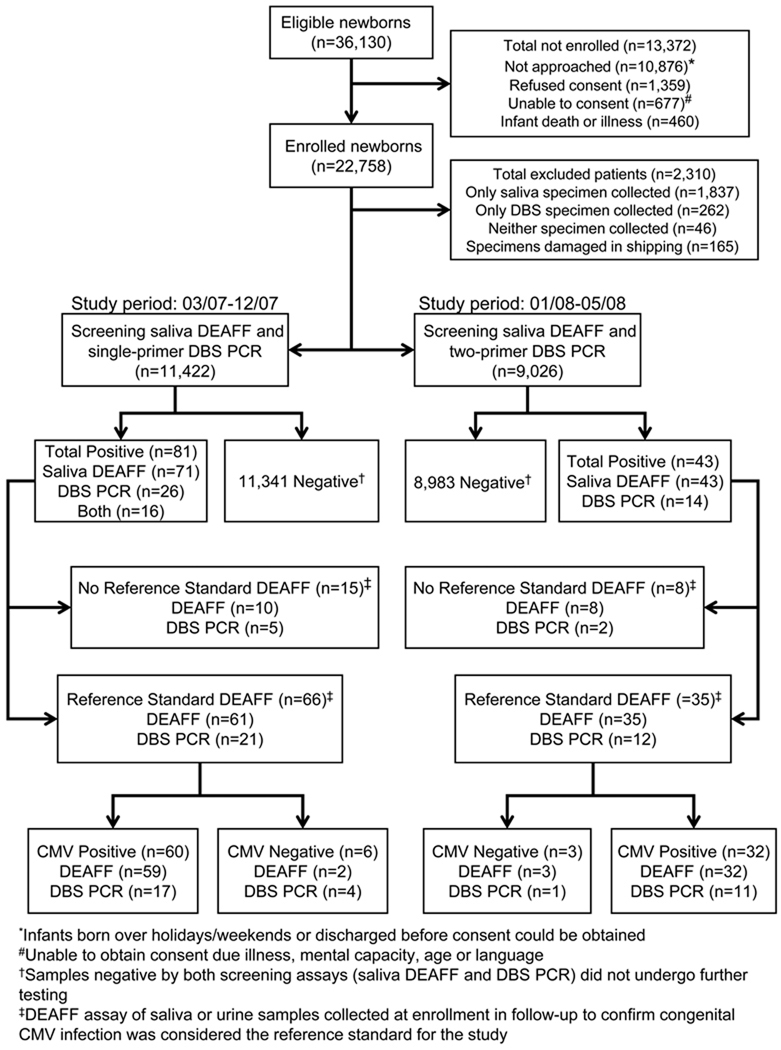
Of the 36,130 eligible infants, 22,758 (63%) infants were enrolled in the study. Although all live born infants were eligible for participation in the CHIMES study, some of the infants born over holidays or weekends and those discharged prior to obtaining consent for participation in the study (n=10,876) were not enrolled in the study. Additional reasons for non-enrollment in the study included: refusal to participate (n=1,359); unable to obtain consent due to maternal factors such as illness, mental capacity, age or language (n=677); and infant death or illness (n=460). Both saliva and DBS specimens were collected from 20,613 (91%) infants, only saliva specimens were collected from 1,837 infants, only DBS specimens were collected from 262 infants, and 46 infants had neither specimen collected (Figure 1). The reasons that both specimens were not available from these newborns included: 1) the infants were unavailable or discharged from the nursery prior to collection (n=1,214); 2) the newborn metabolic screening was completed before infants were enrolled in the study or there was insufficient blood left for the study DBS specimen (n=731); or 3) specimens were mislabeled or misplaced (n=200). The infants (n=2,145) who did not have both specimens collected were more likely to be in the neonatal intensive care unit than infants who had both specimens collected (14.7% vs. 2.9%, Χ2 test = 707.2, p < 0.0001). Of the 20,613 infants who had both specimens collected, saliva specimens from 165 infants could not be tested due to leakage or temperature variations during shipment (Figure 1). Thus, the study population comprises the 20,448 infants who had both saliva and DBS specimens collected and tested.
