Abstract
Solving the phylogeny of the animals with bilateral symmetry has proven difficult. Morphological studies have suggested a variety of alternative hypotheses, of which, Hyman’s Coelomata hypothesis has become the most established. Studies based on 18S rRNA have failed to endorse Coelomata, supporting instead the rearrangement of the protostomes into two new clades: the Lophotrochozoa (including, e.g., the molluscs and the annelids) and the Ecdysozoa (including the Panarthropoda and most pseudocoelomates, such as the nematodes and priapulids). Support for this new animal phylogeny has been attained from expressed sequence tag studies, although these generally have a limited gene sampling. In contrast, deep genomic-scale analyses have often supported Coelomata. However, these studies are problematic due to their limited taxonomic sampling, which could exacerbate tree reconstruction artifacts.
Here, we address both of these sampling limitations; we study the effect of long-branch attraction (LBA) in deep genomic-scale analyses and provide convincing evidence, using both single- and multigene families, that Coelomata is an artifact. We show that optimal outgroup selection is key in avoiding LBA and identify the use of inadequate outgroups as the reason previous deep genomic-scale analyses found strong support for Coelomata.
Keywords: Coelomata, Ecdysozoa, phylogenomics, supertrees, outgroup selection, Bayes factors, supermatrix
Introduction
Bilaterian Phylogenetics
Uncertainty still persists pertaining to the early evolution of the Bilateria; an important group which includes all extant animals with the exclusion of the sponges, the Placozoa, the Cnidaria, and the Ctenophora (see, e.g., Nielsen 2001; Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2009; Sperling et al. 2009). Central to this incertitude are the phylogenetic relationships of the “pseudocoelomates” (sensu Hyman 1940), particularly that of the Nematoda (i.e., the round worms), which remain an issue of debate (Telford et al. 2008).
From a morphological point of view, some of the most prominent features shared by the majority of bilaterians include bilateral symmetry, a pronounced anteroposterior axis and a head with a nervous concentration, that is, a brain (Nielsen 2001). A variety of morphological phylogenies of Bilateria have been proposed since Darwin’s time (Jenner and Schram 1999); however, the dominant view has long been that of Hyman (1940) and her Coelomata hypothesis (see also Halanych 2004; Philippe et al. 2005; Telford et al. 2008). According to Coelomata, Bilateria were classified in three groups: the Acoelomata (Platyhelminthes and Nemertinea), the Pseudocoelomata (Nematoda, Nematomorpha, Rotifera, Priapulida, Kinorhyncha, and Gastrotricha), and the Coelomata (all the other bilaterian phyla, e.g., the Arthropoda, the Mollusca, the Annelida, and the Vertebrata).
The first major challenge to Coelomata came from the analyses of taxon-rich 18S rRNA data sets (Halanych et al. 1995; Aguinaldo et al. 1997), which proposed an alternative division of the Bilateria (with the possible exclusion of the Acoela—see Ruiz-Trillo et al. 1999; Littlewood et al. 2001; Hejnol et al. 2009; but see also Philippe et al. 2007) into the Protostomia and Deuterostomia. The 18S rRNA data further suggested a partitioning of the protostomes into the Lophotrochozoa (Halanych et al. 1995), including, for example, the molluscs and the annelids (i.e., the Eutrochozoa), and the Ecdysozoa (Aguinaldo et al. 1997), including the Panarthropoda and several of Hyman’s Pseudocoelomata. This new animal phylogeny is now generally known and will hereafter be referred to as the Ecdysozoa hypothesis.
Ever since the genomes of the arthropod Drosophila melanogaster (a coelomate protostome), the vertebrate Homo sapiens (a coelomate deuterostome), the nematode Caenorhabditis elegans (a pseudocoelomate protostome), and the fungus Saccharomyces cerevisiae (a nonmetazoan outgroup) became available, many have attempted to test hypotheses of bilaterian relationships using genomic-scale data sets, or in any event, data sets deemed to be of genomic scale at the time they were assembled (Blair et al. 2002; Copley et al. 2004; Dopazo et al. 2004; Wolf et al. 2004; Dopazo H and Dopazo J 2005; Philip et al. 2005; Rogozin et al. 2007, 2008; Zheng et al. 2007). A number of these studies (Copley et al. 2004; Dopazo H and Dopazo J 2005; Irimia et al. 2007; Roy and Irimia 2008; and Belinky et al. 2010) have endorsed Ecdysozoa, however, only that of Dopazo H and Dopazo J (2005) used standard phylogenetic analyses of aligned sequence data.
The majority of published deep genomic-scale analyses have supported Coelomata, leading Lynch (2007), for example, to conclude a literature survey on this argument by claiming: “… [Ecdysozoa] continues to be presented as a fact in many major textbooks, even though phylogenies based on large numbers of protein-coding genes generally either place nematodes on their traditional position or are equivocal on the matter….” Studies supporting Coelomata, however, characteristically suffer from a sparse taxonomic sampling (see also Halanych 2004), which can exacerbate phylogenetic artifacts, particularly long-branch attraction (LBA), in the presence of a fast-evolving species such as C. elegans (e.g., Pisani 2004; Delsuc et al. 2005; Philippe et al. 2005; Jeffroy et al. 2006; Sperling et al. 2009).
Studies conducted using the expressed sequence tags (ESTs) methodology (Philippe et al. 2005, 2009; Dunn et al. 2008; Lartillot and Philippe 2008; and Hejnol et al. 2009), on the other hand, are characterized by a denser taxonomic sampling and generally include more appropriate (animal) outgroups and as such should be less prone to LBA. Accordingly, EST-based studies have recurrently supported Ecdysozoa (see Philippe et al. 2005; Lartillot and Philippe 2008 in particular). However, with the exception of Hejnol et al. (2009), who considered 1,487 genes (but only for a very small subset of the taxa they sampled), EST studies represent shallow genomic sampling (Zilversmit et al. 2002), with Philippe et al. (2005) considering only 146 genes, Dunn et al. (2008) 150 genes, and Philippe et al. (2009) 128 genes. Additionally, EST libraries generated for phylogenetic purposes are generally not normalized (e.g., Dunn et al. 2008; Hejnol et al. 2009), and the protein-coding genes sampled in these studies do not represent a random sample of the genes in the considered genomes. Rather, they correspond to a sample of the most highly expressed genes. This nonrandom sampling is not a problem per se, nevertheless, it does pose the question: what will the outstanding proportion of the animal proteome disclose? To date, the answer has often been that standard sequence analyses of deeply sampled genomic data sets favor Coelomata.
Phylogenomics: Methodological Approaches
From a methodological point of view, two principal approaches are generally employed in phylogenomics: the supertree and the supermatrix approach (Delsuc et al. 2005), with both approaches having different strengths and weaknesses.
In the supertree approach, gene trees are recovered for each individual protein family using the most appropriate phylogenetic method. Gene trees are then combined using one of a number of existing supertree methods (for a brief introduction, see McInerney et al. 2008). Advantages of the supertree approach include: 1) the ability to analyze each gene individually under the best-fitting substitution model, 2) the capacity to amalgamate trees derived from the analysis of both single- and multigene families, and 3) a significant decrease in the computational time necessary to build large phylogenies (facilitating the handling of data sets scoring thousands of genes) for hundreds of taxa (e.g., Pisani et al. 2007). As gene families are first analyzed in isolation, the major limitation of the supertree approach is that the combined trees can be based on relatively small alignments. This can result in significant statistical errors, which may translate into poorly supported phylogenomic supertrees. Filtering strategies, that is, eliminating genes that do not pass the permutation tail probability (PTP) test (Archie 1989) or that do not support the monophyly of universally accepted clades (Pisani et al. 2007), which also serves to alleviate the negative impact of hidden paralogy when analyzing sets of single-gene families, can be used to improve resolution significantly.
