Abstract
To better understand the extent of Class II transposable element activity in mammals, we investigated the mouse lemur, Microcebus murinus, whole genome shotgun (2X) draft assembly. Analysis of this strepsirrhine primate extended previous research that targeted anthropoid primates and found no activity within the last 37 Myr. We tested the hypothesis that members of the piggyBac Class II superfamily have been inactive in the strepsirrhine lineage of primates during the same period. Evidence against this hypothesis was discovered in the form of three nonautonomous piggyBac elements with activity periods within the past 40 Myr and possibly into the very recent past. In addition, a novel family of piggyBac transposons was identified, suggesting introduction via horizontal transfer. A second autonomous element was also found with high similarity to an element recently described from the little brown bat, Myotis lucifugus, further implicating horizontal transfer in the evolution of this genome. These findings indicate a more complex history of transposon activity in mammals rather than a uniform shutdown of Class II transposition, which had been suggested by analyses of more common model organisms.
Keywords: transposon, primate, horizontal transfer, piggyBac
Background
Characterization of the repetitive landscape in mammalian model organisms initially produced findings of a disparity between Class I (retrotransposons) and Class II (DNA transposons) transposable elements (TEs) in terms of their prevalence and activity levels. Human, mouse, rat, opossum, and platypus sequencing projects revealed a general loss of Class II DNA transposon activity, suggesting a general mammalian-wide extinction of these elements (Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2004; Mikkelsen et al. 2007; Warren et al. 2008). A tighter focus on anthropoid primates by Pace and Feschotte (2007) found no signs of Class II transposition younger than 37 Ma. Recently, however, analysis of a vespertilionid bat provided evidence that Class II elements were extremely active in the recent evolutionary past (∼40 Ma to the present) of at least one mammalian lineage (Pritham and Feschotte 2007; Ray et al. 2007, 2008).
Further evidence to reject a general mammalian Class II shutdown hypothesis appeared in the form of SPIN elements from the hAT superfamily (Pace et al. 2008). Horizontal transfer of SPIN TEs within the last 31–46 Myr involving bushbaby, tenrec, and rodent genomes demonstrated the capacity for recent Class II element activity in some mammalian genomes. Novick et al. (2010) substantiated this finding with additional discoveries of hAT families spanning chiropterans, marsupials, reptiles, and primates with no apparent vertical transmission pathway, implicating horizontal transfer as the agent responsible for their presence. Although the continued propagation of a Class II element is thought to rely on its ability to infiltrate new genomes (Brookfield 2005), these were the first identified cases of DNA transposon horizontal transfer involving mammals. Thus, despite their extinction in several model genomes, the continuing role of Class II TEs in mammalian evolution should not be discounted.
Because of their ability to introduce genomic variability, TEs have long been suspected to be powerful agents of evolutionary change (Brosius 1991; Makalowski 2000; Kazazian 2004). For example, increases in TE activity in response to physiological stress may provide the foundation for the punctuated equilibrium model of evolutionary change (Zeh et al. 2009). Numerous other studies have noted a connection between TE transcription and abiotic and biotic stress (Grandbastien 1998; Li et al. 1999; Kalendar et al. 2000; Kimura et al. 2001; van de Lagemaat et al. 2003). The array of prospective genomic changes revolving about the movement of TEs within their host becomes relevant when attempting to elucidate the evolutionary history of the organism itself. As may be observed from the data now available, broad inferences regarding the dynamics of TE activity obtained from model organisms likely does not represent all mammals. Lingering questions addressed by this work include whether the shutdown of Class II TE activity observed in anthropoids extends to all primates, and if recent transpositional activity within mammals is solely from the hAT superfamily. To examine these questions, the whole genome (WGS) draft for the gray mouse lemur, Microcebus murinus, was analyzed for recent DNA transposon activity. As they were shown to be recently active in the bat, Myotis lucifugus (Ray et al. 2008), the non-hAT superfamily, piggyBac, was specifically targeted.
Materials and Methods
Identification of PiggyBac Elements
As shown in figure 1, our search strategy employed methods to recognize both known and novel piggyBac TEs. The WGS draft of M. murinus was provided by the Broad Institute (GenBank accession number ABDC00000000) and obtained in March 2008. An initial survey of known piggyBac elements was performed using the amino acid sequences for 43 autonomous piggyBac coding sequences from RepBase (Jurka et al. 2005) as a query for a local TBlastN search of the WGS. The top 40 nonoverlapping hits (E values ranging from 10−91 to 0) were extracted along with 500 bp of flanking sequence in an effort to determine the element boundaries. Extracted sequences were aligned using a local installation of MUSCLE (Edgar 2004) and used to construct consensus sequences, which were used as queries for a local BlastN search. The top 40 hits for each consensus were extracted, this time with 1,000-bp flanking sequence, and aligned to produce a more accurate consensus. This was reiterated as necessary and the consensus extended further until the boundaries of potential elements were identified. Potential autonomous sequences were searched for open reading frames (ORFs) using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi).