Figure 1.
Flow diagram demonstrating the evaluation of DBS real-time PCR assays for identifying infants with congenital CMV infection. DEAFF, indicates detection of early antigen fluorescent foci; DBS, dried blood spots; PCR, polymerase chain reaction; CMV, cytomegalovirus.
Characteristics of the study population are shown in Table 1. Most (97.1%) of the study infants were from the well baby nurseries. The infants were evenly distributed by sex. Mean maternal age was 27.3 ± 6.1 years. The mean age at enrollment into the follow-up study for confirmation of congenital CMV infection in infants positive by screening saliva DEAFF or DBS PCR was 6.4 ± 6.1 weeks of age. Overall, 92 of the 20,448 (0.45%; 95% CI, 0.36 – 0.55) infants had confirmed congenital CMV infection.
Table 1.
Study characteristics of 20,448 newborns who were tested by saliva DEAFF and DBS PCR assays for CMV infection
| Characteristic | No. (%) |
|---|---|
| Infant Sex | |
| Female | 10,026 (49.0) |
| Male | 10,422 (51.0) |
| Infant Race and Ethnicity | |
| Asian | 1,409 (6.9) |
| African American | 5,526 (27.0) |
| White, Hispanic | 4,765 (23.3) |
| White, Non Hispanic | 7,850 (38.4) |
| Other, including more than one race | 898 (4.4) |
| Maternal Age | |
| Mean ± Standard Deviation | 27.3 ± 6.1 |
| Median (range) | 27 (12–52) |
| Hospital Nursery | |
| Well Baby | 19,858 (97.1) |
| Neonatal Intensive Care | 590 (2.9) |
| Infants with confirmed congenital CMV infection | 92 (0.45) |
DEAFF indicates detection early antigen fluorescent foci; DBS, dried blood spot; PCR, polymerase chain reaction, CMV, cytomegalovirus
Newborn CMV screening with saliva DEAFF and the single-primer DBS PCR assays
Between March and December 2007, 11,422 newborns were screened for congenital CMV infection using saliva DEAFF and the single-primer DBS PCR assay (Figure 1). Eighty-one newborns tested positive for CMV infection by either saliva DEAFF assay (n=71), the DBS PCR assay (n=26), or both methods (n=16). Sixty-six of the 81 (81%) infants who tested positive by either screening method were enrolled in the follow-up study and, of those, 60 children were confirmed to have congenital CMV infection based on the positive reference standard assay, saliva or urine DEAFF. Congenital CMV infection status could not be determined in 15 infants because they were not enrolled in the follow-up study. Reasons for not enrolling in the follow-up study included refusing participation (8), lost to follow-up (6), and relocation (1). Screening saliva DEAFF correctly identified 59 of the 60 (98%) infants with confirmed congenital CMV infection whereas the single-primer DBS PCR only identified 17 of the 60 (28%) infants confirmed to have congenital CMV infection (Table 2). Congenital CMV infection was not confirmed in 2/61 (3%) screening saliva DEAFF positive and 4/21 (19%) DBS PCR positive infants because of the negative reference standard assay. The sensitivity and specificity of the single-primer DBS PCR assay in identifying infants with confirmed congenital CMV infection was 28.3% (95% CI, 17.4 – 41.4%) and 99.9% (95% CI, 99.9 – 100%), respectively. The positive LR for the single-primer DBS PCR assay was 803.7 (95% CI, 278.7 – 2317.9) and the negative LR was 0.7 (95% CI, 0.6 – 0.8). The positive predictive value of the single-primer PCR assay was 80.9% (95% CI, 58.1 – 94.5%) and the negative predictive value was 99.6% (95% CI, 99.5 – 99.7%).
Table 2.