In the supermatrix approach, single-gene alignments are merged into a multiple gene alignment, which is then analyzed using the most appropriate phylogenetic method. The principal merit of this approach is that gene concatenation allows for the minimization of statistical errors, often resulting in well-supported trees (Delsuc et al. 2005). The main shortcomings of this approach are: 1) while it minimizes stochastic errors, it tends to exacerbate systematic ones (e.g., Delsuc et al. 2005; Jeffroy et al. 2006). Although the use of well-performing, parameter-rich models, like categories model (Lartillot and Philippe 2004; Philippe et al. 2007), alleviates this problem, it does not fully eliminate it (e.g., Jeffroy et al. 2006). 2) The supermatrix approach does not lend itself to the integration of multigene families and as such limits the information that can be analyzed to that of single-gene families or in some rare cases (i.e., when the gene phylogeny is well understood) to single paralogy groups within a multigene family (e.g., Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2009). 3) If the number of considered genes, species, or both is considerably large, supermatrix analyses become very difficult to perform due to memory and time constraints (see, e.g., Hejnol et al. 2009). Technological advances should ameliorate this problem, but this limit of the supermatrix approach can be expected to persist for the foreseeable future.
Circumventing LBA
LBA (Felsenstein 1978) is a common phylogenetic artifact (see Brinkmann and Philippe 1999; Pisani 2004; Delsuc et al. 2005; Jeffroy et al. 2006), which can affect every phylogenetic method (Pisani 2004; Delsuc et al. 2005; Jeffroy et al. 2006). Because time and rate are confounded in branch length estimation (e.g., Yang 2006), LBA results in trees in which fast-evolving species are artifactually grouped together or with distantly related taxa (e.g., with the outgroups). Two straightforward approaches to reduce LBA are optimal outgroup selection (to minimize root to tip distances in a phylogeny) and increased taxon sampling (to break long branches), see also Pisani (2004).
Early, deep genomic-scale analyses used fungal outgroups or on occasion even more distantly related outgroups (e.g., Blair et al. 2002). These clearly represent poor choices to investigate the phylogenetic relationships of the Bilateria as they may serve to exacerbate LBA. Dopazo H and Dopazo J (2005) performed standard sequence analyses of a deeply sampled genomic data set using a distant (fungal) outgroup. Realizing that a fungal outgroup might not have been adequate for their analyses, and in the absence of a closer outgroup, these authors used a relative-rate test (for an overview, see Robinson et al. 1998) based approach to identify clock-like genes. Analyses of these genes found support for Ecdysozoa. Although their results are interesting, their approach is not without problems. First, the relative-rate test is not particularly sensitive; a more discriminating approach (i.e., the likelihood ratio test) should have been used instead. In addition, their relative-rate tests were implemented under the simplistic Kimura’s distance in PROTDIST (Felsenstein 2005), which is unlikely to have fit their data well. Finally, these authors considered only homologues of protein-coding genes found in 18 human chromosomes, unnecessarily discarding potentially informative genes not found in this subset of human chromosomes.
The number of complete animal genomes has now increased significantly making the improvement of taxonomic sampling in genomic-scale phylogenetic analyses possible. Recent genome sequencing projects have included that of the cnidarian Nematostella vectensis and the placozoan Trichoplax adherans. Although there is ongoing debate over the phylogenetic relationships of these organisms, there is general agreement that both are nonbilaterian Metazoans (see Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2009; Sperling et al. 2009). Accordingly, N. vectensis and T. adhaerens represent more appropriate outgroups for testing hypotheses of bilaterian evolution than fungi (see also Philippe et al. 2005). We thus avoided gene selection strategies (e.g., Dopazo H and Dopazo J 2005), focusing instead on taxonomic sampling and outgroup selection to test hypotheses of bilaterian evolution.
Maximizing Gene Sampling within a Phylogenomic Approach
The strongest test of a phylogenetic hypothesis is one considering all the relevant information (e.g., Kluge 1989). In phylogenomics, EST studies can maximize taxonomic sampling, whereas studies using complete genomes can maximize gene sampling. Accordingly, a pragmatic solution to the Coelomata versus Ecdysozoa controversy can only be achieved through the congruence of taxonomically well-sampled EST studies and deep genomic-scale analyses.
Here, we performed analyses to maximize gene sampling. We implemented a pluralist approach where phylogenomic trees of Bilateria were generated using supertrees and consensus trees, summarizing both single- and multigene family trees. Because supertrees do not allow for the integration of the subsignals in the data (Pisani and Wilkinson 2002), we augmented our study to include a supermatrix approach, where single-gene families were concatenated and concomitantly analyzed. This was done to confirm the results from the supertree analyses and to provide a statistical test, within a Bayesian framework, of the fit of the considered hypotheses (i.e., Coelomata and Ecdysozoa) to the data.
An experimental approach was used to investigate the support for the considered alternative hypotheses in the light of LBA and to reject the one most likely to be artifactual. In particular, the effect of using fungi, nonbilaterian animals, or both, in order to break long branches, was examined. By comparing our results with those of previous EST studies, we evaluate the congruence between different phylogenomic approaches.
Materials and Methods
Data Collection
Genomic data for 43 eukaryotic species were downloaded from COGENT (http://maine.ebi.ac.uk:8000/services/cogent/), DOE Joint Genome Institute (http://genome.jgi-psf.org/), EMBL-EBI IPI (http://www.ebi.ac.uk/IPI/IPIhelp.html), Ensembl (http://www.ensembl.org/info/data/ftp/index.html), and National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/genomes/).
Experimental Phylogenomics and Data Set Assembly
Rather than simply collecting all available animal genomes and reconstructing yet another metazoan phylogeny, we took an experimental approach. We made the following ad hoc (working) assumption: Coelomata is the true tree and not the result of LBA (our null hypothesis). We predicted what the consequences of this null hypothesis would be, selected a suitable set of complete genomes, and tested whether the predictions derived from our assumption could be met. If our predictions were to be upheld by the data, the null hypothesis was not to be rejected, whereas if overturned, the data would reject the null hypothesis.
Based on our assumption, we first predicted that in sparsely sampled (four taxon) data sets, Coelomata should invariably be recovered irrespective of whether a distant (fungal) or closer (animal) outgroup was used. Conversely, we anticipated that if Coelomata was due to a LBA artifact, then it would only be recovered when using a distant outgroup. We further hypothesized (based again on the postulation that Coelomata is the “bona fide” tree) that Coelomata should continue to be recovered in the presence of an extensive taxonomic sampling, irrespective of the outgroup used. Alternatively, if Coelomata was the result of LBA, we would expect that it should not be recovered if a targeted sampling strategy was adopted to break the long branch connecting the distant (in our case fungal) outgroup and the Bilateria. This could be done by including N. vectensis and/or T. adhaerens in the analyses or by replacing the fungal outgroups with animal outgroups (i.e., N. vectensis and/or T. adhaerens).
We assembled (from our starting set of 43 genomes) five intersecting data sets to test our predictions. Two of these data sets contained a minimal sampling, scoring only four taxa. The remaining three data sets included 41, 42, and 43 species, respectively. The four-taxon data sets were designed to mimic the taxonomic sampling of the earliest phylogenomic studies, whereas the 41, 42, and 43 taxon data sets were constructed to contain the broadest possible sampling of complete animal genomes (for a list of the species in each of the five data sets, see supplementary table S1, Supplementary Material online).
The sparsely sampled data sets were used to investigate, at the most fundamental level, the effect of outgroup choice in phylogenomics. Accordingly, these data sets only differed in the outgroup they included, which was either S. cerevisiae or N. vectensis. In both data sets, the remaining three taxa were H. sapiens, D. melanogaster, and C. elegans. For these sparsely sampled data sets, N. vectensis was preferred over T. adhaerens as outgroup to the Bilateria, as there is little doubt that cnidarians are closer to the Bilateria (Hejnol et al. 2009; Philippe et al. 2009; Sperling et al. 2009).
Similarly, the three densely sampled data sets scored a common set of 40 bilaterian species (see supplementary table S1, Supplementary Material online), to which one to three outgroups were added. The 41-taxon data set only included S. cerevisiae as the outgroup. The 42 species data set contained two animal outgroups (N. vectensis and T. adhaerens) but did not include S. cerevisiae. Finally, the 43 species data set included both the fungal and the animal outgroups (S. cerevisiae, N. vectensis, and T. adhaerens). These densely sampled data sets were used to investigate the effect of using alternative taxon sampling strategies and optimal outgroup selection.
