Abstract
The dynamics of microsatellite, or short tandem repeats (STRs), is well documented for long, polymorphic loci, but much less is known for shorter ones. For example, the issue of a minimum threshold length for DNA slippage remains contentious. Model-fitting methods have generally concluded that slippage only occurs over a threshold length of about eight nucleotides, in contradiction with some direct observations of tandem duplications at shorter repeated sites. Using a comparative analysis of the human and chimpanzee genomes, we examined the mutation patterns at microsatellite loci with lengths as short as one period plus one nucleotide. We found that the rates of tandem insertions and deletions at microsatellite loci strongly deviated from background rates in other parts of the human genome and followed an exponential increase with STR size. More importantly, we detected no lower threshold length for slippage. The rate of tandem duplications at unrepeated sites was higher than expected from random insertions, providing evidence for genome-wide action of indel slippage (an alternative mechanism generating tandem repeats). The rate of point mutations adjacent to STRs did not differ from that estimated elsewhere in the genome, except around dinucleotide loci. Our results suggest that the emergence of STR depends on DNA slippage, indel slippage, and point mutations. We also found that the dynamics of tandem insertions and deletions differed in both rates and size at which these mutations take place. We discuss these results in both evolutionary and mechanistic terms.
Keywords: tandem repeats, comparative genomics, microsatellite emergence, DNA slippage, indel slippage, point mutations, human
Introduction
Microsatellites, or short tandem repeats (STRs), are tandemly repeated DNA sequences with a period of 1 to 6 base pairs (bp). They have been detected in all living organisms (Ellegren 2004; Coenye and Vandamme 2005; Trivedi 2006). An interesting feature is their exceptional polymorphism in natural populations, making them perfect markers in population biology studies (Jarne and Lagoda 1996). This polymorphism results from high mutation rates with up to 10−3 mutations per locus per generation in some eukaryotes (Ellegren 2004). These mutations, which are insertions or deletions of one or more repeats, are thought to essentially result from a molecular process referred to as DNA replication slippage (DNA slippage in what follows; Levinson and Gutman 1987; Ellegren 2000), although recombination events may also be a minor source of microsatellite variability (Richard and Pâques 2000; Kelkar et al. 2008). The mutational dynamics of DNA slippage have been extensively studied for long microsatellites using a variety of approaches (reviewed in Ellegren 2004). These studies have shown that the slippage rate is correlated to STR length (Primmer and Ellegren 1998; Whittaker et al. 2003; Sainudiin et al. 2004). This makes longer microsatellites more variable than shorter ones.
Among the short STRs (below 15–20-bp long), the mechanisms responsible for STR dynamics remain poorly understood, especially when STRs emerge from nonrepeated sequences. The most general model postulates that random point mutations generate very short STRs (sometimes called proto-microsatellites; Jarne et al. 1998). Once a threshold length has been reached, DNA slippage becomes active, whereas the role of point mutations becomes negligible (Messier et al. 1996; Rose and Falush 1998). From a molecular perspective, the threshold length would result from the minimum length of repeated sequences allowing stable misalignment and therefore DNA slippage. Other authors have proposed a more continuous version of this general model, suggesting that slippage occurs even at very short STRs, although at a reduced rate (Pupko and Graur 1999; Noor et al. 2001; Sokol and Williams 2005). Under this scenario, there is no threshold. More recent studies have suggested that a third molecular mechanism, called indel slippage, may contribute to the creation of very short STRs (Dieringer and Schlötterer 2003). Indeed, the majority of small indels (1–4 bp) occurring in the human genome are tandem duplications and deletions (Zhu et al. 2000; Messer and Arndt 2007). As some of these duplications have also been detected at sites with no pre-existing repeats, they constitute a new class of mutations that are not encapsulated by the standard slippage model (Levinson and Gutman 1987). In contrast to DNA slippage, which occurs only in tandem repeats, indel slippage is expected to happen at a constant rate and at random genomic positions (Dieringer and Schlötterer 2003).
The existence of a threshold length for DNA slippage has essentially been inferred from model-fitting methods (Rose and Falush 1998; Sibly et al. 2001; Dieringer and Schlötterer 2003; Lai and Sun 2003). Using these methods, distributions of STR lengths from actual genomes are fitted to length distributions produced by mutation models with specified parameters (e.g., mutation rate). For example, models that assume the only force generating tandem repeats is point mutations produce length distributions that drop off very fast as the probability of reaching more than four or five repeats, even for mono- or dinucleotides, is extremely low (Dieringer and Schlötterer 2003). Applying the point mutation model to the yeast genome, Rose and Falush (1998) showed that the actual distribution of STR lengths departs from the model’s distribution of STR lengths beyond a threshold length of eight nucleotides for mono- to tetranucleotides. This suggests that slippage becomes effective only above this threshold. Subsequent studies have extended this result to other genomes, sometimes proposing a slightly different threshold length, for example, nine nucleotides for mononucleotidic repeats (Sibly et al. 2001; Lai and Sun 2003). However, only one study considered the mechanism of indel slippage on small repeats (Dieringer and Schlöttere 2003). The authors compared the shapes of length distributions when including or excluding indel slippage and observed that the model including indel slippage produce a curve that was more similar to the curves observed in actual genomes than alternative models. However, these simulated distributions were not fitted to actual data. Comparative analyses have also shed some light on the threshold issue. Comparing homologous loci across related species with known evolutionary relationships, this approach has been extensively used to analyze the dynamics of long STRs (Richard and Dujon 1996; Angers and Bernatchez 1997; Dettman and Taylor 2004). The approach has also demonstrated that tandem duplications occur at STR loci shorter than eight nucleotides in swallows (Primmer and Ellegren 1998), fruit flies (Noor et al. 2001), and conifers (Sokol and Williams 2005). These outcomes were interpreted as a result of DNA slippage. Indel slippage, again, was not considered. Moreover, these observations could not be generalized because of the limited number of studied loci.
