Abstract
Eukaryotes arose from prokaryotes, hence the root in the tree of life resides among the prokaryotic domains. The position of the root is still debated, although pinpointing it would aid our understanding of the early evolution of life. Because prokaryote evolution was long viewed as a tree-like process of lineage bifurcations, efforts to identify the most ancient microbial lineage split have traditionally focused on positioning a root on a phylogenetic tree constructed from one or several genes. Such studies have delivered widely conflicting results on the position of the root, this being mainly due to methodological problems inherent to deep gene phylogeny and the workings of lateral gene transfer among prokaryotes over evolutionary time. Here, we report the position of the root determined with whole genome data using network-based procedures that take into account both gene presence or absence and the level of sequence similarity among all individual gene families that are shared across genomes. On the basis of 562,321 protein-coding gene families distributed across 191 genomes, we find that the deepest divide in the prokaryotic world is interdomain, that is, separating the archaebacteria from the eubacteria. This result resonates with some older views but conflicts with the results of most studies over the last decade that have addressed the issue. In particular, several studies have suggested that the molecular distinctness of archaebacteria is not evidence for their antiquity relative to eubacteria but instead stems from some kind of inherently elevated rate of archaebacterial sequence change. Here, we specifically test for such a rate elevation across all prokaryotic lineages through the analysis of all possible quartets among eight genes duplicated in all prokaryotes, hence the last common ancestor thereof. The results show that neither the archaebacteria as a group nor the eubacteria as a group harbor evidence for elevated evolutionary rates in the sampled genes, either in the recent evolutionary past or in their common ancestor. The interdomain prokaryotic position of the root is thus not attributable to lineage-specific rate variation.
Keywords: phylogenies, early evolution, tree of life, microbial genomics, lateral gene transfer
Introduction
Geochemical and isotopic data indicates that life on earth was already flourishing by the time that the oldest known sedimentary rocks had formed some 3.5 Ga (Ueno et al. 2006) and that by about 3.2 Ga prokaryotic communities in anaerobic marine environments looked very much like today’s (Nisbet 2000; Rasmussen 2000; Shen et al. 2001; Brasier et al. 2006; Grassineau et al. 2006). Microfossil data reflect a more or less continuous record of abundant prokaryotic communities from ∼3.5 Ga onward, with eukaryotes appearing later. The presence of diversified and unequivocally eukaryotic cells is documented in sediments ∼1.5 Ga of age (Javaux et al. 2001; Knoll et al. 2006), followed by eukaryotic algae at ∼1.2 Ga (Butterfield 2000). Biomarker evidence once suggested the possible presence of eukaryotes by 2.7 Ga, but the biomarkers were subsequently shown by virtue of their isotope fingerprint not to have arisen within the rocks in which they occur (Fischer 2008; Rasmussen et al. 2008). Accordingly, eukaryotes appear about 2 billion years later in the geological record than do prokaryotes, consistent with the results of recent molecular and genomic investigations indicating that eukaryotes, which ancestrally possess mitochondria, arose from prokaryotes, lineages to which both mitochondria and their host can be traced (Rivera and Lake 2004; Embley and Martin 2006; Pisani et al. 2007; Cox et al. 2008; Koonin 2009). Thus, the first 2 billion years of life on earth, in particular, the very first phases of life’s history, are about prokaryote evolution only. Hence the position of the root in the “tree of life” concerns the deepest divide among the prokaryote groups.
Early efforts to locate the root in the tree of life focused on phylogenies of individual genes (Gogarten et al. 1989; Iwabe et al. 1989; Brown and Doolittle 1995). But with the recognition of lateral gene transfer (LGT) as a widespread and altogether normal mechanism of natural variation affecting prokaryote genome evolution (Doolittle 1999; McInerney and Pisani 2007), concerns became increasingly severe, and well founded, that any individual gene could serve as a reliable proxy for the evolution of a whole genome all the way back to the earliest divergence events in life’s history.