If Coelomata is the correct topology, it should always be recovered in the densely sampled data sets. If Coelomata is a LBA artifact, we expect it to appear only when the fungal outgroup is used in isolation. That is, when the long branch joining the fungi and the Bilateria is present and unbroken. Accordingly, our expectation is that if the data is affected by LBA, Coelomata should be recovered from the 41-taxon data set but not from the 42 and the 43-taxon data sets.
Protein Family Identification
For each sparsely and densely sampled data set, homologous sequences were identified and clustered using the BlastP based, all-versus-all approach of Creevey et al. (2004), Fitzpatrick et al. (2006), and Pisani et al. (2007). For the sparsely sampled data sets, protein families were also identified using the markov cluster (MCL)-based algorithm of Enright et al. (2002). Details of how both protein identification strategies were implemented are reported in the Supplementary online information (SI). As a result, a total of seven initial data sets (four sparsely sampled and three densely sampled ones) were used in this study.
For each of these seven data sets, gene families were partitioned into two groups. Families scoring only one member for any given genome (i.e., the putative single-gene families) were separated from those containing multiple members per genome (i.e., the multigene families). Because phylogenetic analyses can only be performed on gene families that score four or more sequences, only single- and multigene families consisting of a minimum of four sequences were retained for further analysis (for a comparison of the number of single- and multigene families in each of the 7 considered data sets, see table 1).
Table 1.
Progression of Protein Family Numbers At Each Stage of Analysis
| Single-Gene Families |
Multigene Families |
||||||||||
| Data Set | Homology Search | Number of Families | Families with Four or More Taxa | Families Passing the PTP Test | Families Used in Phylogenetic Analysis | Number of Families | Families with Four or More Taxa | Families Passing the PTP Test | Species-Level Trees with Four or More Taxa | Species-Level Trees Passing the GTP-PTP Test | Species-Level Trees Used in Supertree Analyses |
| 40 genomes and fungal outgroup | Creevey et al. (2004) | 82,043 | 3,241 | 2,164 | 2,164 | 26,165 | N/A | N/A | N/A | N/A | N/A |
| 40 genomes and animal outgroups | Creevey et al. (2004) | 86,855 | 3,615 | 2,216 | 2,216 | 25,722 | N/A | N/A | N/A | N/A | N/A |
| 40 genomes, fungal, and animal outgroups | Creevey et al. (2004) | 88,858 | 3,304 | 1,949 | 1,949 | 27,895 | N/A | N/A | N/A | N/A | N/A |
| 3 genomes and fungal outgroup | Creevey et al. (2004) | 16,780 | 201 | 30 | 30 | 7,947 | 4,197 | 3,301 | 917 | 258 | 258 |
| MCL | 6,588 | 254 | 28 | 28 | 8,529 | 5,312 | 4,143 | 1,366 | 392 | 392 | |
| 3 genomes and animal outgroups | Creevey et al. (2004) | 18,094 | 314 | 48 | 48 | 10,146 | 5,269 | 4,328 | 1,923 | 516 | 516 |
| MCL | 6,254 | 332 | 40 | 40 | 9,808 | 6,561 | 4,666 | 2,319 | 682 | 682 | |
NOTE.—na, not applicable; All data sets and protein families were subjected to the same protocol. GTP-PTP test (see text).
Only single-gene families are typically used for phylogenetic reconstruction (e.g., Pisani et al. 2007; Hejnol et al. 2009). This is to minimize the complexity associated with the analysis of multigene data sets and the inclusion of signals representing the relationships of paralogous genes. However, this approach has the disadvantage of considering only a minority of the genes in the genomes, whereas the strongest test of a phylogenetic hypothesis is one considering all relevant information (e.g., Kluge 1989). Only upon the integration of multigene families can such a test be performed. Here, by exploiting the flexibility of the supertree approach, we have combined both single- and multigene families to generate trees based on the deepest possible sample of genomic data. However, due to the volume of multigene families generated, it was not currently practicable to analyze the multigene families in all seven data sets. Owing to their smaller size, the four 4-taxon data sets were selected as exemplar cases for analysis using both single- and multigene families.
Alignment, Curation, and Identification of Gene Families Conveying Significant Hierarchical Signal
All considered single- and multigene families were aligned using ClustalW (Thompson et al. 1994). As the accuracy of traditional multiple sequence alignment software has been questioned (e.g., Löytynoja and Goldman 2008), the single- and multigene families in our four-taxon data sets were also aligned using PRANK (Löytynoja and Goldman 2008). This was done to investigate whether alignment-dependent biases (Löytynoja and Goldman 2008) influenced our results. This experiment was limited to our four-taxon data sets as aligning sequences using PRANK is computationally expensive.
Due to the number of protein families obtained from our data sets, manual curation of alignments was unfeasible. Gblocks (Castresana 2000) was thus used to eliminate highly variable and potentially misaligned regions. Gblocks parameters were set as follows: gapped positions were not eliminated, the minimum block length was set to eight amino acid positions, whereas the maximum number of permitted consecutive nonconserved positions was set to 15 (see also Pisani et al. 2007). Curated alignments were subjected to the PTP test (Archie 1989). This allowed the identification of families conveying significant hierarchical signal (see Pisani et al. 2007). Such families were considered to contain sufficient hierarchical structure to be deemed phylogenetically informative. The PTP test was implemented in PAUP4.0b10 (Swofford 1998). Settings were as follows: 2,000 permutations with heuristic search with one random addition sequence and the MulTrees option set to off. For the PTP test, a probability value P ≤ 0.05 was considered significant. Alignments not passing the PTP test (P ≥ 0.05) were disregarded, as they would not contribute anything except noise to the analyses.
Model Selection and Phylogenetic Analysis
PHYML (Guindon and Gascuel 2003) was used to perform maximum likelihood (ML) phylogenetic analyses of each alignment passing the PTP test. ML analyses were performed under the best-fitting substitution model, as inferred using the Akaike information criterion in Modelgenerator (Keane et al. 2006). For each single- and multigene family tree, support was evaluated using bootstrap (100) replicates.
Single-gene trees were manually inspected to evaluate possible instances of hidden paralogy; trees that failed to recover the monophyly of uncontroversial, universally accepted groups (e.g., Vertebrata or Arthropoda) were excluded from further analyses (see also Pisani et al. 2007).
Supertree and Consensus Tree Analyses
Supertrees represent a generalization of the consensus tree problem in the case of partially overlapping trees (Semple and Steel 2003). Both consensus and supertree methods were used to derive phylogenomic supertrees representing the relationships among the species in our seven starting data sets. The number of gene families considered at each stage of the protocol used to generate the supertrees is reported in table 1.
Deriving Phylogenomic Supertrees for the Four-Taxon Data Sets
For each of the final four-taxon data sets (see fig. 1; eight in total arising from alternative homology assessment and alignment procedures), we derived phylogenomic consensus trees. These were built using 1) the set of all the single-gene families, 2) the set of all the multigene families, and 3) the combined set of all single- and multigene families. Accordingly, a total of 24, four-taxon, phylogenomic trees were derived. Table 1 reports the number of genes used to build each of these trees.
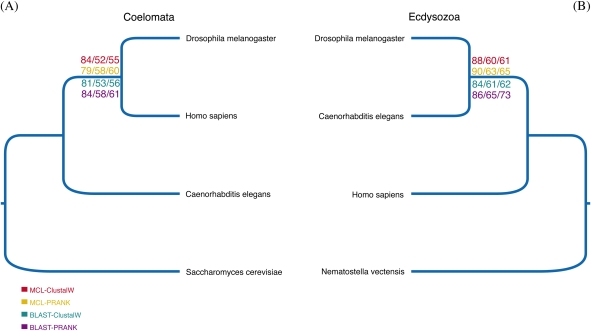
FIG. 1.—
Testing outgroup choice in minimally sampled data sets. Majority rule consensus trees derived from ML gene trees. Bootstrap support from both multigene families and single-gene families is shown for each node. The following core ingroup species are common to all: Homo sapiens, Drosophila melanogaster, and Caenorhabditis elegans. Outgroups used are (A) the yeast Saccharomyces cerevisiae (B) the cnidarian Nematostella vectensis. Bootstrap support values are shown for each combination of protein family identification and alignment method. Bootstrap support is displayed for single-gene families, multigene families, and combined single-gene families and multigene families, respectively.