The issue of the minimal threshold length for DNA slippage thus remains open, with conclusions mainly depending on the investigation method. If a threshold exists for DNA slippage, observations of tandem duplications at very short repeated loci should result from chance insertion or indel slippage. This would agree with the conclusions of model-fitting methods. If there is no threshold, it is not clear why models do not detect the effect of DNA slippage. It is possible that the rate of DNA slippage at very short loci is simply too low to be detected from distributions of existing microsatellites. Moreover, model-fitting methods do not allow one to distinguish between those molecular processes acting on short STRs because these methods work on the net product of various forms of mutations (i.e., point mutation, slippage, and indel slippage).
Here, we address the question of a minimal threshold length for DNA slippage based on a comparative analysis of the whole human and chimpanzee genomes, therefore significantly expanding the number of loci over previous studies. Such a comparative approach is now widely used to analyze various features of genome structure and evolution (Lynch 2007) and has been applied to compare long homologous STR loci between the human and chimpanzee genomes (Webster et al. 2002; Kelkar et al. 2008). We used the genome of the rhesus macaque (Gibbs et al. 2007) as an outgroup to the human and chimpanzee genomes to polarize the direction of mutations. To minimize misinterpretations due to sequencing errors, we focus on mutations in the human genome only because its sequence is of high quality (i.e., we assume that the whole variation is the consequence of mutation events, not of sequencing errors). The large number of mutations screened in this comparison provides statistically reliable results. We analyzed mutations occurring at mono- to hexanucleotides as short as (period + 1) and of length 1–20 bp. This allowed us to estimate divergence rates (which are correlated to mutation rates) for indels and substitutions as a function of STR length.
Methods
Sequences and Alignment
We used the multiple alignment of 27 vertebrate genomes, available at the University of California, Santa Cruz (UCSC) website (http://hgdownload.cse.ucsc.edu/downloads.html) (Hinrichs et al. 2006). This alignment includes the genomes of human (Homo sapiens, version NCBI 36.1, March 2006), chimpanzee (Pan troglodytes, version PanTro 2, March 2006), and rhesus monkey (Macaca mulatta, Baylor College of Medicine Human Genome Sequencing Center, version v.1.0 Mmul_051212, January 2006). Alignment files were downloaded for the whole human genome, and alignment sequences of the three primates were extracted. Alignments were conducted using the MULTIZ algorithm (Blanchette et al. 2004), which first aligned the human and chimpanzee genomes, then aligned the macaque genome on the resulting alignment, and sequentially added the sequences of the other species. Such a method ensures that the human–chimpanzee–macaque alignments were not modified by the addition of other species, to the exception of additional gaps (indels) shared by the three genomes when compared with the other species. These gaps were discarded before analysis.
MULTIZ provides local alignments only derived from a BlastZ computation, and filters these alignments using the “net” approach (Kent et al. 2003). This filter ensures that each nucleotide of a given genome is aligned with a single nucleotide in other genomes and keeps the best alignment (in terms of BlastZ score) when more than a single alignment is possible. A potential drawback of this method is that the best alignments may not be orthologous, for example, in duplicated regions or for transposable elements (TEs). We therefore restricted our analysis to alignments between homologous chromosomes of the human–chimpanzee–macaque genomes (as suggested by the synteny computation provided at the Ensembl website; Hubbard et al. 2007). This represents more than 90% of available alignments.
Alignment errors such as indel misplacement can bias the determination of STRs, especially with score-based alignment methods such as BlastZ (Lunter et al. 2008). However, Lunter et al. (2008) demonstrated that erroneous alignment is negligible for divergences as low as that between the human and chimpanzee genomes.
Locating Mutations and Estimating Divergence Rates
Our study is based on the comparison of divergence rates for indels and substitutions at STR loci between the human genome and a chimpanzee–macaque consensus sequence. We first constructed the filtered sequence of the chimpanzee and macaque genomes using downloaded alignments. Identical sites between the chimpanzee and macaque sequences were retained. Sites that differed, including those with a gap in one of the genomes, were replaced by strings of Ns and not considered in the analysis. Hereafter, we refer to this sequence as the chimpanzee–macaque consensus sequence (CM-cons sequence).
STR lengths are traditionally expressed in repeat numbers, and their periods are often called repeat units (Chambers and MacAvoy 2000). This nomenclature is appropriate for studies on long STRs in which polymorphism is more important than actual length. Here, we focused on very short STRs, where each nucleotide may be of importance for the action of DNA slippage. Therefore, we refer to all STR lengths in nucleotides in what follows, regardless of STR period. The term “period” rather than “repeat unit” will also be exclusively used to prevent any misinterpretation. We restricted our analysis to STRs of length 1–20 bp, and periods of 1–6 (mono- to hexanucleotides).
We defined STRs as all perfectly repeated sequences with length equal or larger than (period + 1) nucleotides. For example, a dinucleotidic STR has a length equal to, or larger than, three nucleotides. This value was preferred to a minimal length of (period × 2) nt, as DNA slippage is theoretically possible even with less than two full repeats. Indeed, the standard slippage model (Levinson and Gutman 1987) requires a single base misalignment for slippage to occur, that is, misalignment of a full repeat is not necessary. STRs should also be maximal (not a subpart of a larger STR with the same motif), and of minimal motif (not a repeat of a shorter motif). Moreover, potential STRs with an “N” either at a flanking site, or at an internal site, were not considered, to avoid misclassifying STRs with regard to length.
Our analysis was based on the calculation of several divergence rates that were defined as the number of mutated positions (nucleotide sites) in the human genome out of the total number of sites of interest. In our analyses, the sites of interest were positions within STRs when we studied indels, and positions adjacent to STRs when we focused on substitutions. Both were allocated to categories depending on the period p and length l of the STR they were associated with. Each category was represented by its total number of nucleotide sites. Mp,l was defined as the number of sites belonging to STRs of period p and length l, and Pp,l as the number of sites adjacent to STRs of period p and length l.