FIG. 1.—
Search strategy to identify piggyBac elements in the Microcebus murinus draft assembly. Initial search programs are shown in rectangles, and methods used to process all output are shown in ovals. For BlastN analyses, up to 40 hits were extracted with flanking sequence and used with MUSCLE to generate a consensus; the process was repeated to extend flanks until TIRs, and nonhomologous flanking sequences were observed.
Two packages were used for the initial search for novel piggyBac TEs. The first analysis, using PILER (Edgar and Myers 2005), was performed to search for recently active TEs of all types in a subset of the WGS comprising ∼37.6 Mb. Minimum length for discovered repetitive families was set to 100 bp and percent identity was set to 95. The output from PILER was organized into families (all sequences with 95% and higher similarity) and superfamilies (sequences from two or more families that exhibited sequence similarity). Each superfamily and family alignment was given a numerical designation. Superfamily and/or family consensus sequences were subjected to CENSOR (Jurka et al. 2005) searches to determine similarity to known repetitive elements in RepBase. The WGS data were then queried using BlastN and the consensus sequences for each presumed element. The top 40 hits obtained (generally E value << 10−5) were extracted along with 500 bp of flanking sequence. Extracted sequences were aligned with MUSCLE, and revised consensus sequences were constructed.
In addition to the PILER analysis, we used RepeatScout (Price et al. 2005) to identify potential TEs in the M. murinus genome. We analyzed 111 Mb of the WGS draft (lmer = 12) to search for potential TEs with a copy number of 100 or more. CENSOR was again used to determine similarity to known elements, and consensus sequences for possible piggyBac elements were obtained as described above using BlastN and MUSCLE.
To identify potential autonomous partners for any nonautonomous elements recovered from the three initial analyses (see fig. 1), a local installation of re-pcr (http://www.ncbi.nlm.nih.gov/sutils/re-pcr/) was used to query the mouse lemur WGS. For each element, queries were designed to include the TTAA target site duplication (TSD) typical of piggyBac transposons, the 13-bp terminal inverted repeats (TIR), and one extra base (i.e., TTAACCCTTTGCACTCGG and TTAACCCTTTGCACTCGC for npiggy1_Mm). Three mismatches and two gaps per primer were allowed, and in silico products from 1,000 to 5,000 bp were extracted. Potential hits were subjected to BlastX searches through National Center for Biotechnology Information (NCBI) using the default settings to search for matches to known piggyBac transposase sequences. Hits were then analyzed for an ORF using ORF Finder. Tentative ORFs were used to query the Microcebus draft 2X assembly in a local BlastN analysis. The top ten hits for each were extracted along with 1,000 bp of flanking sequence and aligned with MUSCLE to generate a consensus sequence. Furthermore, the amino acid sequence of the putative ORF for the newly identified transposon was aligned with a selection of known piggyBac transposases using MUSCLE. Phylogenetic analyses were conducted using MEGA4 (Kumar et al. 2004). A neighbor-joining tree was constructed using the equal input model with 2,000 bootstrap iterations.
Age Analyses
Consensus sequences for each of the reconstructed piggyBac-like families were used to create a custom library for a local installation of RepeatMasker. One quarter of the WGS assembly was masked, and the “.align” output file was analyzed using a custom Perl script, which removes hyper-mutable CpG sites and calculates distances from the consensus sequence using the Kimura 2-parameter model (Kimura 1980). The primate neutral substitution rate μ = 2.5 × 10−9 (Harris et al. 1986) was used to calculate average divergence for each family of elements. Only hits spanning at least 50% of the consensus were included in the analysis. For most of the putative autonomous elements, there were not enough hits within the appropriate size range to allow age estimation of the autonomous elements even after masking the entire WGS. As is often the case, however, there were substantially higher numbers of nonautonomous derivatives. For these nonautonomous elements, the first 100 hits spanning at least 50% of the consensus were extracted using custom Perl scripts and aligned using MUSCLE.