More recently, indels present in seven anciently conserved proteins (IF2, EF-G, Hsp70, HisA, S12, GyrA, PyrD) have been used to infer the position of the root (Lake et al. 2009). This approach first excluded the root from the archaebacteria (Skophammer et al. 2006), then from the Gram-negative eubacteria (Lake et al. 2007) and finally placed it within the eubacteria, on a branch separating the firmicutes and the archaebacteria from all else (Lake et al. 2008). These studies were, however, criticized on the basis that the alignments were problematic (Di Giulio 2007). Furthermore there is the issue that seemingly robust indels can in fact arise independently at the same spots of a protein alignment (and structure) during evolution (Bapteste and Philippe 2002). In addition, the LGT caveat holds for the indel data as well, that is, it is highly questionable whether the evolutionary patterns preserved in the indels of any one gene are indicative for the evolution of the entire genome. Indeed, it is presently difficult at best to muster evidence that any gene has remained immune to lateral transfer over the fullness of geological time (Bapteste et al. 2009). Moreover, the approach to phylogeny using indels, as extensively applied by Gupta and colleagues over the years (Gupta 1998; Gupta and Lorenzini 2007), has the drawback that rather than looking at all the indels, which would contain a large amount of conflicting data, one only looks at a few specifically chosen indels, giving the impression that indel data lack substantial conflict.
Another recent approach to inferring the position of the root in the tree of life entails the logical-parsimonious analysis of characters (Cavalier-Smith 2006b). However, that approach entails a dismissal of molecular data from genomes as inapplicable to the study of microbial evolution because it allows lineage-specific and gene-specific variations of evolutionary rate to be assumed without penalty by invoking “quantum evolution” wherever convenient to account for any observed pattern of sequence similarity or lack thereof (Cavalier-Smith 2010b). As such, the method is independent of tests with evidence founded in gene sequence similarity. It nonetheless places the root within the Chloroflexi, anoxygenic photosynthetic eubacteria (Cavalier-Smith 2010a), and prescribes an origin of the archaebacteria (and eukaryotes) from actinobacteria only 850 Ma (Cavalier-Smith 2006a). That suggestion is distinctly at odds with geochemical evidence for biological methane production >3 Ga (Canfield 2006; Ueno et al. 2006), with biomarker evidence for archaebacteria in 2.7 Ga deposits (Ventura et al. 2007) and is difficult to reconcile with the observation that many archaebacteria inhabit hydrothermal niches that have existed for as long as there has been water on earth (Sleep et al. 2004). It is furthermore at odds with unequivocal microfossil evidence for the existence >850 Ma of eukaryotes (Butterfield 2000; Javaux et al. 2001), which in the neomuran theory are viewed as descendants of the same actinobacterial group as archaebacteria.
Genome-wide data deliver yet other distinctly differing results with respect to the position of the root. Wong et al. (2007), for example, used a combination of data types in an analysis that placed the root close to Methanopyrus within the archaebacteria. That rooting is consistent with isotope evidence for the antiquity of methanogenesis (Ueno et al. 2006). In other work, Zhaxybayeva et al. (2005) analyzed 12 anciently duplicated gene pairs and concluded that the root probably lies between the archaebacteria and the eubacteria but pointed to the caveat that 12 genes might not speak for the whole genome because of LGT and furthermore pointed out a lack of strong phylogenetic signal in their data. Boussau et al. (2008) investigated rRNA phylogeny and about 50 proteins also concluded that the root probably lies between archaebacteria and eubacteria. Indeed, various authors embrace the view that the root lies between archeabacteria and eubacteria because of the few molecular characters that these groups share in common in their genome comparisons (Dagan and Martin 2007; McInerney et al. 2008; Battistuzzi and Hedges 2009; Koonin 2009) but without providing specific molecular analyses to support that view.