Nucleotide sites were allocated to a given STR category using extension procedures derived from Main and Lorentz’s algorithm (Main and Lorentz 1984) (see supplementary fig. 1 for an example). The CM-cons sequence was scanned base to base from left to right. For each position n, the longest STR of period p to which it belongs was found by performing two extension procedures (rightwards and leftwards). The first (rightwards) starts by comparing the nucleotides at positions n and n + p. When they were identical, positions n + 1 and n + p + 1 were compared. This procedure was continued x times until the nucleotides were different. This made the end position of the STR n + p + x. The start position n − y was found by extending rightward from n−1 versus n + p − 1. The STR size was then given by l = p + x + y. A second extension procedure (leftwards) was performed between position n and n−p. We were then faced with three possibilities. 1) When both extension procedures gave the same STR, it was counted only once. The position n was counted in category Mp,l, with l = p + x + y. 2) When STRs had the same size but not the same motif, only one was counted. The position n was counted in Mp,l, with l = p + x + y. 3) When the STRs did not have the same size, n belongs to both STRs and was counted into both STR categories (e.g., the third site of ACATAT was counted in M2,3 for ACA and in M2,4 for ATAT).
A similar procedure was used for detecting and counting sites adjacent to STRs: the length of STRs adjacent to position n was derived by running the extension procedure leftwards (n − 1 versus n − 1 − p) and rightwards (n + 1 versus n + 1 + p) from this position. Several situations could be distinguished: 1) a position flanked by an STR on each side, of different period, motif, or phase was counted for both STRs. For example, C in TTTCGAGA was counted in categories P1,3 (for TTT) and P2,4 (for GAGA). 2) A site flanked by two, or more, STRs on the same side was also counted for both STR categories. For example, G into TAAATAAAG was counted in P1,3 and P4,8. 3) A site flanked on both sides by two STRs of the same period, motif, and phase was counted only once, and its length was considered equal to the sum of both STR lengths (e.g., the A site in GTGTATGT was counted in P2,7). This principle also applied to sites flanked by an STR on one side and a base in agreement with the motif and phase on the other side (e.g., A in GTGTGTAT), as well as to sites where a mutation might create an STR larger than (period + 1) bp (e.g. T in CAGTAG was counted in P3,5, or A in TAT counted in P1,2).
Note that, when following this definition and procedure, each position was counted at most once for STRs of a given motif size. In other words, the same sequence may be interpreted as several STRs displaying distinct motifs of different size (e.g., TAAATAAA is a TAAA repeated twice, although the sequence also includes two mononucleotidic STRs of motif A). These STRs indeed overlap in sequence, but were treated independently. In our example, position 4 belongs to the mononucleotide AAA, as well as to (TAAA)2 (see also supplementary fig. 1). This position was counted twice (mononucleotides of length 3 and tetranucleotide of length 8). However, the two categories were analyzed separately, and allowing for overlapping in the counting procedure did not bias our results. A second methodological point is that our detection method returns short perfect STRs that are parts of longer, imperfect, or compound ones as lonesome microsatellites. However, previous studies including imperfect microsatellites showed that the fraction of short perfect STRs decreases exponentially with STR length (Leclercq et al. 2007). In other words, the number of short STRs of a given length included in longer imperfect ones is far from the number of solitary short loci of the same length, and we assumed that they have little influence on divergence rates (defined below).
Once nucleotides had been allocated to a given category, we looked for mutations occurring at this position in the human genome compared with the CM-cons sequence. For example, when the human sequence CCATATTAG was aligned to CGATA-TAG in the CM-cons sequence, a substitution (G to C) and a 1-bp insertion (T) were counted. These mutations were counted per category. This produced a number Ip,l,s of insertions (respectively, Dp,l,s for deletions) of size s that occurred within STRs of period p and length l, and a number Sp,l of substitutions occurring at sites adjacent to STRs of period p and length l. Ip,l,s and Dp,l,s were divided by Mp,l to obtain the divergence rate at STR sites for insertions and deletions, respectively, and Sp,l was divided by Pp,l to obtain the divergence rate for substitutions at sites adjacent to STRs. Indels were defined as focal when p = s, mutiple of focal when s was a multiple of p, and nonfocal otherwise.
Our categorization procedure was not restricted with regard to STR length. For example, when no repeat was present for period p at, or adjacent to, a given position, this position was counted in the category Mp,p, and Pp,p, respectively. As we defined STRs as repeated sequences of length at least (period + 1), divergences calculated for Mp,p, and Pp,p were referred to as no-repeat reference (NR) divergence rates. These values were used as divergence estimates free of STR effect.
Focal insertions occurring at STR sites result in tandem duplications when insertions are identical to the motif studied. The fraction of duplications among insertions was calculated for all lengths studied by comparing the inserted motif with those bounding the insertion site. We also estimated the expected rate at which duplications might occur at random (outside STRs) as a function of the motif considered, its size, and the GC rate of the human genome (see supplementary table 2 for details). The expected values are 0.258, 0.067, 0.017, 0.0044, 0.0011, and 0.0003 for insertions of size 1–6, respectively.
Calculation of Confidence Intervals
In the analyses we separately considered insertions, deletions, and substitutions, and each was categorized according to both motif size and STR length. Confidence intervals (CIs) were built as follows for each category (see supplementary fig. 1): mutations of a given category were randomly distributed (drawn without replacement) across 100 independent “boxes.” Divergence rates were recalculated for each box, giving a distribution of 100 divergence rates for each category. The 95% CI was given by the 94 less extreme divergence rates. CI for the proportions of duplications among insertions were built similarly.