Visual analysis revealed several obvious subfamily groupings with each group sharing distinct features, including indels and sequence differences. Analysis of members from distinct subfamilies would artificially inflate the estimated ages. Thus, any set of five or more sequences sharing multiple features (indels and substitutions) clearly distinguishing them from the consensus was considered a separate subfamily and excluded from the distance analysis.
Comparative Analyses
Computational as well as polymerase chain reaction (PCR)-based approaches were employed to further investigate the relative periods of activity for each family of elements (fig. 2). First, we sought computational evidence of transposon mobilization among M. murinus and the Northern greater galago (Otolemur garnettii). The M. murinus database was queried using the consensus sequences for each element via BlastN. The top ten full-length insertions from each family were extracted along with 500 bp of flanking sequences. If substantial flanking sequence was not available due to the fragmented nature of the assembly, the next available hit was used until a total of ten Blast probes were collected per element. The resulting extracts were then used as queries for a local BlastN analysis of the O. garnettii genome (AAQR00000000). For example, sequences containing npiggy1_Mm loci + 500 bp of each flank identified in M. murinus were used as Blast queries when searching the current draft of O. garnettii. Hits were extracted and aligned with their respective query sequences to determine the presence or absence of the relevant transposon in O. garnettii (supplementary material, Supplementary Material online).
FIG. 2.—
Summary of comparative analyses to determine lineage specificity of selected elements. Individual piggyBac insertion loci recovered from Microcebus murinus were used as probes to query the Otolemur garnettii WGS and also to design primers for PCR-based analyses of Lemur catta, Cheirogaleus medius, and M. murinus (fig. 7). Additionally, multiple primer combinations were designed to amplify the piggyBac1_Mm ORF as per figure 8.
Taxa more recently diverged from the M. murinus lineage, Lemur catta, and Cheirogaleus medius, were then interrogated via PCR to test for recent activity. Briefly, the consensus sequence for npiggy1_Mm (estimated to be the most recently active, see Results) was used as a BlastN query of the draft 2X M. murinus assembly in order to identify specific insertion loci. The top ten hits were extracted along with 500 bp of flanking sequences, and oligonucleotide primers (Table 1) were designed to amplify the orthologous loci in a panel of primate DNAs. The panel consisted of L. catta (Coriell Institute for Medical Research, NG07099A), C. medius (Coriell, PR00794), and M. murinus (San Diego Frozen Zoo, KB6993). DNA from M. murinus and C. medius was limited and was subjected to whole genome amplification using the GenomiPhi kit (GE Healthcare) as per the manufacturer’s protocol. Twenty-five microliter PCR amplifications were performed under the following conditions: 10–50 ng template DNA, 7 pM of each oligonucleotide primer, 200 mM deoxynucleotide triphosphates, in 50 mM KCl, 10 mM Tris–HCl (pH 8.4), 2.0 mM MgCl2, and Taq DNA polymerase (1.25 units). An initial denaturation at 94 °C for 2 min was followed by 30–32 cycles of 94 °C for 15 s, the appropriate annealing temperature for 15 s, and 72 °C for 1 min and 10 s. A final incubation at 72 °C for 5 min prepared the fragments for cloning. PCR products were cloned using the TOPO-TA cloning kit (Invitrogen), and inserts were sequenced using chain termination sequencing on an ABI 3130xl Genetic Analyzer. Sequences were aligned with the original computationally identified orthologous locus from M. murinus and the npiggy1_Mm consensus sequence. All sequences generated for this work have been deposited in GenBank under accession numbers HM133643-HM133648.
Table 1.
Insertion Coordinates of npiggy1_Mm Elements and the Oligonucleotide Primers Designed to Amplify Them in the Primate Panel Described
| Contig ID, Location | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
| 8835, 3183-3822 | ACTACCACCCCAGACATTGC | TGTTCTCTTGAGTGTTTTCTATTTGG |
| 9360, 909-1549 | TACAAATGGAAGCCCACACA | TATGCCATGTGAACCTCCAA |
| 9997, 5791-6430 | GGGAGTTAAGAGGCAGTAGTGG | GCCACCAACTTTATGAGCAGA |
| 10547, 1506-2143 | GAAGCCAGGAAAGCTGCTAA | GTTGGTAATGCAGGGCAGAG |
| 28035, 3749-4388 | TGGTAGCTCACATTACTTGCTGA | TACCCACTCCCCATTTCTCT |
| 77903, 3811-4450 | TAAATGGCCCCATATGCTGT | TGCTGCTCCTGATTTCTGAC |
| 82968, 3459-4098 | GGTCCAAGATGGCAACACTT | AATCCTCCTTTGGGAAAAGC |
To test the taxonomic distribution of piggyBac1_Mm, a novel, autonomous piggyBac family (see Results), we designed an additional four oligonucleotide primers to amplify three overlapping fragments internal to its presumed ORF. The primers were as follows: piggyBac1_Mm_1086+, CTTGCAGAGTTATTGGTCCATGG; piggyBac1_Mm_1571+, GACAGGTATTACACTAGTGTCACTC; piggyBac1_Mm_1614−, CTGTCAAGTGTGTTTTTTCCTTG; and piggyBac1_Mm_2077−, CCATCTCTGAATTCTCCAACAAGATC. These primers were tested on the panel described above using similar reaction conditions.