Utility of the two DBS real-time PCR assays to identify infants with confirmed congenital CMV infection.
| Single-primer DBS PCR | Two-primer DBS PCR | |||||
|---|---|---|---|---|---|---|
| Congenital CMV Infection | + | − | Total | + | − | Total |
| + | 17 | 43 | 60 | 11 | 21 | 32 |
| − | 4 | 11,343 | 11,347 | 1 | 8,985 | 8,986 |
| Total | 21 | 11,386 | 11,407 | 12 | 9,006 | 9,018 |
| Sensitivity | 28.3% (17.4 – 41.4%) | 34.4% (18.6 – 53.2%) | ||||
| Specificity | 99.9% (99.9 – 100%) | 99.9% (99.9 – 100%) | ||||
| Positive Likelihood Ratio | 803.7 (278.7 – 2,317.9) | 3088.9 (410.8 – 23,226.7) | ||||
| Negative Likelihood Ratio | 0.7 (0.6 – 0.8) | 0.7 (0.5 – 0.8) | ||||
| Positive Predictive Value | 80.9% (58.1 – 94.5%) | 91.7% (61.5 – 99.8%) | ||||
| Negative Predictive Value | 99.6% (99.5 – 99.7%) | 99.8% (99.6 – 99.9%) | ||||
DBS indicates dried blood spots; PCR, polymerase chain reaction; CMV, cytomegalovirus
Newborn screening with saliva DEAFF and the two-primer DBS PCR assay
During the study period between January and May, 2008, 9,026 newborns were screened for congenital CMV infection using saliva DEAFF and the two-primer DBS PCR assay (Figure 1). Forty-three newborns tested positive for CMV infection by either saliva DEAFF assay (n=43) or the DBS PCR assay (n=14). Thirty-five of the 43 (81%) infants who screened positive were enrolled in the follow-up study and, of those, 32 children were confirmed to have congenital CMV infection based on a positive reference standard DEAFF assay (Figure 1). Congenital infection status could not be determined in 8 infants since they did not enroll in the follow-up study. Reasons for not enrolling in the follow-up study included refusing participation (4), lost to follow-up (2), death (1), and relocation (1). Screening saliva DEAFF correctly identified all 32 infants who were confirmed to have congenital CMV infection, whereas the two-primer DBS PCR identified only 11 of the 32 infants confirmed to have congenital CMV infection (Table 2). Congenital CMV infection was not confirmed in 3/35 screening saliva DEAFF positive and 1/12 screening DBS PCR positive infants because the reference standard assay was negative. The sensitivity and specificity of the two-primer DBS PCR assay for the detection of infants with confirmed congenital CMV infection were 34.4% (95% CI, 18.6 – 53.2%) and 99.9% (95% CI, 99.9 – 100%), respectively. The positive LR for the two-primer DBS PCR assay was 3088.9 (95% CI, 410.8 – 23226.7) and the negative LR was 0.7 (95% CI, 0.5 – 0.8). The positive predictive value of the two-primer assay was calculated to be 91.7% (95% CI, 61.5 – 99.8%) and the negative predictive value was 99.8% (95% CI, 99.6 – 99.9%).
Extraction methods
Of the 71 DBS samples from saliva positive infants, 29 (41%) robot-extracted samples were positive for CMV DNA, whereas only 19 (29%) of column-extracted samples were positive (Χ2 test=3.14; p=0.08) (eTable 1). In addition, in 185 randomly selected DBS specimens from CMV-negative infants, the amount of genomic DNA obtained using robotic extraction (0.86 ± 0.46 µg/mL) and with a commercial column kit (0.78 ± 0.44 µg/mL) was similar (t(368)=− 1.58, p=0.11) as measured by amplifying the RNase P gene (eAppendix).
In 86 infants with confirmed congenital CMV infection, 40 (47%) were positive on the two-primer PCR and 30 (35%) were positive by the nested PCR assay (Χ2 test=2.41; p=0.12). Both methods failed to identify 48% (41/86) who were confirmed CMV positive (eTable 2).