Results
No Threshold Length for Slippage Mutations
We analyzed the whole-genome multiple alignment provided by the UCSC to retrieve mutations that occurred at (or adjacent to) very short human STRs since the human–chimpanzee divergence. The raw counts upon which divergence rates were estimated are provided in supplementary table 3, and summarized in table 1 for indels. We highlight that the number of STRs and mutation events detected is large enough to ensure statistical relevance when calculating divergence rates at short loci. However, this number dramatically decreases with length, and the divergences estimated at the longest STRs (larger than 12–15 bp) should be considered with caution. We first estimated the divergence rates for focal indels (i.e. with length equal to the STR period) occurring in STRs as short as (period + 1) present in the CM-cons sequence. Our results showed a clear exponential increase in divergence rates with STR length for all STR periods (fig. 1). Linear regression models were fitted on the relationship between size and log-divergence (from 1 to 10 nt for mononucleotides and from n to 15 for all other n nucleotides), and the part of variance explained by the models (r2) exceeded 92% in all cases. The increase was very strong for all periods, as indel rates gained more than two orders of magnitude over the range of STR lengths studied (1–20 bp). For all periods, the increase in divergence rate for insertions started from (period + 1), and did not show any lower bound threshold (fig. 1a). The deletion rates also increased from (period+1) for mono- and dinucleotidic sites. However, no increase was detected from (period) to (period+1) for trinucleotides and larger periods, and this was followed by a step increase from (period+2) (fig. 1b).
Table 1.
A Summary of Some Raw Counts used in this Study
| Coverage (nt) | Insertions | Deletions | |
| Mononucleotide | |||
| No repeat | 1641434130 | 143600 | 383306 |
| 10 nt | 105270 | 1685 | 2997 |
| 20 nt | 5440 | 27 | 114 |
| Dinucleotide | |||
| No repeat | 911725879 | 73516 | 200491 |
| 10 nt | 281748 | 369 | 471 |
| 20 nt | 1001 | 4 | 14 |
| Trinucleotide | |||
| No repeat | 1529675024 | 126273 | 340746 |
| 10 nt | 1140837 | 224 | 1183 |
| 20 nt | 2527 | 6 | 21 |
| Tetranucleotide | |||
| No repeat | 1552650634 | 117391 | 320009 |
| 10 nt | 3845566 | 546 | 2792 |
| 20 nt | 6500 | 20 | 24 |
| Pentanucleotide | |||
| No repeat | 1653486490 | 148139 | 400736 |
| 10 nt | 14490438 | 1403 | 5737 |
| 20 nt | 8054 | 20 | 28 |
| Hexanucleotide | |||
| No repeat | 1541103809 | 138927 | 352123 |
| 10 nt | 48631861 | 4992 | 13900 |
| 20 nt | 7562 | 17 | 27 |
The number of nucleotides analyzed for unrepeated, 10-bp length, and 20-bp length mono- to hexanucleotides found in the CM-cons sequence (see Methods) is reported in the first column. The overall number of insertions and deletions detected at these sites in the human genome are displayed in the second and third columns. The no repeat category corresponds to events with size equal to that of the motif considered (e.g., 2-bp length for dinucleotides). See supplementary table 3 for more detailed distributions of insertions and deletions.
FIG. 1.—
Divergence rates (log scale) for human focal (a) insertions and (b) deletions at STR loci of period 1–6 and length 1–20. Large black squares represent the NR divergence rate for the curves they are associated with. CIs were not displayed for the sake of clarity.
To confirm that the increase in focal insertions is the consequence of tandem duplications and not due to the insertion of random nucleotides, we calculated the proportion of duplications among insertions. This proportion showed a clear significant deviation from the expectation under random motif insertion and increased with STR length for all periods (fig. 2). This proportion was far larger than the expectation even when no repeat pre-exists (white circle in fig. 2), that is, when DNA slippage is not possible. For example, the proportion of duplications among mononucleotidic insertions was 3.5 times larger than expected by random. For di-, tetra- and hexanucleotidic insertions, the rate of duplications was 1, 2, and 3 orders of magnitude higher, respectively. This strongly suggests that tandem duplications without repeats do not occur under random point mutation only, but presumably results from the action of indel slippage. Interestingly, proportions also depend on the size of the inserted motif in a nonlinear way: duplications represented 90% of 1-bp insertions, a proportion reduced to 65% for 2- to 4-bp insertions, and to about 41% for 5- to 6-bp insertions. Note that the NR divergence rate for focal insertions (Fig. 1a, black squares) was also almost constant for di, tri- and tetranucleotides, whereas it was larger for mononucleotides and lower for penta- and hexanucleotides.
FIG. 2.—
Proportion of observed tandem duplications among focal insertions at STR loci of period 1–6 and length 1–20. The white circles in each histogram represents the NR proportion. Black circles are proportions for STRs of length from (period+1) to 20. 95% CIs are represented as vertical, shaded areas. Horizontal, red bars represent the expected proportions when inserted motifs are random, and depend on motif size and composition (see Methods for details). For tetra- to hexanucleotides, these bars are indistinguishable from the abscissa line.
Our analysis also indicated that divergence rates for both insertions and deletions at mononucleotidic STRs follow a more than exponential increase with STR length, to reach a maximum value around 10- to 11-bp STR before slightly decreasing (fig. 1). The same behavior was observed for insertions at dinucleotide loci, except that we observed a stabilization rather than a decrease for loci larger than 12 bp. An increase of divergence rates for indels was also observed for “multiple of focal” indels, beginning though at larger STR lengths than for focal indels (supplementary fig. 4).
A comparison of insertion and deletion rates showed that deletions were on average 1.3–3 times more frequent than insertions, for a given STR length and period (fig. 1). For STR lengths with small CIs (≤10–12 bp, CI not shown), deletion rates were almost always larger than insertion rates, with a maximum of 3.82 deletions for one insertion at 5-bp dinucleotidic loci. Five- to 6-bp mononucleotides were exceptions with insertion rates trespassing the deletion rate, a tendency that reversed at larger lengths.