Further analyses were performed to locate instances of the new M. murinus TEs in lineages outside Strepsirrhini. A library containing all piggyBac elements identified in M. murinus were checked against RepBase to determine similarity to other known elements. A local BlastN search of a subset of genomic databases (table 2) was carried out; hits of E value < 10−20 were extracted and aligned with MUSCLE. Consensus sequences of the alignments were then aligned with the corresponding transposon from M. murinus. TEs were also used in a more expansive BlastN search through NCBI against NR and WGS databases, excluding M. murinus.
Table 2.
Each Genome Listed Below Was Queried Using BlastN and a Custom Microcebus murinus DNA Transposon Library to Assay for Potential Cases of Horizontal Transfer
| Genome | ID | Fold-Coverage | Genome | ID | Fold-Coverage |
| Anolis | AAWZ | 6.85 | Myotis | AAPE | 2 |
| Callithrix | ACFV | 6 | Ochotona | AAYZ | 2 |
| Canis | canFam2 | 7.6 | Oryctolagus | AAGW | 7.5 |
| Carollia | 138695 | (6,606,146 bp) | Otolemur | AAQR | 2 |
| Cavia | AAKN | 6.8 | Pan | AACZ | 6 |
| Dasypus | AAGV | 2 | Petromyzon | petMar1 | 5.9 |
| Echinops | AAIY | 2 | Pongo | ABGA | 6 |
| Erinaceus | AANN | 2 | Pteropus | ABRP | 2 |
| Equus | AAWR | 6.8 | Rhinolophus | 59479 | (40,249,618 bp) |
| Felis | felCat3 | 2 | Sorex | AALT | 2 |
| Homo | ABBA | NA | Spermophilus | AAQQ | 2 |
| Loxodonta | AAGU | 2 | Taeniopygia | ABQF | 6 |
| Macaca | AANU | 6 | Tupaia | AAPY | 2 |
| Microcebus | ABDC | 1.9 | Tursiops | ABRN | 2 |
| Monodelphis | AAFR | 6.8 |
NOTE.—Depending on the source, GenBank accession numbers, UCSC genome assembly IDs, or NCBI taxon IDs are provided. For the bats Carollia perspicillata and Rhinolophus ferrumenquinum, data from the National Institutes of Health Comparative Vertebrate Sequencing Database were used and the data represent only a small portion of the genome. The number of bases queried are provided for these taxa. NA, not applicable.
Results
Identification of PiggyBac Elements
All elements described herein have been named according to standard principles (Wicker et al. 2007) and deposited in RepBase (http://www.girinst.org/repbase/index.html). Final alignments and the resulting consensus sequences are available as supplementary material (Supplementary Material online). The top 40 hits found during the TBlastN search using known piggyBac coding sequences (fig. 1) were all to piggyBac2_ML (M. lucifugus) with E values ranging from 10−91 to 0. The alignments from M. murinus fell into three groups, which yielded the consensus sequences piggyBac2_Mm, piggyBac2a_Mm, and piggyBac2b_Mm. All displayed characteristic TTAA TSDs, shared 15-bp TIRs, and an ORF region. PiggyBac2a_Mm and piggyBac2b_Mm differ from one another only by a 44-bp indel, with the former spanning a total length of 1,043 bp, whereas the latter is 999 bp. A single full-length piggyBac2_Mm was not recovered but instead the consensus was reconstructed from seven overlapping contigs to produce a 2,211-bp sequence with a 1,839-bp ORF. A 765-bp ORF was also identified in piggyBac2a_Mm and piggyBac2b_Mm. All three elements and their structures relative to the 2,639 bp piggyBac2_ML are shown in figure 3. As seen in the figure, piggyBac2_Mm harbors the entire 1,752-bp ORF from piggyBac2_ML of M. lucifugus.