Specific attempts to root the tree of life through data analyses deliver conflicting results, although most commonly a eubacterial root (Gogarten et al. 1989; Lake et al. 2009). Particularly problematic with any rooting of the tree of life within the eubacteria, however, is that the archaebacteria—which 1) generally share very few genes with eubacteria (Snel et al. 1999; Graham et al. 2000), 2) have different plasma membrane and cell wall chemistries than eubacteria (Martin and König 1996; Claus et al. 2005; Engelhardt 2007), 3) have different machineries of DNA maintenance than eubacteria (Chong et al. 2000; Frols et al. 2009), 4) employ many different cofactors than eubacteria (Dimarco et al. 1990; Deppenmeier 2002; Fujihashi et al. 2007), and 5) have different core promotor and RNA polymerase structures than eubacteria (Bell and Jackson 2001)—assume the status of a derived group of eubacteria in such schemes. Importantly, all current eubacterial root views (Lake et al. 2008; Cavalier-Smith 2010b) invoke the hitherto untested corollary assumption that there is some form of systematic acceleration in the evolutionary rate of sequence change within the archaebacterial lineage. Such studies are furthermore based on a few specifically chosen characters, not whole genome data.
Genomes sequences should contain more evidence addressing the deepest divide among prokaryotes than just a few genes do. The root inferred from whole genomes should correspond to the bipartition separating those genomes that share the fewest genes in common and the least sequence similarity. That root should, in turn, correspond to the most ancient, in terms of geological time, split in the prokaryotic world, barring the existence of lineage-specific rate fluctuations across that divide, a hefty caveat. Here, we pinpoint the most ancient prokaryote genome divergence on the basis of whole genome data. By analyzing gene distribution patterns, we reconstruct a phylogenetic network of 191 prokaryotes. Using the midpoint rooting approach (Farris 1972), we then identify the root position within the network. Furthermore, we show through quartet analysis of the eight ancient paralogous genes that arose by duplication in the prokaryote common ancestor that the position of the root so identified cannot be attributed to lineage-specific increases in rates of sequence change.
Materials and Methods
Orthologous Protein Families
Completely sequenced prokaryotic genomes were downloaded from the National Center for Biotechnology Information (NCBI) Website (http://www.ncbi.nlm.nih.gov/; genomes available at August 2005). For each species, only the strain with the largest number of genes was used. Of 191 genomes (562,321 proteins) in the data, 22 are archaebacterial and 169 are eubacterial. All proteins in the 191 genomes were clustered by similarity into gene families using reciprocal best Blast hit (BBH) approach (Tatusov et al. 1997). Each protein was Blasted against each of the genomes. Pairs of proteins that resulted as reciprocal BBHs of E value < 10−10 were aligned using ClustalW (Thompson et al. 1994) to obtain amino acid identities. Protein pairs with ≥30% amino acid identity where clustered into protein families of ≥2 members using the Markov cluster algorithm (MCL; Enright et al. 2002) setting the inflation parameter, I, to 2.0. For the comparison of gene distribution patterns over different protein similarity thresholds, six additional sets of protein families were clustered using ascending threshold (Ti, where i = {35,40,45,50,55,60}) for the percent amino acid identity between protein pairs that are included in the analysis. Protein families reconstructed by the MCL algorithm include both orthologous and paralogous proteins. Because, in this study, we are interested in orthologous proteins only, we sorted out the paralogous genes from the protein families. To distinguish between orthologs and paralogs, we used the number of reciprocal BBHs for each gene within a family. In the case of multiple genes for a genome in a certain protein family, orthologs are expected to have more reciprocal BBHs in other genomes than paralogs. Thus, for each genome, only the protein with the maximum number of reciprocal BBHs is considered.
Splits Network
Protein families from each protein similarity threshold were compared with the protein families reconstructed under a 5% higher threshold. Proteins that are included within one family at a certain threshold may be clustered into one or more families at the higher threshold. The first case indicates a conservation of the family and the latter indicates one split or more. Thus, for each of the families in the higher threshold (those that comprise proteins clustered into a single family at the lower threshold), a new split is recorded in a binary pattern that includes 191 digits; if the protein family includes a protein from genome i then digit xi in its corresponding pattern is “1,” otherwise it is “0.” Species that are not represented in the protein family are coded as “?”. All splits for a certain threshold were then summarized by a splits network using SplitsTree (Huson and Bryant 2006).