Unbiased Point Mutation Around Microsatellite Loci
The divergence rates for substitutions at sites adjacent to STR loci did not vary with STR length and were equal to the NR divergence rate for almost all STR periods (fig. 3). The 0.6% divergence recovered here for all periods is in good agreement with previous reports of divergence rate for substitutions between the human and chimpanzee genomes (Mikkelsen et al. 2005; Taylor et al. 2006). (Our estimates are for mutations that occurred in the human genome only. Values should be doubled to obtain divergence rates between the human and chimpanzee genomes, assuming equal mutation rates in these two genomes.) The only exception was for substitutions adjacent to dinucleotidic loci, where we observed a divergence rate of 0.65% for 3-bp loci, increasing up to 1% for 9-bp loci. These values were significantly, although weakly, higher than the NR divergence rate. Note also that the mean values for STRs larger than 10 bp had large CIs (fig. 3) and should therefore be considered with extreme caution.
FIG. 3.—
Divergence rate (log scale) for human substitutions at sites adjacent to STR loci of period 1–6 and length 1–20. 95% CIs are represented with dashed lines. Horizontal red lines give the NR divergence rates for each period.
Because point mutations are not restricted to substitutions, we also conducted an analysis of nonfocal indel mutations (of size different from, and not multiple of, the period) occurring at STR loci. Divergence rates for nonfocal indels were almost constant and equal to the NR divergence rate for all STR lengths and periods, for most indel sizes considered (examples are given for di- and trinucleotidic loci in fig. 4). However, there were some exceptions. The rate of 3-bp deletions, for example, was significantly larger than the NR divergence rate at dinucleotidic loci shorter than 5 bp, though identical to the NR divergence rate for larger loci (fig. 4). Other exceptions were 1-bp deletions at dinucleotidic STRs, which occurred at constant but lower rate than the NR divergence rate, and 2-bp deletions at trinucleotidic sites, which occurred at a constant but higher rate than the NR divergence rate. Importantly, these exceptions were limited to deletions and did not seem to show any consistent pattern. This analysis cannot be conducted on mononucleotides, for which all indels are focal or multiple of focal.
FIG. 4.—
Divergence rate (log scale) for human nonfocal insertions and deletions at di- and trinucleotides of length 2–10. Nonfocal indels have size of 1, 3, and 5 bp for dinucleotides, and 1, 2, 4, and 5 bp for trinucleotides. CIs are given as shaded areas. A, B, and C in the right-hand panels refer to NR divergence rates for 1-bp (A) and 3-bp (B) deletions at dinucleotidic loci and 2-bp deletions at trinucleotidic loci (C). The red lines indicate the upper and lower limits of their CIs. The four other NR divergence rates were not displayed because they were included in the CIs of the relevant nonfocal deletions. Values for loci larger than 10 bp were not depicted because of too large confidence intervals.
Discussion
The DNA slippage model assumes that slippage can be initiated at very short size—indeed at (period + 1) nucleotides, the shortest size at which the polymerase can be misled at replication (and not at, or beyond, two full copies as often mentioned in the literature). Our study of orthologous STRs shows that the tandem duplication rate increases continuously as a function of STR length, beginning with the shortest possible length of (period+1) bp (figs. 1 and 2). The relationship is exponential and valid for all period sizes, that is, for mono- to hexanucleotidic STRs. Tandem deletion rates are influenced in the same manner, with the slight difference that the effect starts at a length of (period+2) for tri- to hexanucleotidic STRs. Thus, tandem duplications/deletions act on short STRs and there is no evidence of a minimum threshold length.
Tandem duplications and deletions may in principle result from two processes, DNA slippage and indel slippage, and their respective influences have to be evaluated. Comparing repeated and nonrepeated sites should help us in this endeavor. At nonrepeated sites, the rate of tandem duplication was significantly higher than expected: The observed tandem duplications cannot be created by a series of point insertions and therefore reflect indel slippage alone. The rate of tandem duplications within STRs is larger than the rate at nonrepeated sites for a given period (compare white and black circles in fig. 2). The traditional view attributes this pattern to DNA slippage. Our work suggests that DNA slippage acts continuously on STRs of length ≥ (period + 1), and can expand a proto-STR as soon as a single base is repeated. This contradicts the conclusions of previous research based on model-fitting approaches concerning the existence of a threshold size for DNA slippage. It remains of course possible that indel slippage also occurs within STRs, as discussed below.
This opens the question of why model-fitting methods predict a threshold length from STR distributions. One explanation may reside in the power of the models to detect low slippage rates. Model-fitting methods are based on the comparison between actual length distributions of microsatellites in genomic data and theoretical distributions generated through point mutations alone. Our approach allowed for a direct estimate of the respective rates of substitutions and indels, and we found a slippage-induced divergence (focal indels) at least 100 times lower than that caused by random substitutions at very short STR loci (compare fig. 1 and fig. 3). Expansions resulting from slippage are therefore hidden by the bulk of random expansions by substitutions, and cannot be detected through the method implemented in previous works.
Our results also provide some insights on the mutational processes at sites adjacent to short STRs. Substitutions at those sites occur at random (as they do in other parts of genomes) regardless of STR size. This is a classical assumption of models describing size variation of STRs (Rose and Falush 1998; Sainudiin et al. 2004; Buschiazzo and Gemmel 2006) that had not yet been evaluated properly. We detected some exceptions though, with a positive influence of STR length on the substitution rate at sites flanking dinucleotides. Such a relationship has already been observed at AC loci (Brohede and Ellegren 1999), with a substitution rate at bases adjacent to AC microsatellites that is higher than the background rate in humans. Dinucleotide loci also show periodic patterning in their flanking regions, caused by nonrandom association with other dinucleotidic repeats (Vowles and Amos 2004; Varela et al. 2008). This association was explained by an increased substitution rate at microsatellite boundaries. Although similar patterns can be obtained in simulations of microsatellite sequence evolution under random substitutions (Webster and Hagberg 2007), our data confirm the existence of a small mutation bias in the vicinity of dinucleotides.