FIG. 3.—
Schematic of piggyBac2_ML from Myotis lucifugus (top) and three similar piggyBac elements from Microcebus murinus. Deletions and duplications relative to M. lucifugus are indicated for any difference greater than 3 bp. The 1,752-bp ORF is shown for M. lucifugus in lighter shading.
As would be expected from a primate, results from the PILER analysis recovered mostly retrotransposons, primarily L1 and Alu. However, DNA transposon families were also evident from CENSOR hits to representatives of the hAT (hobo/activator/Tam) and Tc1/Mariner superfamilies. Although no members from the piggyBac superfamily were immediately noted, an initially unidentified superfamily was recognized as a probable piggyBac due to its TTAA TSDs. The consensus sequence was short (240 bp) and therefore likely a nonautonomous variant npiggy1_Mm. Out of 91 hits obtained from RepeatScout output, two exhibited piggyBac-like characteristics, npiggy2_Mm (348 bp) and npiggy3_Mm (276 bp). The three nonautonomous families do not share TIRs, suggesting that each is mobilized by a different autonomous partner. The unique TIRs were used in primers for re-pcr, leading to the discovery of a potential autonomous partner for npiggy1_Mm, piggyBac1_Mm, an element not recovered as part of our survey using known piggyBac transposases and therefore likely to be novel.
PiggyBac1_Mm was reconstructed from fragments identified during the re-pcr analysis. The putative autonomous element extends 2,527 bp and harbors a 1,311-bp ORF (436 aa). The size of the ORF falls short when compared with other piggyBac elements, such as those in M. lucifugus (573 aa and 583 aa; Ray et al. 2008) and Uribo elements in Xenopus (594 aa and 589 aa; Hikosaka et al. 2007). The limited size may be an artifact of an inaccurate consensus sequence. The ORF may have not been correctly reconstructed due to its rather limited representation in the genome (BlastN analysis of the WGS using the consensus only resulted in five significant hits with E value of 10−50 or better for the region upstream of the ORF described) and the actual start codon could be further upstream. Additionally, full-length autonomous elements are usually several kbp and can be difficult to piece back together when the genome has not been fully assembled. The average contig for the WGS is only 2,800 bp.
Despite these problems, the amino acid alignment with other known transposases in figure 4 shows the presence of conserved motifs thought to be involved in transposition (Keith et al. 2008). Interestingly, even with these hallmarks of piggyBac transpositional capability, the Neighbor-Joining tree (fig. 5) offers no support for a relationship to any of the known piggyBac ORFs used in the analysis. Instead, the low bootstrap values indicate that piggyBac1_Mm is unique and appears to be a novel family.
FIG. 4.—
Portion of an amino acid alignment of piggyBac1_Mm and other representative piggyBac elements. The alignment includes the Trichoplusia ni element that has been shown to catalyze transposition. Conserved motifs among the transposase sequences are shaded. Numbers and arrows indicate amino acid residue positions in the presumed piggyBac1_Mm ORF that is described in the text. The complete alignment is available as Supplementary Material online.
FIG. 5.—
Results of ORF phylogenetic analysis. Terminal nodes for all known piggyBac transposases are consensus sequences from RepBase (element name followed by genus in which it was identified) or GenBank (accession number followed by genus in which it was identified). Consensus sequences for piggyBac1_Mm and piggyBac2_Mm (boxed) were generated as described in the text.
RepeatMasker analysis showed high representation within the M. murinus genome for the three nonautonomous elements. The most copies (reported only for hits >100 bp) were recovered for npiggy2_Mm, with 3,780 hits amounting to 0.059% of the entire 1.85 Gb WGS. This was followed by npiggy3_Mm with 2,850 hits (0.032%) and npiggy1_Mm with 943 hits (0.011%). PiggyBac1_Mm was present in 501 copies, or 0.008% coverage of the WGS, but the piggyBac2_Mm TEs were much more limited with only 16 hits identified. The shorter versions, piggyBac2a_Mm and piggyBac2b_Mm, were found with 38 and 47 copies, respectively. The last three each amounted to roughly 0.001%. In all, these elements comprised approximately 0.114% of the WGS assembly.