Each of the eight single-gene family based, four-taxon phylogenomic trees (see fig. 1) were built as follows: 1) the 100 bootstrap ML trees generated for each single-gene family in that data set were pooled to generate a single bootstrap tree file. 2) The trees in the pooled, bootstrap tree file were summarized using the majority rule consensus tree method (Margush and McMorris 1981), as implemented in the software Consense (Felsenstein 2005). This was possible as all considered bootstrap trees were on the same taxon set (i.e., they were fully overlapping). As these phylogenomic trees were derived from pooling trees obtained from the individual bootstrap replicates, assessment of the support for the clades in these trees was straightforward because the four-taxon phylogenomic trees were also bootstrap consensus trees.
Each of the eight multigene family-based phylogenomic trees (see fig. 1) were derived as follows: 1) for each considered multigene family, the 100 bootstrap ML trees were used to generate reconciled species trees. This was done using the duplication only, gene tree parsimony (GTP) method (e.g., Cotton and Page 2004) as implemented in the software DupTree (Wehe et al. 2008), with the nogenetree option turned on, using a partial queue based heuristic search (see supplementary fig. SI1, Supplementary Material online for an exemplar multigene family and the corresponding GTP-derived species tree). 2) The resulting species trees (one per bootstrap ML tree) were pooled into a single file. 3) The pooled, bootstrap (species) trees were summarized using the majority rule consensus method (as implemented in the software Consense), thus generating a bootstrap consensus phylogenomic tree. Also, in this case, the use of the majority rule consensus method could be implemented, as all the bootstrap species trees were on the same taxa set.
Each of the eight combined multigene family and single-gene family phylogenomic trees (see fig. 1) were derived as follows: the corresponding sets of individual bootstrap trees (obtained from the ML analyses of the single-gene families) and the species trees derived from the DupTree analysis of the bootstrap trees from the multigene families (see above) were pooled into a single file. Trees in the pooled file were summarized using the majority rule consensus method to derive a bootstrap consensus phylogenomic tree.
The GTP-PTP Test
Not all of our multigene families were used for phylogenetic reconstruction (i.e., some families, despite passing the PTP test, were deemed not viable). An additional PTP test was developed to evaluate whether the duplication history of each considered multigene family was phylogenetically informative. To implement the GTP-PTP test, for each optimal multigene family tree derived using PHYML, 100 permuted trees were generated. This was done by randomly swapping the labels associated with the terminal nodes of the optimal multigene family tree, whereas maintaining the unlabeled phylogenetic history as fixed. This is similar to the YAPTP test of Creevey et al. (2004). Each permuted tree was used to infer a species phylogeny using the GTP method (as implemented in DupTree). The score of each GTP reconstruction was recorded, and these values were compared against the GTP score of the species history derived from the original (unpermuted) multigene family tree. Families were retained for phylogenetic analysis when the species history derived from the unpermuted tree was significantly better than those obtained from the GTP analysis of the permuted trees. For these analyses, the significance level was set to P ≤ 0.01. PERL scripts to implement the GTP-PTP are available upon request.
The species phylogeny embedded in multigene families failing to pass the GTP-PTP test has essentially been erased due to a complex gene deletion/duplication history. These multigene families can only contribute noise to the analyses and were thus not used for phylogenetic reconstruction.
Deriving Phylogenomic Trees for the 41, 42, and 43-Taxa Data Sets
Because genes do not have a universal distribution, in the case of the 41, 42, and 43 species data sets, single-gene families could score in the range of 4–41, 4–42, or 4–43 sequences, respectively. That is, unlike the four-taxon data sets, single-gene family trees in these data sets are partially, rather than fully, overlapping. Accordingly, gene trees derived from protein families identified in these larger data sets could not be summarized using a standard consensus method. Instead a supertree approach was used to derive phylogenomic supertrees for these data sets.
For each of the three densely sampled data sets, consensus supertrees were generated as follows: 1) the bootstrap trees obtained from the ML analysis of each considered single-gene family were pooled into one single data set. 2) Input tree bootstrapping (Creevey et al. 2004; Burleigh et al. 2006; Moore et al. 2006; Pisani et al. 2007) of the pooled trees was used to generate 100 pseudoreplicate data sets. 3) For each pseudoreplicate data set, supertrees were derived using the matrix representation with parsimony (MRP) method (Baum 1992; Ragan 1992). To do so, for each pseudoreplicate data set, a standard MRP matrix was generated using CLANN (Creevy and McInerney 2005). This matrix was then analyzed using maximum parsimony in PAUP (Swofford 1998) to generate the MRP supertrees. For the parsimony analysis, 100 heuristic searches were performed with random sequence addition and tree bisection and reconnection branch swapping. 4) The supertrees derived from the analysis of each pseudoreplicate data set were summarized using the majority rule consensus method, generating a majority rule consensus genomic supertree in which support for the clades recovered was expressed as their bootstrap support.
Supermatrix Analysis
For each of the 41, 42, and 43 taxon data sets, a superalignment of the single-gene families that passed the PTP test was generated. However, only families that contained at least one nematode sequence were concatenated. This was done to reduce the dimensions of the superalignment (thus making it more manageable) whereas retaining all the information that could possibly bear on the phylogenetic position of the Nematoda. The three concatenated data sets generated in this way were thus subsamples of our complete data sets and scored: 43392 amino acid positions (41-taxon data set), 38701 amino acid positions (42-taxon data set), and 25857 amino acid positions (43-taxon data set). Because the considered genes are not universally distributed, there was a significant amount of missing data in each alignment.
Phylogenetic analyses of the three data sets were performed in Phylobayes (Lartillot and Philippe 2004) under the CAT + G model. We selected CAT as it has been shown (e.g., Philippe et al. 2007; Sperling et al. 2009) that this model provides a better fit to data in comparison with ordinary general time reversible models (e.g., Whelan and Goldman model [WAG] or mechanistic general time revisible [GTR]). We also tested the use of CAT-GTR but under this model we could not reach convergence.
For each data set, two independent runs were performed. Convergence was tested using the bpcomp program (which is part of phylobayes). Two runs were considered to have converged when the maximum difference in observed bipartitions dropped below 0.2.
BFs: Testing Coelomata and Ecdysozoa in a Bayesian Framework
Bayes factors (BFs) are general statistical tools that can be used, within a Bayesian framework, to compare alternative models—for example, the trees representing the relationships for a group of taxa (see Sperling et al. 2009) and evaluate the weight of evidence in favor of one of the compared models (and hence against the alternative one). To calculate BFs for each considered data set, we ran two constrained Bayesian analyses using MrBayes (Ronquist and Huelsenbeck 2003). Each of these analyses could only visit trees compatible with one of the two compared hypotheses (i.e., Ecdysozoa or Coelomata). For each of the two constrained analyses, two runs of one chain were run for 1,000,000 generations (sampling every 100 generations). A burn in of 500,000 generations was considered for all analyses. All analyses were performed under WAG + G. This is not ideal, but we could not perform BF analyses under CAT, as the current Phylobayes output is not suitable for estimating BFs (see also Sperling et al. 2009), while running our analyses under GTR in MrBayes was not feasible because of time limitations.
BFs were calculated in Tracer 1.4.1 (Rambaut and Drummond 2007) using, for each constrained analysis, the trace files from the run of highest harmonic mean. Standard errors around the estimated BF were calculated using the bootstrap (1,000 replicates). BFs were interpreted according to the table of Kass and Raftery (1995).