COMMENT
This study demonstrates that real-time PCR analysis of DBS has low sensitivity for correctly identifying infants with congenital CMV infection. These results have major public health implications because they indicate that such methods as currently performed will not be suitable for the mass screening of newborns for congenital CMV infection, the most common non-genetic cause of deafness in the U.S. Our data indicate that up to 80% of infants with congenital CMV infections could be missed, even when using sensitive, two-primer DBS real-time PCR assays for detection of congenital CMV infection. The high positive LRs for both the single-primer and the two-primer PCR assays provides strong evidence that a positive DBS PCR result using these assays will identify infants with congenital CMV infection. However, the negative LRs for both PCR assays are not sufficiently small enough to rule out congenital CMV infection in newborns with a negative DBS PCR result.
PCR testing of peripheral blood has been widely used as a standard diagnostic method to detect invasive CMV infections in immunocompromised individuals including allograft recipients and patients with AIDS.30, 31 This utility, together with the results of several studies that reported successful identification of infants with congenital CMV infection by DBS PCR, has led to anticipation that DBS PCR methods would become valuable tools in newborn CMV screening.12–15, 19–21 However, the pathogenesis of congenital CMV infection is likely to be different from that in immunocompromised hosts since such patients usually experience acute CMV infection or symptomatic reactivation shortly before blood CMV PCR testing, whereas congenitally infected infants may have acquired CMV infection months before birth and thus are no longer viremic when tested as newborns.
Our screening study in which the two DBS real-time PCR assays were directly and prospectively compared to a reference standard for identification of infants with congenital CMV infection provides important test measures of the utility of DBS PCR. Several previous reports have demonstrated that newborns with congenital CMV infection can be identified with varying degrees of success by testing DBS using different PCR methods.12, 15, 32, 33 However, the prospective studies that confirmed CMV infection following identification of CMV DNA in DBS did not determine the number of false negatives (infants with congenital CMV infection who tested negative on DBS PCR). Having the complete denominator, as provided by this study, is essential to determine the utility of DBS PCR as a newborn CMV screening method.
The low sensitivity of our DBS PCR methods could possibly be explained by one of several factors: 1) the methodology used for DNA extraction; 2) the real-time PCR techniques; or 3) the possibility that not all infants with congenital CMV infection have detectable CMV DNA in their blood at birth. To evaluate extraction methods, we compared the ability of the two-primer DBS real-time PCR to detect CMV DNA in DBS samples processed with the robot-extraction protocol used in this study and the column-extraction method and found no difference. Of the 71 DBS samples from saliva-positive infants, 29 (41%) robot-extracted samples were positive for CMV DNA whereas only 19 (29%) column-extracted samples were positive (Χ2 test=3.15; p=0.08) (eTable 1). In addition, the amount of genomic DNA obtained using the robotic extraction protocol and with a commercial column kit was similar as measured by amplifying the RNase P gene.
A number of amplification methods including qualitative, quantitative, and real-time PCR protocols with different primers, probes, and cycling parameters have been reported with varying performance characteristics.11–14, 20, 21, 33, 34 The single-primer real-time PCR assay used in this study was developed in our Central Laboratory.26, 27 A number of newborn CMV screening studies in which DBS samples were tested using a nested PCR protocol report a sensitivity of the DBS PCR assay approaching 100% in some populations.12, 19, 20 However, these studies did not include a direct comparison of the DBS PCR results with a standard culture-based assay. Further, in a more recent study in which different laboratories were given similar sample panels, the sensitivity of CMV PCR methodologies has been shown to vary from laboratory to laboratory.35 A comparison of our two-primer real-time PCR assay with a nested PCR protocol demonstrated that the two-primer PCR had a higher sensitivity than the nested PCR but neither method identified most of the infants with congenital CMV infection in the study population.
A limitation of our study is that the 20,324 infants who were negative on both screening assays, saliva DEAFF and DBS PCR, were not enrolled in follow-up to rule out congenital CMV infection using the reference standard method (saliva or urine DEAFF), resulting in the possibility that some CMV-infected newborns may have been missed by the saliva DEAFF screening assay. However, it is unlikely that the screening saliva DEAFF assay missed significant number of infants with congenital CMV infections. The saliva DEAFF assay used in our study was adapted from the shell vial assay which was shown to be as sensitive and specific as the conventional tube culture method and, thus, considered a standard method for the diagnosis of CMV infections in a variety of clinical settings.36, 37 In addition, the saliva DEAFF assay we used has been demonstrated to be ≥98% sensitive in identifying infants with congenital CMV infection.24 Finally, the results of our study showed that 99% (91/92) of infants with confirmed congenital CMV infection were positive by the screening saliva DEAFF assay.