Age Analyses
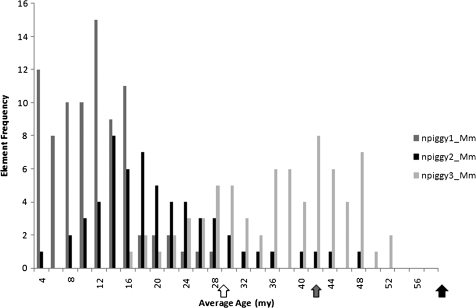
The high copy number of the three nonautonomous piggyBacs identified in M. murinus provided sufficient data for their age estimations. All displayed relatively recent activity, <40 Myr (table 3). It should be noted that piggyBac2a_Mm and piggyBac2b_Mm have limited representation in the genome; as a result, these estimates of their activity periods should be taken with caution. The larger piggyBac1_Mm and piggyBac2_Mm were not present in copy numbers large enough to allow age analysis. Figure 6 illustrates the recent peaks of activity for the nonautonomous TEs. Of particular interest is npiggy1_Mm, whose histogram suggests activity up to and including as little as 4 Ma. As denoted by the arrows in figure 6, some activity appears to have spanned the same period during which the Microcebus lineage diverged from Cheirogaleus and Lemur. Once available, these genomes should be the subject of additional analyses.
Table 3.
Divergence Values for Selected PiggyBac Elements
| Family | n | Average Divergence | Estimated Average Age |
| npiggy1_Mm | 84 | 0.026 ± 0.001 | 10–11 |
| npiggy2_Mm | 61 | 0.053 ± 0.004 | 20–23 |
| npiggy3_Mm | 73 | 0.091 ± 0.003 | 35–38 |
| piggyBac2a_Mm | 13 | 0.04 ± 0.005 | 14–18 |
| piggyBac2b_Mm | 37 | 0.039 ± 0.003 | 15–17 |
NOTE.—Sequences spanned at least 50% of the consensus size and showed no evidence of belonging to a separate subfamily. The K2P nucleotide substitution model was used, and CpG sites were excluded. Estimated ages were determined using the primate neutral mutation rate (μ = 2.5 × 10−9). Few or no elements spanning at least 50% of the consensus were not recovered for piggyBac1_Mm or piggyBac2_Mm. As a result, these were excluded.
FIG. 6.—
Histogram showing element frequency over estimated age distributions for the nonautonomous piggyBac TEs. The presumed dates of the Microcebus/Cheirogaleus, Microcebus/Lemur, and Microcebus/Otolemur divergences are indicated by white, gray, and black arrows, respectively.
Comparative Analyses
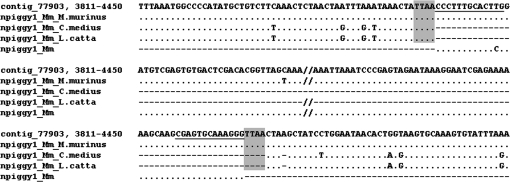
Computational analysis using full-length insertion loci from M. murinus as queries yielded “empty” loci in O. garnetti for npiggy1_Mm and npiggy2_Mm (i.e., the insertion was not present at the presumed orthologous location). For the PCR-based analyses, the more recent activity of npiggy1_Mm made it the most suitable marker for testing whether transposition has occurred in the Microcebus genome before or after the hypothesized divergences with L. catta and C. medius. Seven primer pairs for npiggy1_Mm loci provided evidence for insertions specific to mouse lemur (i.e., in the form of “filled” bands in M. murinus vs. empty bands in L. catta and C. medius [data not shown]). Figure 7 shows the unambiguous presence of npiggy1_Mm and the TTAA TSDs in the mouse lemur only for sequences generated from the PCR amplicons (see supplementary material, Supplementary Material online). PCR-based analyses of the ORF for piggyBac1_Mm, the likely autonomous partner of npiggy1_Mm, provided evidence that piggyBac1_Mm is absent from the genomes of L. catta and C. medius (fig. 8).
FIG. 7.—
Example alignment of a mouse lemur-specific Class II insertion. The WGS contig sequence is at the top with comparisons with experimentally derived sequences from Microcebus murinus, Cheirogaleus medius, and Lemur catta below. The bottom sequence is the consensus of npiggy1_Mm. TIRs are underlined, and TSDs are shaded.
FIG. 8.—
PCR amplification of piggyBac1_Mm ORF fragments from lemuriform primates. At the bottom of the figure, relative primer locations are provided on a simplified map of piggyBac1_Mm.