Results
Four-Taxon Data Sets
The four species data sets were analyzed to assess at a very basic level the effect of outgroup selection in phylogenomics. The first interesting result we obtained from these analyses was that only a somewhat diminutive number of single-gene families conveying a significant amount of phylogenetic information could be identified (see table 1). This was not fully unforeseen as the stringency of the PTP test increases as the number of considered species decreases. More families were found when N. vectensis was used as an outgroup instead of S. cerevisiae; however, the difference was small (from 31 to 48). The number of single-gene families passing the PTP test in the four-taxon data sets did not change significantly when either an alternative homology assignment strategy or alignment software was used (see table 1), suggesting that the small number of single-gene families arising from these analyses does not stem from methodological biases. It merely implies that when only 4 taxa are considered, there are very few, universally distributed single-gene families conveying significant phylogenetic information pertinent to testing hypotheses of bilaterian relationships. The number of multigene families (see table 1) passing all of our quality checks is also quite low but significantly higher than the equivalent number of single-gene families. This was to be expected as there are far more multigene families than single-gene families in the average animal genome. However, interestingly, we noted that although the number of phylogenetically informative multigene families identified if S. cerevisiae is used as outgroup is 258 (using the Creevey et al. 2004 homology assessment strategy) or 392 (using MCL), the number of phylogenetically informative multigene families identified when N. vectensis is the outgroup is 516 (using the Creevey et al. 2004 homology assessment strategy) or 682 (using MCL), that is approximately twice as many. This strongly implies that using closer outgroups is key to maximizing the amount of phylogenetic information and increasing the signal to noise ratio in phylogenomic data sets.
Phylogenomic trees derived from single-gene families passing the PTP test showed that when S. cerevisiae was used as an outgroup, support was found for Coelomata (see fig. 1). This result holds true irrespective of the protein family identification method used and of the alignment software used (see fig. 1 and table 2). When only multigene families are used similar results are found, although there is a significant decrease in the level of support observed (fig. 1 and table 2). Finally, in the phylogenomic, trees obtained when both the single-gene families and the multigene families were considered concurrently the support for Coelomata ranges between 55% and 61% depending on the clustering method and alignment software used (fig. 1 and table 2). This represents a marked decrease in the support for Coelomata. Similar results were obtained in the study of Philippe et al. (2005), although based solely on single-gene families.
Table 2.
Percentage Bootstrap Support for Each Hypothesis (Coelomata, Ecdysozoa, or the Alternative Topology) Arising from the Analysis of the Sparsely Sampled Data Sets
| Percent Support for Each Hypothesis Under Each Alignment Protocol |
||||||||
| ClustalW |
PRANK |
|||||||
| Data Set | Homology Search | Gene Families | Coelomata | Ecdysozoa | Vertebrata–Nematoda | Coelomata | Ecdysozoa | Vertebrata–Nematoda |
| Fungal outgroup | Creevey et al. (2004) | Single | 81 | 9 | 10 | 84 | 6 | 10 |
| Multi | 53 | 26 | 21 | 58 | 23 | 19 | ||
| Single + multi | 56 | 24 | 20 | 61 | 20 | 19 | ||
| MCL | Single | 84 | 6 | 10 | 79 | 7 | 14 | |
| Multi | 52 | 26 | 21 | 58 | 22 | 20 | ||
| Single + multi | 55 | 25 | 20 | 60 | 20 | 20 | ||
| Animal outgroup | Creevey et al. (2004) | Single | 14 | 84 | 2 | 6 | 86 | 8 |
| Multi | 21 | 61 | 18 | 18 | 65 | 17 | ||
| Single + multi | 20 | 62 | 17 | 13 | 73 | 14 | ||
| MCL | Single | 9 | 88 | 3 | 7 | 90 | 3 | |
| Multi | 21 | 60 | 19 | 19 | 63 | 18 | ||
| Single + multi | 20 | 61 | 18 | 18 | 65 | 17 | ||
When the cnidarian N. vectensis is used as an outgroup, Coelomata is no longer recovered. Instead, a nematode–arthropod clade emerges, supported most strongly in the analysis of the single-gene families (bootstrap proportion [BP] = 90%; fig. 1 and table 2). Support for Ecdysoza arising from the analysis of single-gene families and multigene families, both in isolation and when combined, ranges from 60% to 90% (fig. 1 and table 2).
It is important to note that when multigene families are used, we observe a general decrease in support for the nodes in the recovered tree, irrespective of whether a fungal or animal outgroup is used. This suggests that multigene families contain more noise than single-gene families. Or more likely that the approach used to infer species trees from the multigene family trees (i.e., duplication only GTP) is not ideal and cannot completely eliminate the paralogy signal. It is to be expected that the development of more refined methods for inferring species trees from multigene family trees will alleviate this problem in the future.
Analyses of the four-taxon data sets illustrate that when a closer outgroup is used sequence analyses with a deep genomic sampling support Ecdysozoa. Conversely, Coelomata is found only when a distant outgroup is used, thus failing to uphold our predictions. The recovery of Coelomata can be better viewed as inconsistent (i.e., “strongly supported but erroneous” Philippe et al. 2005), arising from the selection of a distant outgroup. In the presence of a distantly related outgroup like S. cerevisiae (which probably shared a last common ancestor with the Bilateria one billion years ago; see Peterson et al. 2008; Sperling et al. 2010), the rapidly evolving nematode C. elegans is placed at the base of the tree, close to the outgroup. When in its stead, a closer outgroup (N. vectensis), which probably shared a last common ancestor with the Bilateria only ≈ 670 MYA (Peterson et al. 2008; Sperling et al. 2010) is used, C. elegans emerges as the sister group of the arthropod D. melanogaster and thus as an Ecdysozoan. This strongly implies the recovery of Coelomata to be the result of a tree reconstruction artifact.
Densely Sampled Data Sets
Although the small data sets demonstrate at the most fundamental level the effects of outgroup selection, they still consider only a scant taxonomic sampling. These analyses allow us to reject our null hypothesis (i.e., Coelomata is the true tree) but only relative to small data sets. To test the validity of these results in a more practicable context, we turned our attention to data sets with a broader taxonomic sampling.
Three experiments were performed. In the first, a data set in which taxon sampling was incremented from 4 to 41 species was used. Saccharomyces cerevisiae was maintained as the outgroup, whereas all supplementary taxa included were Bilaterian. That is, no attempt at breaking the putative long branch between the fungi and the Bilateria was made. In the second experiment, a data set sampling 43 taxa was used. This data set was designed to contain the full complement of taxa from the first data set but additionally included T. adhaerens and N. vectensis. Here S. cerevisiae, T. adhaerens, and N. vectensis were simultaneously used as outgroups for the Bilateria. The branch joining the fungi and the Bilateria was still present, but now it was split into three shorter branches, allowing us to investigate the effect of targeted taxon sampling. Finally, the third data set sampled 42 genomes. All metazoan genomes used to generate the first two data sets were retained, whereas S. cerevisiae was removed. Excluding S. cerevisiae eliminates the long branch joining the fungi and the Bilateria, thus allowing the investigation of using only nonbilaterian metazoans (T. adhaerens and N. vectensis) as outgroups.
The analysis of the data set generated for experiment one resulted in 2,164 single-gene families passing the PTP test. Results of an input tree bootstrapping supertree analysis of the ML bootstrap trees generated for these families is reported in figure 2A and shows the placement of the Nematoda as the sister group of all the other Bilateria, that is, 100% support for Coelomata. This tree also displays monophyletic Deuterostomia, Arthropoda and, interestingly, Eutrochozoa. (BP = 98%, 100%, and 100%, respectively). The BF analysis shows that the data fit the Coelomata tree better than the Ecdysozoa tree, thus decisively discriminating against Ecdysozoa: log10-BF = 10.792 (±0.29).
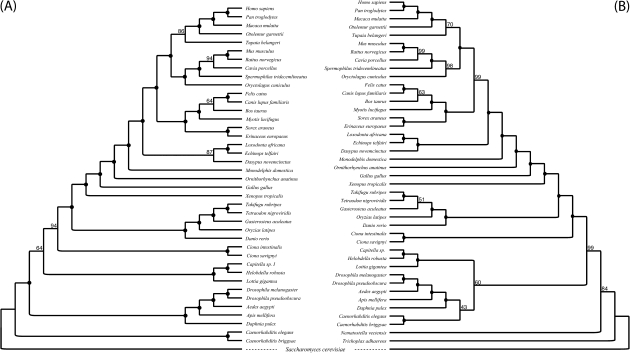
FIG. 2.—
Phylogenomic supertrees of the Bilateria. (A) A tree derived using only the fungal outgroup. This tree is based on 2,164 from 41 species. (B) A tree derived using fungal and animal (nonbilaterian) outgroups. This tree is based on 1,949 genes from 43 species. The monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia is recovered in (B), whereas (A) supports Coelomata. Numbers at the nodes represent bootstrap support. Full circles indicate 100% bootstrap support for a node.