Another possible limitation is the relative overrepresentation of African Americans in our study population which could make the finding of this study less generalizable to other populations. Although African American infants have a greater risk of infection, there is no scientific evidence that the clinical course or the sensitivity of diagnostic assays differs by race.22, 23 However, the overrepresentation of African Americans may have influenced the prevalence of congenital CMV infection in our study. For populations with differing prevalences of congenital CMV infection than we found in this study, the predictive values calculated for the DBS PCR assays would not be appropriate since predictive values are dependent on the underlying prevalence of disease in the population.
Previous studies observed that some infants with clinically apparent or symptomatic congenital CMV infection had no detectable CMV DNA in whole blood samples obtained in the neonatal period.26, 27, 38 These findings argue strongly that the low sensitivity of our DBS PCR methods is most likely not due to our assay performance, but to the absence of detectable CMV DNA in the peripheral blood of some newborns with congenital CMV infection. This may be particularly true for those infants with clinically inapparent infection. Since nearly all infants with symptomatic infection will be identified on routine clinical examination and about 10 to 15% of infants with asymptomatic or clinically inapparent congenital CMV infection develop hearing loss, it is critical that an ideal CMV screening method identify most newborns with asymptomatic congenital CMV infection.
In summary, the results of this large, prospective newborn CMV screening study that included a direct comparison of the real-time PCR assays with the culture-based method on saliva samples demonstrated that real-time DBS PCR assays are not suitable for screening newborns for congenital CMV infection since they miss approximately two-thirds of the infections. As the disease burden from congenital CMV infection remains a significant public health problem, there continues to be a need to identify the large number of infants with clinically inapparent congenital CMV infection early in life. The results of our study underscore the need for further evaluation of high throughput methods performed on saliva or other samples that can be adapted to large-scale newborn CMV screening.
ACKNOWLEDGEMENTS
Funding/Support: This study was supported by a contract (N01 DC50008) from the National Institute on Deafness and Other Communication Disorders.
Role of the sponsor: The National Institutes on Deafness and Other Communication Disorders provided overall oversight for the design and conduct of the study. The funding organization had no role in the collection, management, analysis, and interpretation of the data, and in preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs. Boppana & Fowler had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Boppana, Fowler
Acquisition of data: Boppana, Ross, Novak, Shimamura, Tolan, Palmer, Ahmed, Michaels, Sánchez, Bernstein, Fowler
Analysis and interpretation of data: Boppana, Ross, Novak, Shimamura, Britt, Tolan, Palmer, Ahmed, Michaels, Sánchez, Bernstein, Fowler
Critical revision of the manuscript for important intellectual content: Boppana, Ross, Novak, Shimamura, Britt, Tolan, Palmer, Ahmed, Michaels, Sánchez, Bernstein, Fowler
Statistical Analysis: Fowler
Obtained funding: Boppana, Britt, Fowler
Administrative, technical, or material support: Boppana, Ross, Novak, Shimamura, Britt, Tolan, Palmer, Ahmed, Michaels, Sánchez, Bernstein, Fowler
Study supervision: Boppana, Fowler
Financial Disclosures: None of the authors had potential conflicts of interest, including specific financial interests and relationships and affiliations (other than those listed in the title page of the manuscript) relevant to the subject of the manuscript.