Finally, BlastN analyses of the genomic databases shown in table 2 revealed that piggyBac2_Mm elements from M. murinus are nearly identical (E value = 0, coverage = 94%, identity = 96%) to piggyBac2_ML from the little brown bat (M. lucifugus). Furthermore, the phylogenetic analysis resulted in a node grouping the ORFs of these two elements with 100% bootstrap support (fig. 5). Some sequence similarity was also indicated in the tenrec WGS, although it was over a smaller portion of the element (Echinops telfairi, E value = 2 × 10−102, coverage = 43%, identity = 80%). However, no evidence of this same family of elements was found in any of the other genomes surveyed, which may indicate a horizontal transfer event rather than vertical transmission to explain the presence of piggyBac2_Mm in the gray mouse lemur and the little brown bat. There was no evidence of piggyBac1_Mm in any of the surveyed data, including M. lucifugus.
Discussion
Members of the piggyBac superfamily were found to have been active within the recent past in the lineage of M. murinus. Low divergence levels among elements with shared sequence characteristics and a likely case of horizontal transfer are all evidence for Class II activity in M. murinus within the past 30 Myr and possibly ongoing. Our age estimates (table 3) show that several piggyBac elements reached their activity peaks after the period during which DNA transposon activity had become extinct in multiple other mammals. These ages may be subject to error because the mutation rate we employed has not been thoroughly calibrated for the mouse lemur lineage and because of the stochastic nature of random mutation resulting in some sequences with more or fewer mutations than others of the same age. However, when considered in conjunction with the lineage-specific insertions found for M. murinus, the evidence indicates that Class II elements were active after the divergence from both Lemur and Cheirogaleus, whose last common ancestors with M. murinus were approximately 42 and 29 Ma (Yoder and Yang 2004; Steiper and Young 2006), respectively, and likely much more recently. At least one of the three nonautonomous elements exhibit M. murinus-specific insertions, and the ORFs of putative autonomous elements were not identified in related primates.
We also identified a novel family of elements, piggyBac1_Mm. This is confirmed by the lack of similarity of the consensus to known elements in RepBase or GenBank. Despite this overall lack of sequence similarity to other representatives of the superfamily, piggyBac1_Mm exhibits many of the conserved amino acid motifs typical of them. Also interesting is the observation that piggyBac1_Mm is not identifiable in the other primate genomes surveyed. Nor, for that manner, is it identifiable in any of the genomes surveyed. This lineage-specific distribution suggests a relatively recent invasion to the M. murinus genome, at the very least, after its divergence with C. medius ∼29 Ma (fig. 6). Introduction into the genome via horizontal transfer is the most likely explanation but without any evidence of additional taxa harboring the element family, it is unclear what the source might be. Likewise, npiggy1_Mm (a likely nonautonomous partner of piggyBac1_Mm) and npiggy2_Mm were not recovered in any other genomes during the comparative analyses, suggesting lineage specificity.
The taxonomic distribution of piggyBac2_Mm is also of note and likely a clear case of introduction to the genome via horizontal transfer. This element is essentially identical to piggyBac2_ML in the little brown bat and exhibits some similarity to sequences found in tenrec but is absent from the bushbaby, O. garnettii, and all of the other genomes surveyed for this project. Both the tenrec and little brown bat have been implicated in horizontal transfer events previously (Pace et al. 2008; Ray et al. 2008; Novick et al. 2010) and may be taxa with a higher propensity for intergenomic exchange. It is possible of course that the level of sequence similarity can be explained by vertical inheritance from a common ancestor of bats (90+ Ma; Hedges and Kumar 2003) and/or afrotherians (100+ Ma; Hedges and Kumar 2003; Springer et al. 2003) followed by purifying selection and the cleansing of any evidence of these elements from many of the other genomes listed in table 2. A more parsimonious scenario, however, is that the elements were introduced into all three taxa via horizontal transfer and subsequently expanded within each genome.
Recent discoveries of horizontal transfer events in mammals have been described for members of the hAT superfamily (Pace et al. 2008; Novick et al. 2010). To our knowledge, however, this is the first documented case of horizontal transfer of piggyBac elements in mammals. The piggyBac superfamily has shown itself as a robust vector for gene transformation in insects (Sarkar et al. 2003) as well as for human gene therapy research (Feschotte 2006). Microcebus murinus is an established model organism for biomedical research in aging and Alzheimer’s disease (Eichler and Dejong 2002). Thus, the discovery of relatively recent DNA transposon activity and novel primate-specific piggyBac elements in a primate genome adds a potential new facet for gene therapy research. PiggyBac elements from the moth Trichoplusia ni were proposed as efficient vectors for directed mutation in mice and humans (Ding et al. 2005). However, some concern revolved around the lack of understanding of specific host/transposon interactions in mammals (Feschotte 2006). For instance, target site preferences within the mammalian genome could influence their effectiveness and have implications for safety. If it is possible to utilize native mammalian piggyBacs, however, these problems may be more easily avoided. Thus, these elements may represent valuable future tools for researchers interested in the genetic manipulation of primates and other mammals.