When S. cerevisiae, T. adhaerens, and the Cnidarian N. vectensis were concurrently used as outgroups, we found a total of 1,949 single-gene families that conveyed significant phylogenetic signal (see table 1). When these gene families were used for supertree reconstruction, Ecdysozoa was recovered but with very low support (BS = 43%; see fig. 2B). Bilateria finds significant support in this analysis (BP = 99%) and is partitioned into Protostomia and Deuterostomia. Monophyly of the Eumetazoa is also supported (BP = 85%), whereas support for Protostomia is not very high (BP = 60%). Inspection of the partition table for this bootstrap analysis shows that Coelomata is still recovered, albeit with minimal support (BP = 13%). This is suggestive of an enduring LBA effect. LBA is obviously reduced when the additional animal outgroups are included in the analyses to the point where the Ecdysozoa tree is the most commonly recovered in the individual bootstrap replicates. However, the reduction of the LBA effect is not significant enough to completely exclude Coelomata from the set of possible solutions. Interestingly, BFs still favor Coelomata with respect to Ecdysozoa (at the least under WAG + G): log10-BF = 6.67 (±0.59). However, in agreement with the results of the bootstrap analysis, which suggest that the LBA effect was indeed reduced when nonbilaterian animals were included in the sample, the weight of the evidence in favor of Coelomata is now greatly decreased (by 4.122 points in a log10 scale). That is, when the fungi–Bilateria branch is broken Coelomata is still favored but the data fits the tree ∼13,243 times less well than it did when the branch was not interrupted.
In the third experiment, S. cerevisiae was interchanged with two animal outgroups (T. adhaerens and N. vectensis). With this specific taxonomic sampling, we recovered a total of 2,216 single-gene families conveying significant phylogenetic signal. Their analysis recovered a phylogenomic supertree supporting all major, recognized groups (Protostomia, Deuterostomia, Euthrocozoa, and Arthropoda). Additionally this analysis found significant support for Ecdysozoa (BS = 90%) within Protostomia (see fig. 3), with the BF now decisively discriminating against Coelomata: log10-BF = 90.811 (±0.977). If one compares the fit of the Ecdysozoa tree to the data set where S. cerevisiae is the only outgroup, with the fit of the same tree to the data set where only the animal outgroups were used, a dramatic change (∼10100) in the BF in favor of Ecdysozoa is observed. This clearly highlights the major role played by outgroup selection in phylogenomics.
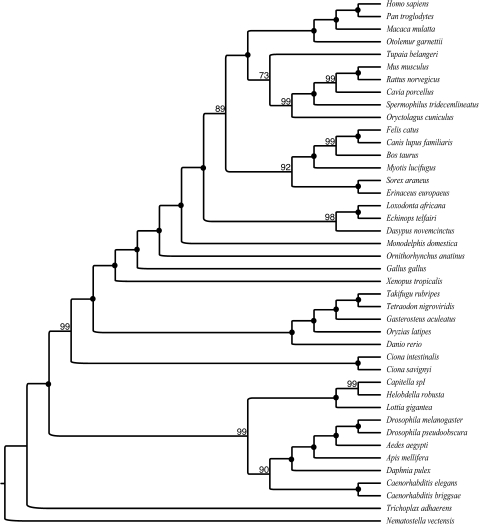
FIG. 3.—
Phylogenomic supertree of the Bilateria recovered using only animal (nonbilaterian) outgroups. This tree is based on 2,216 genes from 42 species. High support for the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia can be observed. Numbers at the nodes represent bootstrap support. Full circles indicate 100% bootstrap support for a node.
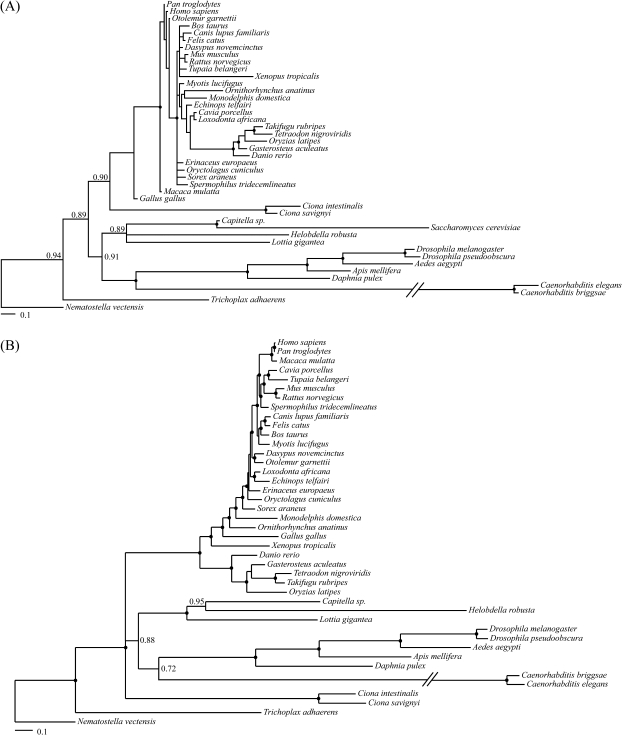
These results are finally confirmed by our supermatrix analyses. In these analyses, when S. cerevisiae was used as the only outgroup, convergence could not be reached and the resulting phylogeny (not shown) was nonsensical. When all outgroups were included (fig. 4A), Ecdysozoa was recovered, but the effect of LBA was still evident. If one was to root the tree using N. vectensis to better pinpoint the LBA effect, a tree essentially consistent with the new animal phylogeny was recovered. However, in this rooted tree, S. cerevisiae is incorrectly clustered within Protostomia. If the tree is correctly rooted using S. cerevisiae (not shown), the Lophotrochozoa would be incorrectly attracted toward the root. This result, which was somewhat unexpected, is probably a partial consequence of our gene subsampling strategy, in which we maximized information bearing on the relationships of the Nematoda, while ignoring the Lophotrochozoa and the Deuterostomia (see Materials and Methods); however, it is also clearly telling of an enduring LBA effect. Finally when only the animal outgroups are used (fig. 4B), the Ecdysozoa tree is recovered. In figure 4B, support for the Urochordata as members of the Deuterostomia is not significant, and this group is thus collapsed into a polytomy. We conjecture that this result is also most likely an effect of our gene subsampling strategy (see above). This is confirmed by the supertree analysis of our full data sets in which support for monophyletic Deuterostomia varies between 94% and 100% depending on the outgroup used (see figs. 2 and 3). Notably, a similar effect was observed in the EST study of Hejnol et al. (2009) in which Urochordata became unstable when gene sampling was reduced; see supplementary figure S1 (Supplementary Material online) Hejnol et al. (2009).
FIG. 4.—
Results of the supermatrix analyses. (A) The effect of LBA is obvious if one roots the tree using Nematostella vectensis, as a tree essentially consistent with the new animal phylogeny is recovered, but Saccharomyces cerevisiae is incorrectly nested within the Protostomia. (B) A tree illustrating that Ecdysozoa is easily recovered when analyses are performed using only nonbilaterian animals as outgroups. Numbers at the nodes represent posterior probabilities. Full circles indicate a posterior probability of 1. Posterior probabilities lower than 1 have only been reported for nodes that are relevant to the Ecdysozoa versus Coelomata problem. Urochordata is collapsed in a basal polytomy because the posterior probability of Deuterostomia is less than 0.5.