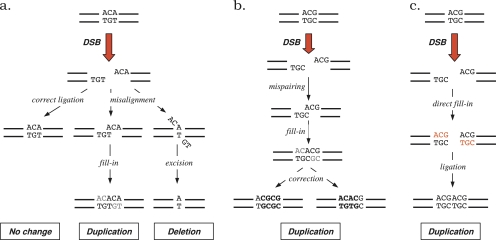
Another important result of our study is the confirmation at wide genomic scale of the occurrence of indel slippage (Zhu et al. 2000). Messer and Arndt (2007) proposed that indel slippage might be explained from a molecular perspective through nonhomologous end joining (NHEJ) repair. NHEJ is a by-product of the standard ligation of complementary overhanging DNA ends dedicated to repair double-strand breaks (DSB; Paques and Haber 1999). The complementarity of overhanging ends ensure correct ligation in most cases, but misalignment may occur which induce tandem duplications or deletions of a few bases (fig. 5a). However, NHEJ-induced indels cannot explain tandem duplications at unrepeated sites as they require microhomologies. A solution, though, is that NHEJ may provoke mispairings stable enough to allow for full repair of DNA breaks (Fig. 5b). The mismatch would then be corrected by the mismatch repair system, leading to a small tandem duplication without any preexisting repeat. Tandem duplications may also be the consequence of an alternative NHEJ process that includes direct fill-in of complementary ends, followed by ligation of double-stranded ends (Roth et al. 1985) (Fig. 5c). Our results show that the indel-slippage rate does not vary linearly with the size of the duplicated fragment, decreasing from mono- to hexanucleotides. Mononucleotide duplication is extremely frequent. Duplications occur at about the same rate for di- to tetranucleotides, and at a similar, though lower, rate for penta- and hexanucleotides (fig. 2). Such a variation might result from preferential size for DNA cleavage. DNA breaks might more often include 2-bp cleavage, whereas 3- to 5-bp cleavages may occur at a similar rate, as well as 6–7 bp, assuming that mispaired NHEJ is the major source of indel slippage (fig. 5b). Cleavage sizes should be reduced by one nucleotide in the case of NHEJ with direct fill-in of complementary ends (Fig. 5c). Chemicals (toxic agents) and radiations (UV, IR, X-rays) are natural sources of DSB, although the biochemical processes involved are not well understood. We are thus unable to provide some ground at the molecular level to such a variation in cleavage size nor to discriminate between the two NHEJ processes possibly involved in indel slippage.
FIG. 5.—
Models of indel slippage induced by NHEJ. Ligation of complementary ends is a molecular mechanism dedicated to repair DSB in all living organisms. It is an efficient, error-free process. (a) NHEJ occurs when complementary strands misalign during ligation, because of microhomologies in the cleaved sequence. This process can lead to tandem duplications or deletions of a few bases, depending on the misalignment (Paques and Haber 1999). (b) Mispairing during ligation can lead to tandem duplications of a few bases in the absence of microhomology. The duplicated motif depends on the direction of the mismatch correction. The size of the duplicated fragment is shorter by one nucleotide than the cleavage size. (c) An alternative NHEJ process includes direct fill-in of complementary ends, followed by a ligation of double-strand ends (Roth et al. 1985). This process leads to tandem duplication of the cleaved bases, even in the absence of microhomology.
Our study also opens the interesting possibility that indel slippage may not only be a source of duplications from nonrepeated sequences, but might also affect size variation in STR as a function of STR length. We found an exponential, or nearly exponential, increase in the divergence rate through insertions with STR length. This confirms observations at long microsatellite loci, from both mutation analysis in yeast (Wierdl et al. 1997) and pedigree and comparative genomics analyses in humans (Leopoldino and Pena 2003; Kelkar et al. 2008). Model-fitting methods also showed that nonlinear increases in DNA slippage rate with STR length provide a better fit to empirical distributions than linear increases (Calabrese and Durrett 2003; Whittaker et al. 2003). In the standard DNA slippage model, the slippage probability should be multiplied by the number of sites where slippage can occur (i.e., STR length) leading to a linear relationship between mutation rate and STR length. To explain the observed exponential relationship, some authors proposed a less efficient proofreading exonuclease activity in longer repeats (Wierdl et al. 1997), or an interference between the slippage loop and the DNA polymerase (Kelkar et al. 2008). Another possibility is that the rate of both DNA slippage and indel slippage increases with STR length. This may happen if STR length affects the rate of DSB events. STRs can block the replication machinery in a length-dependent manner (Samadashwily et al. 1997; Hile and Eckert 2004), which is known to induce DNA breaks (Michel et al. 2001; Saintigny et al. 2001). However, these breaks occur beyond the replication fork on single-stranded DNA and are repaired through homologous recombination (Jakupciak and Wells 2000; Michel et al. 2001). This does not exclude that alternative undiscovered mechanisms promote NHEJ in microsatellites.
Our study also produces three intriguing results. The first intriguing result is the plateauing curves of focal indel rates for long mono- and dinucleotides (fig. 1). This is unexpected because the mutation rate is known to increase with STR length. It is precisely this increase that might artifactually create these plateaus. Given that alignment procedures are unable to distinguish between indels that occurred at distinct sites in the same STR, two independent indels will be returned either as a single larger one (when both are expansions or contractions), or as no variation (when one is a contraction and the other an expansion). This might increase the number of both multiple of focal mutations and no mutation for a given length (the latter situation is referred to as homoplasy; Estoup et al. 2002; Dettman and Taylor 2004). These issues should be dealt with in future studies of loci mutating at high rates based on a comparative approach.
The second intriguing result is the fact that the increase of deletion rates seems to be initiated at (period+2) for tri- to hexanucleotides, whereas the increase is initiated at (period+1) for all insertions and for deletions at shorter motifs (fig. 1). This is in line with the idea that slippage-induced insertions and deletions in STRs are not governed by the same processes (Ellegren 2004). One reason might be that the DNA strand involved in slippage differs between insertions and deletions. Insertions result from a loop in the neosynthetized strand, whereas deletions derive from a loop in the template strand. Our data suggest that at least two correct nucleotidic bonds on the template strand are required to ensure efficient elongation of the neosynthetized strand for tri- to hexanucleotidic motifs (3- to 6-bp loops). This might be explained by the stronger biophysical effort supported by the template strand at the replication fork, as it is constrained in both 5′ and 3′ by the rest of the chromosome, whereas the neosynthesized strand is constrained only in 5′ (Hardy et al. 2004). Here, we focused on the period only, but this threshold might also depend on the slipping motif, as G–C bonds are known to be more stable than A–T ones. However, our protocol was not dedicated to explore this possibility.