In conclusion, the recent activity of several piggyBac elements in the M. murinus genome readily illustrates how DNA transposition might still continue in mammalian genomes through lateral transfer. The expansive activity profile for the three nonautonomous TEs described demonstrates that elements have continued to expand throughout the past 40 Myr. Furthermore, npiggy1_Mm shows activity patterns suggesting that it may currently still be actively transposing in M. murinus. Finally, the successful invasion and expansion of piggyBac and hAT elements into primate and other mammalian genomes via horizontal transfer suggests that our knowledge of the impact of DNA transposons on mammalian genome evolution in general and primate genome evolution in particular is far from complete. Thus, it would be wise not to discount the potential impacts of Class II elements when considering the large numbers of mammalian genomes still to be sequenced.
Supplementary Material
Supplementary materials are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank the Broad Institute Genome Sequencing Platform and Genome Sequencing and Analysis Program, F. Di Palma and Kerstin Lindblad-Toh for making the data for M. murinus and O. garnettii available. M. Batzer, J. Walker (Louisiana State University), and O. Ryder (Zoological Society of San Diego) kindly provided DNA from M. murinus and C. medius. T. Disotell and L. Pozzi (New York University) provided insightful discussion on strepsirrhine phylogeny. This work was supported by the Eberly College of Arts and Sciences at West Virginia University (to D.A.R.). Approved for publication as Journal Article N0 J-11774 of the Mississippi Agricultural and Forestry Experimental Station, Mississippi State University.
References
- Brookfield JF. The ecology of the genome—mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–136. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- Brosius J. Retroposons–seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Ding S, et al. Efficient transposition of the piggyBac (pb) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Myers EW. Piler: identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Dejong PJ. Biomedical applications and studies of molecular evolution: a proposal for a primate genomic library resource. Genome Res. 2002;12:673–678. doi: 10.1101/gr.250102. [DOI] [PubMed] [Google Scholar]
- Feschotte C. The piggyBac transposon holds promise for human gene therapy. Proc Natl Acad Sci U S A. 2006;103:14981–14982. doi: 10.1073/pnas.0607282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Grandbastien MA. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- Harris S, Thackeray JR, Jeffreys AJ, Weiss ML. Nucleotide sequence analysis of the lemur β-globin gene family: evidence for major rate fluctuations in globin polypeptide evolution. Mol Biol Evol. 1986;3:465–484. doi: 10.1093/oxfordjournals.molbev.a040415. [DOI] [PubMed] [Google Scholar]
- Hedges S, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Hikosaka A, Kobayashi T, Saito Y, Kawahara A. Evolution of the xenopus piggyBac transposon family TxpB: domesticated and untamed strategies of transposon subfamilies. Mol Biol Evol. 2007;24:2648–2656. doi: 10.1093/molbev/msm191. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by bare-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci U S A. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Keith JH, Schaeper CA, Fraser TS, Fraser MJ., Jr Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol Biol. 2008;9:73. doi: 10.1186/1471-2199-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura RH, Choudary PV, Stone KK, Schmid CW. Stress induction of Bm1 RNA in silkworm larvae: SINEs, an unusual class of stress genes. Cell Stress Chaperones. 2001;6:263–272. doi: 10.1379/1466-1268(2001)006<0263:siobri>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- Makalowski W. Genomic scrap yard: how genomes utilize all that junk. Gene. 2000;259:61–67. doi: 10.1016/s0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2010;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Evolution. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 2007;17:422–432. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Pagan HJT, Thompson ML, Stevens RD. Bats with hats: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- Sarkar A, et al. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol Genet Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the cretaceous-tertiary boundary. Proc Natl Acad Sci U S A. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiper ME, Young NM. Primate molecular divergence dates. Mol Phylogenet Evol. 2006;41:384–394. doi: 10.1016/j.ympev.2006.05.021. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Yang Z. Divergence dates for Malagasy lemurs estimated from multiple gene loci: geological and evolutionary context. Mol Ecol. 2004;13:757–773. doi: 10.1046/j.1365-294x.2004.02106.x. [DOI] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA, Ishida Y. Transposable elements and an epigenetic basis for punctuated equilibria. Bioessays. 2009;31:715–726. doi: 10.1002/bies.200900026. [DOI] [PubMed] [Google Scholar]