Midpoint Rooting in Splits Network
The root within the splits network was located by adapting the midpoint rooting approach in phylogenies (Farris 1972). This method assumes that all lineages evolve at roughly similar rates. In a phylogeny, the root is located half way along the path connecting the pair of taxa that are furthest apart in the tree (fig. 1a). Here, the distance between two taxa in the tree is measured according to “phenetic distance,” the length of the path (i.e., the sum of split weights) from one taxon to the other in the tree.
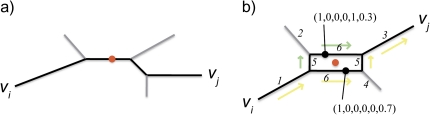
FIG. 1.—
Midpoint rooting trees and networks. (a) The pathway from node vi to vj and the midpoint (red circle) in a phylogenetic tree is shown. (b) An illustration of the procedure used to root a split network. The two most distant taxa are vi and vj. There are two shortest paths between these two taxa (colored arrows) and two midpoints. The numbering of the splits is indicated, noting that the two central splits are associated with two edges each. The vector encoding is made with reference to taxon vi. The encoding for the root is (1,0,0,0.5,0.5), which corresponds to the center of the central box (red circle).
In a split network, there can be multiple paths between any two nodes, and the phenetic distance between two nodes in a split network is therefore defined as the length of the “shortest” path connecting the nodes. As well, there can be multiple shortest paths between two nodes, giving multiple possible midpoint locations (fig. 1b).
To locate the root of the split network, a pair of taxa at maximum phenetic distance is identified. Ties can be broken arbitrarily: any pair with the maximum distance will give the same root location. Once a pair is selected, the set of path midpoints half way between the two taxa is obtained. An arbitrary reference taxon v is selected, and the splits in the network are numbered 1, 2, … , m. The location of each midpoint node x is then encoded as a vector (x1, x2, … , xm) of length m where xi = 1 if the shortest paths from v to x traverse an edge labeled by split i and xi = 0 if they don’t. In a split network, all the shortest paths between any two nodes will cross over edges labeled by the same set of splits (Dress and Huson 2004). This encoding is extended to locations along edges or within boxes by allowing the components of the vector to take on fractional values between 0 and 1. Let (z1, z2, … , zm) be the average of the midpoint location vectors; this is the location vector for the root. To determine the position of the root in the network, a path is traced starting from v and using edges labeled by splits i for which zi = 1 (and never two edges with the same labels). The fraction components of the location vector then determine the position of the root along the edge or within a box.
It can be shown that in any planar drawing of the split network, the position of the root in the plane will be exactly the average of the positions of the midpoint. Also, when the split network is actually a tree, this network root will be exactly the midpoint root.
The robustness of the midpoint network root was tested using a type of jackknife resampling approach. By this approach, the most distant pair of taxa is excluded from the splits network, and the midpoint root is recalculated. This procedure was repeated until the root was no longer found between archaebacteria and eubacteria. We note that if a large number of pairs need to be removed to modify the position of the root then that position will also be stable if random taxa are removed according to a statistical jacknife procedure.
Test of the Global Clock Assumption
Ancient paralogous genes were identified by their four-letter synonym within NCBI’s genome annotations (ptt files). Genomes for which proteins were not found using the four-letter synonym were searched by reciprocal BBH procedure using an already identified protein from the same lineage (see below) as a query and the genome in question as subject. The annotation of proteins identified this way was double-checked manually. The taxonomic classification of the 191 species is done by NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy). For species within Firmicutes or Proteobacteria phyla, the lineage is defined as the taxonomic class otherwise it is defined as the taxonomic phylum.