Discussion
Phylogenomics in a Pluralist Context
ESTs provide an excellent means of increasing taxon sampling and have been shown to produce highly resolved, well-supported phylogenies (e.g., Philippe et al. 2005, 2009; Dunn et al. 2008; Hejnol et al. 2009). However, EST studies consider only a shallow sampling of genomic content and include a large amount of missing data, the effect of which has never been thoroughly investigated. For Coelomata to be robustly rejected, EST data, although obviously important, cannot be considered sufficient: accord between taxonomically rich EST studies, and gene-rich deep-scale analyses must be reached. With the wealth of genomic data that is currently available, arising from an ever-increasing number of sequencing projects, coupled with advances in sequencing technologies, taxon sampling is becoming less of a limitation for deep genomic-scale phylogenetic analyses. In short, we now have at our disposal the data to conduct extensive, experimental phylogenomic studies of metazoan evolution.
Supertree methods offer an ideal solution for the reconstruction of large-scale phylogenies based upon complete genomes, as they provide a means of overcoming the limits of gene concatenation-based approaches. Gene concatenation methods, at present, do not allow for the easy amalgamation of thousands of genes. Supertrees (and in the four taxon case consensus methods), implementing a divide and conquer strategy, facilitate the analysis of entire genomes for many taxa by coalescing the results of multiple subanalyses to attain a global solution (Wilkinson and Cotton 2006). However, supermatrix approaches also have important advantages, particularly as they overcome the most important limitation of supertrees, that is that the latter do not allow hidden subsignals to interact and thus lack total evidence like properties (Pisani and Wilkinson 2002). In addition, supermatrix approaches allow for the use of statistical tools (like BFs) to test alternative phylogenetic hypotheses. Bearing in mind that both approaches have highly desirable and significantly different properties, we therefore opted for a pluralist, supertree/consensus tree and supermatrix approach in our study.
Our four-taxon analyses show that multigene families can be appropriately treated to derive species phylogenies and suitably included in a consensus tree (if all considered gene families are universally distributed) or supertree (if the gene families are not universally distributed) analyses. In particular, we show that all our consensus supertrees (including those that sample multigene families) continue to support Ecdysozoa, a result that is further confirmed by our supermatrix analyses.
Supertrees have previously been employed to address the phylogenetic position of the nematodes (Philip et al. 2005). Although carefully conducted, using the best methods and data available at that time, this analysis did contain (by the authors’ own admission) a very limited sampling of just 10 genomes. In particular, a noticeable problem that Philip et al. (2005) faced was the absence of an adequate outgroup (i.e., nonbilaterian metazoan genomes). As postulated by Philip et al. (2005), in time, an increased sampling could well serve to alter their results. In line with that prediction, our supertree analyses performed using appropriate outgroups and a significantly increased taxon (and gene in the case of the four-taxon data sets) sampling has revealed an alternative topology (see figs. 2B, 3, and 4B). Our results suggest that the study of Philip et al. (2005) and indeed other genomic-scale analyses (e.g., Blair et al. 2002; Wolf et al. 2004) may have been influenced by systematic errors arising from poor outgroup choice, sparse taxon sampling, and hidden paralogy.
Circumventing Systematic Errors
Our study illustrates the importance of outgroup choice in phylogenomic-scale studies. We see that the use of a distant outgroup has a marked effect, irrespective of whether ingroup sampling is spare or dense. We found, like in other studies (Philippe et al. 2005; Rota-Stabelli and Telford 2008), that outgroup choice completely alters the resulting topology, consequently lending analogous support to competing hypothesis. The recovery of the Coelomata topology can be considered a LBA artifact brought about by the use of a divergent outgroup. Comparison of BF values gives an indication of the strength of the bias and of how difficult it is to limit its effects. Our results also reject the contention of Rosenberg and Kumar (2001) and Rokas and Carroll (2005) that poor taxon sampling is irrelevant as long as enough genes are considered.
Our densely sampled data sets illustrate that optimal outgroup selection is more important than targeted taxon sampling in avoiding LBA artifacts. If a distant outgroup (S. cerevisiae) is included in the analysis, targeted taxon sampling (i.e., breaking the long Bilateria–fungi branch), does not completely eradicate (as shown most powerfully by the BF analyses) LBA. Only upon the exclusion of S. cerevisiae do the BFs show a radical decrease in fit of the Coelomata tree. Optimal outgroup selection is a rarely addressed topic in phylogenetics and phylogenomics, and one has to bear in mind that the optimal outgroup for a given data set is not necessarily the closest one (for an interesting example, see Rota-Stabelli and Telford 2008). Aside from LBA, another important source of phylogenetic artifacts is gene (or amino acid) composition bias, and one should thus try to select outgroups that simultaneously minimize the likelihood of both artifacts occurring.
Stringency and the Selection of Families for Phylogenetic Reconstruction
When analyzing a small selection of genomes we could not identify a number of single-gene families comparable with those identified by, for example, Blair et al. (2002). Disparity between our study and that of Blair et al. (2002) is particularly striking when comparing their four-taxon data set to our data set including S. cerevisiae. Although the ultimate results of both data sets are congruent, that is, both data sets support Coelomata; our analysis considers 70% less single-gene families than Blair et al. (2002). Failure of these data sets to have correlating numbers of single-gene families merits discussion. We suggest that the observed difference can partially be explained by the use of different outgroups. Blair et al. (2002) primarily used a plant outgroup and only in cases where plant genes were not available was a fungal outgroup used. However, this difference can also be accounted for by the implementation of measures to assess data quality in our study. Under our protocol, a gene family was only considered for phylogenetic analysis if it demonstrated significant clustering signal. Our approach thus ensured that noisy families or families devoid of clustering signal were eliminated from our analysis. It is interesting to note that prior to this filtering stage, the number of single (four-taxon)- gene families identified in our study was twice the number identified by Blair et al. (2002).
Conclusions
The Ecdysozoa hypothesis has accumulated significant support in recent years (Philippe et al. 2005; Irimia et al. 2007; Bourlat et al. 2008; Dunn et al. 2008; Lartillot and Philippe 2008; Telford et al. 2008), particularly from the analyses of EST data sets. To supplement this amassment of evidence, here, we present support for Ecdysozoa from genomic-scale data sets. From these, overall, Ecdysozoa represents the most cogent hypothesis. It is supported from the analyses of both single-gene families and multigene families, once suitable outgroups are considered. Coelomata, on the other hand, is only supported upon the inclusion of a distantly related outgroup, which suggests that this topology is systematically generated by a LBA artifact.
Our results, based on arguably the deepest gene sampling of the Bilateria to date, present overwhelming support for Ecdysozoa and clearly illustrate that it is the use of a distant outgroup that mislead previous analyses. Taken in combination with results from the aforementioned EST studies, it now appears that all aspects of molecular-based phylogenetics support the rejection of Coelomata. Although lack of unambiguous morphological support for Ecdysozoa persists as a moot point, in the light of overwhelming molecular evidence and lack of morphological evidence conclusively discrediting Ecdysozoa, is it now finally time to shed the notion of Coelomata?
Supplementary Material
Supplementary figure SI1 and table S1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
This study was supported by an “IRCSET EMBARK Initiative” PhD scholarship awarded to T.A.H., partially supported by a Science Foundation Ireland Research Frontiers Programme grant awarded to D.P. (08-RFP-EOB1595), by the High Performance Computing (HPC) Facility at NUI Maynooth, and by the Irish Centre for High-End Computing (ICHEC). We would like to thank Omar Rota-Stabelli, James O. McInerney, and David Fitzpatrick. We also thank the three anonymous reviewers for their suggestions and criticisms, which significantly helped improving the quality of this paper.