Online-only material: eAppendix and eTables 1 and 2 are available at http://www.jama.com
Previous Presentation: This study was presented in part at the Pediatric Academic Societies Annual Meeting, May 4, 2009, Baltimore, MD (Abstract #4115.1)
Additional Contributions
We are indebted to our medical and nursing colleagues and the infants and their parents who agreed to take part in this study. We would like to thank the following members of the CHIMES study team for their contributions:
University of Alabama at Birmingham Health System: Nitin Arora, MBBS, MPH, Amita Bey, MPH, Belinda Blackstone, MS, CCC-A, Jennifer Blumenthal, Valisa Brown, MPH, Alice Brumbach, MSN, Nazma Chowdhury, MBBS, PhD, Steven Febres-Cordero, Monique Jackson, Mirjam Kempf, PhD, David Kimberlin, MD, Noelle Le Lievre, Faye McCollister, EdD, Emily Mixon, MPH, Misty Purser, Julie Woodruff, AuD.
Carolinas Medical Center: Edie Cox, AuD, Julie Courtney, Nubia Flores, Molly Ricart, Lisa Schneider, AuD, Jennifer West
Children’s Hospital of Pittsburgh of UPMC: Jena Colaberardino, Noreen Jeffrey, Anne Maracek, Gretchen E. Probst, MAT, CCC-A, Cheryl Rosenberg, Diane Sabo, PhD.
Saint Peter’s University Hospital: Melissa Calderon, Maria Class, Kristina Feja, MD, Marci Schwab, AuD.
University of Cincinnati and Cincinnati Children's Hospital Medical Center: Daniel Choo, MD, Linda Jamison, Patty Kern, Kurt Schibler, MD, Maureen Sullivan-Mahoney, Au.D, Stacie Wethington.
University of Mississippi Medical Center: Kathy Irving, AuD, Delia Owens, Suzanne Roark, AuD, Mindy Ware.
University of Texas Southwestern Medical Center at Dallas, Parkland Health & Hospital System and Children's Medical Center Dallas: Cathy Boatman, MS, CIMI, Jessica Esquivel, Gregory L. Jackson, MD, MBA, Kathy Katz-Gaynor, April Liehr, Asuncion Mejías, MD, Kristine E. Owen, AuD, CCC-A, Peter S. Roland, MD, Oscar Rosado, MD, Angela G. Shoup, PhD, David Sosa, Jessica Santoyo, Elizabeth K. Stehel, MD, Lizette Torres, Fiker Zeray, RN, PNP.
REFERENCES
- 1.Dahle AFKB, Wright JD, Boppana SB, Britt WJ, Pass RF. Longitudinal investigation of hearing disorders in children with congenital cytomegalovirus. J. Am Acad Audiol. 2000;11:283–290. [PubMed] [Google Scholar]
- 2.Demmler GJ. Infectious Diseases Society of America and Centers for Disease Control. Summary of a workshop on surveillance for congenital cytomegalovirus disease. Rev Infect Dis. 1991;13:315–329. doi: 10.1093/clinids/13.2.315. [DOI] [PubMed] [Google Scholar]
- 3.Morton CC, Nance WE. Newborn hearing screening -- a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 4.Ross SA, Fowler KB, Guha A, et al. Hearing loss in children with congenital cytomegalovirus infection born to mothers with preexisting immunity. J Pediatr. 2006;148:332–336. doi: 10.1016/j.jpeds.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Stehel EK, Shoup AG, Owen KE, et al. Newborn hearing screening and detection of congenital cytomegalovirus infection. Pediatrics. 2008;121:970–975. doi: 10.1542/peds.2006-3441. [DOI] [PubMed] [Google Scholar]
- 6.Stagno S. Cytomegalovirus. In: Remington JS, Klein JO, Wilson CB, Baker CJ, editors. Infectious Diseases of the Fetus and Newborn Infant. Philadelphia, PA: W.B. Saunders Company; 2006. pp. 389–424. [Google Scholar]
- 7.Boppana SB, Pass RF, Britt WJ, Stagno S, Alford CA. Symptomatic congenital cytomegalovirus infection: neonatal morbidity and mortality. Pediatr Infect Dis J. 1992 February;11:93–99. doi: 10.1097/00006454-199202000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Fowler KB, McCollister FP, Dahle AJ, Boppana SB, Britt WJ, Pass RF. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J Pediatr. 1997;130:624–630. doi: 10.1016/s0022-3476(97)70248-8. [DOI] [PubMed] [Google Scholar]