Quartets of ancient paralogs (fig. 2a) were assembled from the sequences of two ancient paralogs from two different lineages for all possible species pairs. Sequence alignments were reconstructed using ClustalW (Thompson et al. 1994). Sequence alignment reliability was tested using the HoT procedure (Landan and Graur 2007), and only alignments with a sum-of-pairs score >80% were included in the analysis. Phylogenetic trees reconstructed from quartets of ancient paralogs may result in three possible topologies (fig. 2b). The most likely tree topology for each quartet was tested with the SH test (Shimodaira and Hasegawa 1999) using ProML of PHYLIP (Felsenstein 1996). Only quartets of vertical topology (tvert) were considered for further analysis.
FIG. 2.—
Ancient paralogs quartet analysis. (a) Ancient paralogs are defined as paralogous proteins that were duplicated in the common ancestor of archaebacteria and eubacteria. (b) For a phylogenetic tree of four OTUs (operational taxonomic units), there are three possible topologies. No LGT among the major taxa results in topology tvert (in black), whereas evolution by LGT may result in any of the other two topologies (in gray). (c) Here, we tested two different rate models for the tvert topology: in the rglobal model all OTUs evolve in the same rates. In the rlineage model, OTUs from the same lineage evolve in the same rate, which differs between the lineages.
Different models of evolutionary rate variation along the branches were tested using the PAML package (Yang 2007). Each quartet was first tested for global molecular clock model (rglobal), assuming equal rates on all branches (fig. 2c), using the null hypothesis H0: all branches evolve with rate r1. This model has three parameters corresponding to the n − 1 interior nodes in a rooted tree, whereas the alternative hypothesis H1 assumes different rates for all five branches in an unrooted tree and therefore has five parameters for a tree of four taxa (Yoder and Yang 2000). The maximum log-likelihood values under both models (l0 and l1, respectively) are estimated with CodeML, and twice the log-likelihood difference, 2Δl = 2(l1 − l0) was compared with a χ2 distribution with degrees of freedom (df) = 2 to test whether the global clock hypothesis is rejected (Yang 1998). Quartets for which the global clock hypothesis was rejected were subsequently tested for a lineage-specific rate (rlineage) assuming different rates between the two lineages and equal rates between each paralogs pair (fig. 2c). The null hypothesis in this case is H0: branches α1, β1 evolve with rate r1 and branches α2, β2 evolve with rate r2. The alternative hypothesis H1 assumes the free-rate model again with its five parameters. Because the lineage-specific rate model has two free parameters less than the free-rate model, we analogously compare 2Δl = 2(l1 − l0) with a χ2 distribution with df = 2 to test whether the null hypothesis is rejected.
Results and Discussion
Splits Networks for Prokaryotic Genomes
We clustered the 562,321 protein-coding sequences that occur among 191 completely sequenced prokaryotic genomes from 15 higher level taxa with standard procedures (Enright et al. 2002) into groups based upon sequence similarity threshold. The clustering threshold corresponds to a value of amino acid identity, T30 designating the 30% threshold, for example, indicating that each protein in the cluster at T30 must share at least 30% amino acid identity with one other member (not with all other members) in the cluster. Depending upon the threshold set for the clustering procedure, these proteins fall into comparatively few large and inclusive families of distantly related sequences, or many smaller families whose members share high sequence identity. For example, clustering using T30 results in 57,743 families, 103 of them are nearly universal, including ≥90% of the species and 39,781 families include between 2 and 4 species.
Across different clustering thresholds Ti of increasing stringency (in 5% increments, e.g., T30, T35, T40, etc.), a given family will tend to break apart into two or more separate families, each containing a smaller number of more highly conserved sequences at higher values of Ti. Depending upon the distribution of sequence similarities within a given family, an individual increase in clustering stringency, ΔTi, may or may not introduce such a split within the protein family. Each new split within the family, termed here a protein family split, corresponds to a split among the strains (genomes) in which the family is present, termed here a genome split. The set of all genome splits can be readily converted into networks using NeighborNet (Bryant and Moulton 2004) in SplitsTree (Huson and Bryant 2006), which constructs phylogenetic networks based on the Neighbor-Joining algorithm (Saitou and Nei 1987). In the resulting networks, splits separating the genome set reflect overall sequence similarity between members of all protein families shared across the corresponding genomes, regardless of whether that similarity stems from vertical descent, differential loss, or LGT.