The third intriguing result is that deletions were twice as common as insertions at all STR periods and lengths, as previously noted (Kvikstad et al. 2007; Messer and Arndt 2007). In other words, DNA and/or indel slippage seem to preferentially contract rather than expand short microsatellites in the human genome. It is therefore difficult to understand how long microsatellites arise from random sequences since their increase in size should be countered by deletions. We envisage two explanations. The first explanation is based on the fact that long, A-rich microsatellites might derive from poly-A tails inserted with some families of TEs, especially LINEs and SINEs (Nadir et al. 1996; Buschiazzo and Gemmel 2006). However, several arguments suggest that these “adopted” STRs do not represent the main fraction of long STRs. For example, the association between TEs and STRs is mainly restricted to A-rich microsatellites in humans (Jurka and Pethiyagoda, 1995; Nadir et al. 1996), which is not true of long microsatellites in general. In addition, a large fraction of long STR loci (even A-rich ones) are not associated to TEs in the human genome. Finally, many genomes with limited numbers of TEs harbor long microsatellites (e.g., Neurospora crassa; Galagan et al. 2003; Leclercq et al. 2007).
A second explanation for how long microsatellites arise from random sequences, despite the higher rate of contractions compared with expansions, is based on the classical model of STR expansion. The divergence rates for insertions and deletions estimated here are average values of distributions. It is therefore quite possible that some STRs tend to expand, whereas the majority of repeated sequences are stuck to small sizes because deletions override insertions. A bias toward expansions has indeed been demonstrated experimentally at loci larger than about 10 repeats (Weber and Wong 1993; Primmer et al. 1996; Wierdl et al., 1997; Xu et al. 2000). The reasons for such variability in slippage rate remain elusive. A recent study (Kelkar et al. 2008) conducted on long orthologous microsatellites from the human and chimpanzee genomes concluded that STR intrinsic features (length and period) are much better indicators of STR variability than extrinsic factors (GC and recombination rates, distance to telomere, Alu and L1 contents). The role of intrinsic features can be discarded because the difference in slippage between deletions and insertions holds whatever the STR length and period (fig. 1). However, the motif itself may be a source of bias, which may explain the differences in genomic distributions among motifs (e.g., underrepresentation of GC-rich STRs; Toth et al., 2000; Katti et al. 2001). The influence of STR motif on the direction of mutation still has to be investigated.
Supplementary material
Supplementary figs. 1 and 4 and tables 2 and 3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank L. Duret for the initial thought about the method, G-F. Richard, E. Desmarais, and three anonymous referees for helpful comments on the manuscript, and R. Relyea for streamlining our English. The authors are supported by research grants from the “Action Concertée Incitative—Informatique, Mathématiques, Physique pour la Biologie” and from the BioSTIC-LR program. S.L. is supported by a fellowship from the Ministère Français de l'Education Nationale.
References
- Angers B, Bernatchez L. Complex evolution of a salmonid microsatellite locus and its consequences in inferring allelic divergence from size information. Mol Biol Evol. 1997;14(3):230–238. doi: 10.1093/oxfordjournals.molbev.a025759. [DOI] [PubMed] [Google Scholar]
- Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14(4):708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohede J, Ellegren H. Microsatellite evolution: polarity of substitutions within repeats and neutrality of flanking sequences. Proc Biol Sci. 1999;266(1421):825–833. doi: 10.1098/rspb.1999.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006;28(10):1040–1050. doi: 10.1002/bies.20470. [DOI] [PubMed] [Google Scholar]
- Calabrese P, Durrett R. Dinucleotide repeats in the Drosophila and human genomes have complex, length-dependent mutation processes. Mol Biol Evol. 2003;20(5):715–725. doi: 10.1093/molbev/msg084. [DOI] [PubMed] [Google Scholar]
- Chambers GK, MacAvoy ES. Microsatellites: consensus and controversy. Comp Biochem Physiol B Biochem Mol Biol. 2000;126(4):455–476. doi: 10.1016/s0305-0491(00)00233-9. [DOI] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. Characterization of mononucleotide repeats in sequenced prokaryotic genomes. DNA Res. 2005;12(4):221–233. doi: 10.1093/dnares/dsi009. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Taylor JW. Mutation and evolution of microsatellite loci in Neurospora. Genetics. 2004;168(3):1231–1248. doi: 10.1534/genetics.104.029322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieringer D, Schlötterer C. Two distinct modes of microsatellite mutation processes: evidence from the complete genomic sequences of nine species. Genome Res. 2003;13(10):2242–2251. doi: 10.1101/gr.1416703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 2000;16:551–558. doi: 10.1016/s0168-9525(00)02139-9. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet J. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422(6934):859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Hardy CD, Crisona NJ, Stone MD, Cozzarelli NR. Disentangling DNA during replication: a tale of two strands. Philos Trans R Soc Lond B Biol Sci. 2004;359(1441):39–47. doi: 10.1098/rstb.2003.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J Mol Biol. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006;34(database issue):D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(database issue):D610–D7. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarne P, David P, Viard F. Microsatellites, transposable elements and the X chromosome. Mol Biol Evol. 1998;15(1):28–34. doi: 10.1093/oxfordjournals.molbev.a025844. [DOI] [PubMed] [Google Scholar]