- 9.Williamson WD, Demmler GJ, Percy AK, Catlin FI. Progressive hearing loss in infants with asymptomatic congenital cytomegalovirus infection. Pediatrics. 1992;90:862–866. [PubMed] [Google Scholar]
- 10.Joint Committee on Infant Hearing. Year 2007 Position Statement: Principles and Guidelines for Early Hearing Detection and Intervention Programs. Pediatrics. 2007;120:898–921. doi: 10.1542/peds.2007-2333. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto AY, Mussi-Pinhata MM, Pinto PCG, Figueiredo LTM, Jorge SM. Usefulness of blood and urine samples collected on filter paper in detecting cytomegalovirus by the polymerase chain reaction technique. J Virol Meth. 2001;97:159–164. doi: 10.1016/s0166-0934(01)00347-0. [DOI] [PubMed] [Google Scholar]
- 12.Barbi M, Binda S, Primache V, et al. Cytomegalovirus DNA detection in Guthrie cards: a powerful tool for diagnosing congenital infection. J Clin Virol. 2000;17:159–165. doi: 10.1016/s1386-6532(00)00089-5. [DOI] [PubMed] [Google Scholar]
- 13.Johansson PJ, Jonsson M, Ahlfors K, Ivarsson SA, Svanberg L, Guthenberg C. Retrospective diagnosis of congenital cytomegalovirus infection performed by polymerase chain reaction in blood stored on filter paper. Scan J Infect Dis. 1997;29:465–468. doi: 10.3109/00365549709011855. [DOI] [PubMed] [Google Scholar]
- 14.Scanga L, Chiang S, Powell C, et al. Diagnosis of human congenital cytomegalovirus infection by amplification of viral DNA from dried blood spots on perinatal cards. J Mol Diagn. 2006;8:240–245. doi: 10.2353/jmoldx.2006.050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagishi Y, Miyagawa H, Wada K, et al. CMV DNA detection in dried blood spots for diagnosing congenital CMV infection in Japan. J Med Virol. 2006;78:923–925. doi: 10.1002/jmv.20642. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa H, Suzutami T, Baba K, et al. Etiology of severe sensorineural hearing loss in children: independent impact of cytomegalovirus infection and GJB2 mutations. J Infect Dis. 2007;15:782–788. doi: 10.1086/511981. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto AY, Mussi-Pinhata MM, Marin LJ, Brito RM, Oliviera PF, Coelho TB. Is saliva as reliable as urine for detection of cytomegalovirus DNA for neonatal screening of congenital CMV infection? J Clin Virol. 2006;36:228–230. doi: 10.1016/j.jcv.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Mussi-Pinhata MM, Yamamoto AY, Moura Britto RM, et al. Birth prevalence and natural history of congenital cytomegalovirus infection in a highly seroimmune population. Clin Infect Dis. 2009;15:522–528. doi: 10.1086/600882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbi M, Binda S, Primache V, Clerici D. Congenital cytomegalovirus infection in a northern Italian region. Eur J Epidemiol. 1998;14:791–796. doi: 10.1023/a:1007554726449. [DOI] [PubMed] [Google Scholar]
- 20.Barbi M, Binda S, Primache V, Corbetta C. Diagnosis of congenital cytomegalovirus infection by detection of viral DNA in dried blood spots. Clin Diagn Virol. 1996;6:27–32. doi: 10.1016/0928-0197(96)00228-0. [DOI] [PubMed] [Google Scholar]
- 21.Shibata M, Takano H, Hironaka T, Hirai K. Detection of cytomegalovirus DNA in dried newborn blood filter paper. J Virol Meth. 1994;46:279–285. doi: 10.1016/0166-0934(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 22.Fowler KB, Stagno S, Pass RF. Maternal age and congenital cytomegalovirus infection: screening of two diverse newborn populations, 1980–1990. J Infect Dis. 1993;168:552–556. doi: 10.1093/infdis/168.3.552. [DOI] [PubMed] [Google Scholar]
- 23.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007;17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 24.Balcarek KB, Warren W, Smith RJ, Lyon MD, Pass RF. Neonatal screening for congenital cytomegalovirus infection by detection of virus in saliva. J Infect Dis. 1993;167:1433–1436. doi: 10.1093/infdis/167.6.1433. [DOI] [PubMed] [Google Scholar]