References
- Aguinaldo AMA, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Archie JW. A randomization test for phylogenetic information in systematic data. Syst Zool. 1989;38:219–225. [Google Scholar]
- Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Belinky F, Cohen O, Huchon D. Large-scale parsimony analysis of metazoan indels in protein-coding genes. Mol Biol Evol. 2010;27:441–451. doi: 10.1093/molbev/msp263. [DOI] [PubMed] [Google Scholar]
- Blair JE, Ikeo K, Gojobori T, Hedges SB. The evolutionary position of nematodes. BMC Evol Biol. 2002;2:7. doi: 10.1186/1471-2148-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlat SJ, Nielsen C, Economou AE, Telford MJ. Testing the new animal phylogeny: a phylum level molecular analysis of the animal kingdom. Mol Phylogenet Evol. 2008;49:23–31. doi: 10.1016/j.ympev.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artefacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- Burleigh JG, Driskell AC, Sanderson MJ. Supertree bootstrapping methods for assessing phylogenetic variation among genes in genome-scale data sets. Syst Biol. 2006;55:426–440. doi: 10.1080/10635150500541722. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Copley RR, Aloy P, Russell RB, Telford MJ. Systematic searches for molecular synapomorphies in model metazoan genomes give some support for Ecdysozoa after accounting for the idiosyncrasies of Caenorhabditis elegans. Evol Dev. 2004;6:164–169. doi: 10.1111/j.1525-142X.2004.04021.x. [DOI] [PubMed] [Google Scholar]
- Cotton JA, Page RDM. Tangled trees from molecular markers: reconciling conflict between phylogenies to build molecular supertrees. In: Bininda-Emonds ORP, editor. Phylogenetic supertrees: combining information to reveal the tree of life. Dordrecht (The Netherlands): Kluwer Academic; 2004. pp. 107–125. [Google Scholar]
- Creevey CJ, et al. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc Biol Sci. 2004;271:2551–2558. doi: 10.1098/rspb.2004.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevy CJ, McInerney JO. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21:390–392. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Dopazo H, Dopazo J. Genome-scale evidence of the nematode-arthropod clade. Genome Biol. 2005;6:R41. doi: 10.1186/gb-2005-6-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo H, Santoyo J, Dopazo J. Phylogenomics and the number of characters required for obtaining an accurate phylogeny of eukaryote model species. Bioinformatics. 2004;20:i116–i121. doi: 10.1093/bioinformatics/bth902. [DOI] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.67. Seattle (WA): Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Halanych KM. The new view of animal phylogeny. Annu Rev Ecol Evol Sys. 2004;35:229–256. [Google Scholar]
- Halanych KM, et al. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc B Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LH. The invertebrates. Vol. 1: Protozoa through Ctenophora. New York: McGraw-Hill; 1940. [Google Scholar]
- Irimia M, Maeso I, Penny D, Garcia-Fernàndez J, Roy SW. Rare coding sequence changes are consistent with Ecdysozoa, not Coelomata. Mol Biol Evol. 2007;24:1604–1607. doi: 10.1093/molbev/msm105. [DOI] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Jenner RA, Schram FR. The grand game of metazoan phylogeny: rules and strategies. Biol Rev Camb Philos Soc. 1999;74:121–142. [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Keane TM, Creevy CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge AG. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes) Syst Zool. 1989;38:7–25. [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of the Bilateria. Philos Trans R Soc Lond B. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood DT, Olsen PD, Telford MJ, Herniou EA, Riutort M. Elongation factor 1-alpha sequences alone do not assist in resolving the position of the acoela within the metazoa. Mol Biol Evol. 2001;18:437–442. doi: 10.1093/oxfordjournals.molbev.a003821. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- Margush T, McMorris FR. Consensus n-trees. Bull Math Biol. 1981;43:239–244. [Google Scholar]
- McInerney JO, Cotton JA, Pisani D. The prokaryotic tree of life: past, present… and future? Trends Ecol Evol. 2008;23:276–281. doi: 10.1016/j.tree.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Moore BR, Smith SA, Donoghue MJ. Increasing data transparency and estimating phylogenetic uncertainty in supertrees: approaches using nonparametric bootstrapping. Syst Biol. 2006;55:662–676. doi: 10.1080/10635150600920693. [DOI] [PubMed] [Google Scholar]
- Nielsen C. Animal evolution, interrelationships of the living phyla. Oxford: Oxford University Press; 2001. [Google Scholar]
- Peterson KJ, Cotton JA, Gehling JG, Pisani D. The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos Trans R Soc Lond B Biol Sci. 2008;363:1435–1443. doi: 10.1098/rstb.2007.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip GK, Creevy CJ, McInerney JO. The Opisthokonta and the Ecdysozoa may not be clades: stronger support for the grouping of plant and animal than for animal and fungi and stronger support got the Coelomata than Ecdysozoa. Mol Biol Evol. 2005;22:1175–1184. doi: 10.1093/molbev/msi102. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Martinez P, Riutort M, Baguñà J. Acoel flatworms are not platyhelminthes: evidence from phylogenomics. PLoS One. 2007;2:e717. doi: 10.1371/journal.pone.0000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multi gene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Pisani D. Identifying and removing fast-evolving sites using compatibility analysis: an example from the Arthropoda. Syst Biol. 2004;53:978–989. doi: 10.1080/10635150490888877. [DOI] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- Pisani D, Wilkinson M. Matrix representation with parsimony, taxonomic congruence, and total evidence. Syst Biol. 2002;51:151–155. doi: 10.1080/106351502753475925. [DOI] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007. [Internet]. Available from: http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Robinson M, Gouy M, Gautier C, Mouchiroud D. Sensitivity of the relative-rate test to taxonomic sampling. Mol Biol Evol. 1998;15:1091–1098. doi: 10.1093/oxfordjournals.molbev.a026016. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Thomson K, Csürös M, Carmel L, Koonin EV. Homoplasy in genome-wide analysis of rare amino acid replacements: the molecular-evolutionary basis for Vavilov's law of homologous series. Biol Direct. 2008;3:7. doi: 10.1186/1745-6150-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Carmel L, Koonin EV. Analysis of rare amino acid replacements supports the Coelomata clade. Mol Biol Evol. 2007;24:2594–2597. doi: 10.1093/molbev/msm218. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proc Natl Acad Sci U S A. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O, Telford MJ. A multi criterion approach for the selection of optimal outgroups in phylogeny: recovering some support for the Mandibulata over Myriochelata using mitogenomics. Mol Phylogenet Evol. 2008;48:103–111. doi: 10.1016/j.ympev.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Rare genomic characters do not support Coelomata: intron loss/gain. Mol Biol Evol. 2008;25:620–623. doi: 10.1093/molbev/msn035. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DTJ, Herniou EA, Baguna J. Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- Semple C, Steel M. Phylogenetics. New York: Oxford University Press; 2003. [Google Scholar]
- Sperling EA, Peterson KJ, Pisani D. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Mol Biol Evol. 2009;26:2261–2274. doi: 10.1093/molbev/msp148. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Robinson JM, Pisani D, Peterson KJ. Where's the glass? Biomarkers, molecular clocks and microRNAs suggest a 200 million year missing precambrian fossil record of siliceous sponge spicules. Geobiology. 2010;8:24–36. doi: 10.1111/j.1472-4669.2009.00225.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sunderland (MA): Sinauer Associates Inc.; 1998. [Google Scholar]
- Telford MJ, Bourlat SJ, Economou A, Papillon D, Rota-Stabelli O. The evolution of the Ecdysozoa. Philos Trans R Soc Lond B Biol Sci. 2008;363:1529–1537. doi: 10.1098/rstb.2007.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, Cotton JA. Supertree methods for building the tree of life: divide-and-conquer approaches to large phylogenetic problems. In: Hodkinson T, Parnell J, Waldren S, editors. Towards the tree of life: taxonomy and systematics of large and species rich taxa. Systematic Association special volume. CRC Press; 2006. pp. 61–75. [Google Scholar]
- Wolf YI, Rogozin IB, Koonin EV. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Computational molecular evolution. Oxford series in ecology and evolution. New York: Oxford University Press; 2006. [Google Scholar]
- Zheng J, Rogozin IB, Koonin EV, Przytycka TM. Support for the Coelomata clade of animals from a rigorous analysis of the pattern of intron conservation. Mol Biol Evol. 2007;24:2583–2592. doi: 10.1093/molbev/msm207. [DOI] [PubMed] [Google Scholar]
- Zilversmit M, O'Grady P, Desalle R. Shallow genomics, phylogenetics, and evolution in the family Drosophilidae. Pac Symp Biocomput. 2002;7:512–523. doi: 10.1142/9789812799623_0048. [DOI] [PubMed] [Google Scholar]