- Jarne P, Lagoda PJL. Microsatellites, from molecules to populations and back. Trends Ecol Evol. 1996;11(10):424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- Jakupciak JP, Wells RD. Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life. 2000;50:355–359. doi: 10.1080/713803749. [DOI] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40(2):120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 2001;18(7):1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18(1):30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100(20):11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvikstad EM, Tyekucheva S, Chiaromonte F, Makova KD. A macaque’s-eye view of human insertions and deletions: differences in mechanisms. PLoS Comput Biol. 2007;3(9):1772–1782. doi: 10.1371/journal.pcbi.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YL, Sun FZ. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol Biol Evol. 2003;20(12):2123–2131. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Rivals E, Jarne P. Detecting microsatellites within genomes: significant variation among algorithms. BMC Bioinformatics. 2007;8:125. doi: 10.1186/1471-2105-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopoldino AM, Pena SD. The mutational spectrum of human autosomal tetranucleotide microsatellites. Hum Mutat. 2003;21(1):71–9. doi: 10.1002/humu.10153. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage m13 in Escherichia coli k-12. Nucleic Acids Res. 1987;15(13):5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Rocco A, Mimouni N, Heger A, Caldeira A, Hein J. Uncertainty in homology inferences: assessing and improving genomic sequence alignment. Genome Res. 2008;18(2):298–309. doi: 10.1101/gr.6725608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates, Inc; 2007. [Google Scholar]
- Main M, Lorentz R. An o(n log n) algorithm for finding all repetitions in a string. J Algorithms. 1984;5:422–432. [Google Scholar]
- Messer PW, Arndt PF. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol Biol Evol. 2007;24(5):1190–1197. doi: 10.1093/molbev/msm035. [DOI] [PubMed] [Google Scholar]
- Messier W, Li SH, Stewart CB. The birth of microsatellites. Nature. 1996;381(6582):483. doi: 10.1038/381483a0. [DOI] [PubMed] [Google Scholar]
- Michel B, et al. Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci U S A. 2001;98:8181–8. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Nadir E, Margalit H, Gallily T, Ben-Sasson SA. Microsatellite spreading in the human genome: evolutionary mechanisms and structural implications. Proc Natl Acad Sci U S A. 1996;93(13):6470–6475. doi: 10.1073/pnas.93.13.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Kliman RM, Machado CA. Evolutionary history of microsatellites in the obscura group of Drosophila. Mol Biol Evol. 2001;18(4):551–556. doi: 10.1093/oxfordjournals.molbev.a003834. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63(2):349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer CR, Ellegren H. Patterns of molecular evolution in avian microsatellites. Mol Biol Evol. 1998;15(8):997–1008. doi: 10.1093/oxfordjournals.molbev.a026015. [DOI] [PubMed] [Google Scholar]
- Primmer CR, Saino N, Moller AP, Ellegren H. Directional evolution in germline microsatellite mutations. Nat Genet. 1996;13(4):391–393. doi: 10.1038/ng0896-391. [DOI] [PubMed] [Google Scholar]
- Pupko T, Graur D. Evolution of microsatellites in the yeast Saccharomyces cerevisiae: role of length and number of repeated units. J Mol Evol. 1999;48(3):313–316. doi: 10.1007/pl00006474. [DOI] [PubMed] [Google Scholar]
- Richard GF, Dujon B. Distribution and variability of trinucleotide repeats in the genome of the yeast Saccharomyces cerevisiae. Gene. 1996;174(1):165–174. doi: 10.1016/0378-1119(96)00514-8. [DOI] [PubMed] [Google Scholar]
- Richard GF, Pâques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000;1:122–126. doi: 10.1093/embo-reports/kvd031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose O, Falush D. A threshold size for microsatellite expansion. Mol Biol Evol. 1998;15(5):613–615. doi: 10.1093/oxfordjournals.molbev.a025964. [DOI] [PubMed] [Google Scholar]
- Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainudiin R, Durrett RT, Aquadro CF, Nielsen R. Microsatellite mutation models: insights from a comparison of humans and chimpanzees. Genetics. 2004;168(1):383–395. doi: 10.1534/genetics.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- Sibly RM, Whittaker JC, Talbot M. A maximum-likelihood approach to fitting equilibrium models of microsatellite evolution. Mol Biol Evol. 2001;18(3):413–417. doi: 10.1093/oxfordjournals.molbev.a003817. [DOI] [PubMed] [Google Scholar]
- Sokol KA, Williams CG. Evolution of a triplet repeat in a conifer. Genome. 2005;48(3):417–426. doi: 10.1139/g05-004. [DOI] [PubMed] [Google Scholar]
- Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD. Strong and weak male mutation bias at different sites in the primate genomes: insights from the human-chimpanzee comparison. Mol Biol Evol. 2006;23(3):565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10(7):967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S. Comparison of simple sequence repeats in 19 Archaea. Genet Mol Res. 2006;5(4):741–772. [PubMed] [Google Scholar]
- Varela MA, Sanmiguel R, Gonzalez-Tizon A, Martinez-Lage A. Heterogeneous nature and distribution of interruptions in dinucleotides may indicate the existence of biased substitutions underlying microsatellite evolution. J Mol Evol. 2008;66:575–580. doi: 10.1007/s00239-008-9107-3. [DOI] [PubMed] [Google Scholar]
- Vowles EJ, Amos W. Evidence for widespread convergent evolution around human microsatellites. PLoS Biol. 2004;2:E199. doi: 10.1371/journal.pbio.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2(8):1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Webster MT, Hagberg J. Is there evidence for convergent evolution around human microsatellites? Mol Biol Evol. 2007;24:1097–1100. doi: 10.1093/molbev/msm051. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Ellegren H. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc Natl Acad Sci U S A. 2002;99(13):8748–8753. doi: 10.1073/pnas.122067599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker JC, et al. Likelihood-based estimation of microsatellite mutation rates. Genetics. 2003;164(2):781–787. doi: 10.1093/genetics/164.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146(3):769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Peng M, Fang Z. The direction of microsatellite mutations is dependent upon allele length. Nat Genet. 2000;24(4):396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Strassmann JE, Queller DC. Insertions, substitutions, and the origin of microsatellites. Genet Res. 2000;76(3):227–236. doi: 10.1017/s001667230000478x. [DOI] [PubMed] [Google Scholar]