- 25.Boppana SB, Smith R, Stagno S, Britt WJ. Evaluation of a microtiter plate fluorescent antibody assay for rapid detection of human cytomegalovirus infections. J Clin Microbiol. 1992;30:721–723. doi: 10.1128/jcm.30.3.721-723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boppana SB, Fowler KB, Pass RF, et al. Congenital cytomegalovirus infection: The association between virus burden in infancy and hearing loss. J Pediatr. 2005;146:817–823. doi: 10.1016/j.jpeds.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 27.Ross SA, Novak Z, Fowler KB, Arora N, Britt WJ, Boppana SB. Cytomegalovirus blood viral load and hearing loss in young children with congenital infection. Pediatr Infect Dis J. 2009;28:588–592. doi: 10.1097/INF.0b013e3181979a27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford RD, Cloud G, Lakeman AD, et al. Detection of cytomegalovirus (CMV) DNA by polymerase chain reaction is associated with hearing loss in newborns with symptomatic congenital CMV infection involving the central nervous system. J Infect Dis. 2005;191:227–233. doi: 10.1086/426456. [DOI] [PubMed] [Google Scholar]
- 29.Simel DL, Samsa GP, Matchar DB. Likelihood ratios with confidence: sample size estimation for diagnostic studies. J Clin Epidemiol. 1991;44:763–770. doi: 10.1016/0895-4356(91)90128-v. [DOI] [PubMed] [Google Scholar]
- 30.Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. The Lancet. 2000;355:2032–2036. doi: 10.1016/S0140-6736(00)02350-3. [DOI] [PubMed] [Google Scholar]
- 31.Spector SA, Hsia K, Crager M, Pilcher M, Cabral S, Stempien MJ. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J Virol. 1999;73:7027–7030. doi: 10.1128/jvi.73.8.7027-7030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbi M, Binda S, Caroppo S, et al. Multicity Italian study of congenital cytomegalovirus infection. Pediatr Infect Dis J. 2006;25:156–159. doi: 10.1097/01.inf.0000199261.98769.29. [DOI] [PubMed] [Google Scholar]
- 33.Engman ML, Malm G, Engstrom L, et al. Congenital CMV infection: prevalence in newborns and the impact on hearing deficit. Scand J Infect Dis. 2008;40:935–942. doi: 10.1080/00365540802308431. [DOI] [PubMed] [Google Scholar]
- 34.Soetens O, Vauloup-Fellous C, Foulon I, et al. Evaluation of different cytomegalovirus (CMV) DNA PCR protocols for analysis of dried blood spots from consecutive cases of neonates with congenital CMV infections. J Clin Microbiol. 2008;46:943–946. doi: 10.1128/JCM.01391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbi M, MacKay WG, Binda S, van Loon AM. External quality assessment of cytomegalovirus DNA detection in dried blood spots. BMC Microbiol. 2008;8:2. doi: 10.1186/1471-2180-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gleaves CA, Smith TF, Shuster EA, Pearson GR. Comparison of standard tube and shell vial cell culture techniques for the detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 1985;21:217–221. doi: 10.1128/jcm.21.2.217-221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiele GM, Bicak MS, Young AE, Kinsey J, White RJ, Purtillo DT. Rapid detection of cytomegalovirus by tissue culture, centrifugation and immunofluorescence with a monoclonal antibody to an early nuclear antigen. J Virol Methods. 1987;16:327–338. doi: 10.1016/0166-0934(87)90018-8. [DOI] [PubMed] [Google Scholar]
- 38.Kimberlin DW, Acosta EP, Sanchez PJ, et al. Pharmacokinetic and pharmacodynamic assessment of oral valganciclovir in the treatment of symptomatic congenital cytomegalovirus disease. J Infect Dis. 2008;197:836–845. doi: 10.1086/528376. [DOI] [PubMed] [Google Scholar]