At the amino acid identity threshold of 25% (T25), the 562,321 proteins fall into 53,429 families of ≥2 proteins. Of those, only 3,832 (7% of the total) of the families have members occurring in both archaebacteria and eubacteria. The fraction of protein families with this broad distribution decreases with the increase of protein similarity threshold, down to 172 (0.2%) in T60. The fraction of archaebacterial-specific proteins remains almost constant (10–11%) across values of Ti, whereas the proportion of eubacterial-specific proteins increases from 83% to 90%. The proportion of group-specific protein families increases with the protein similarity threshold in most groups (e.g., Actinobacteria and α-Proteobacteria), whereas in Cyanobacteria this proportion is almost constant (supplementary table S1, Supplementary Material online). Hence, reconstruction of protein families using ascending amino acid identity threshold generally yields more exclusive protein families of increasingly narrow taxonomic range. Moreover, when increasing the protein similarity threshold, inclusive protein families (e.g., proteobacterial specific) split into more exclusive protein families (e.g., α- and β-Proteobacteria).
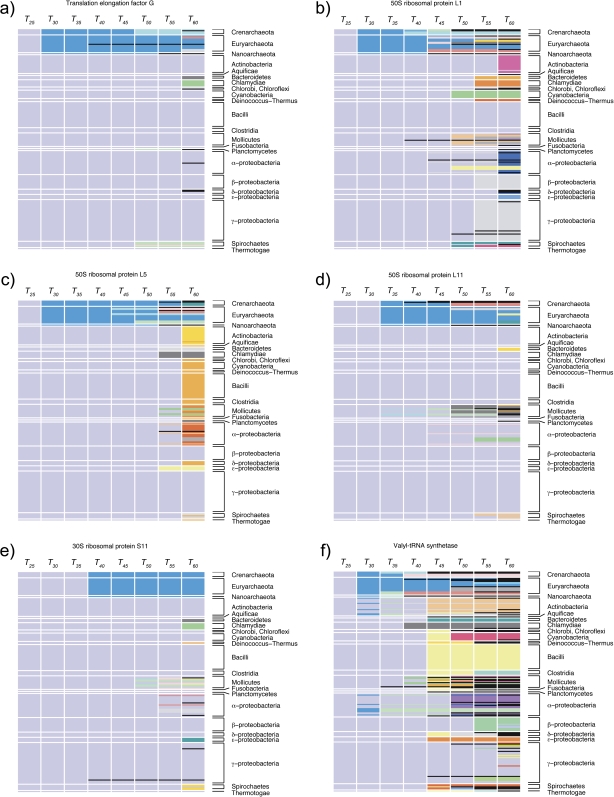
The set of all protein family splits was then extracted by comparison of families clustered at incrementally increased thresholds. To illustrate, at T25 only six protein families are present in all 191 species in the data set (fig. 3). Three of them—translation elongation factor G (fig. 3a), ribosomal protein L1 (fig. 3b), and ribosomal protein L5 (fig. 3c)—split into two protein families: one archaebacterial specific and one eubacterial specific. Only two of these families are still universal at T30: ribosomal proteins L11 (fig. 3d) and S11 (fig. 3e). The last family, valyl-tRNA synthetase (ValRS), splits at T30 into one eubacterial-specific family and one including all archaebacteria, five Actinobacteria, and seven α-Proteobacteria (fig. 3f). At T35, the latter ValRS family splits into three families, two of them containing archaebacteria only and one including the Thermoplasmatales (Euryarchaeota), Actinobacteria, and α-Proteobacteria. At T40, the latter family splits into three families specific to Thermoplasmata, Actinobacteria, and α-Proteobacteria, respectively. These splits are the result of lateral transfer of ValRS genes from archaeabacteria to α-Proteobacteia and Actinobacteria (Raoult et al. 2003), followed by vertical descent within these groups.
FIG. 3.—
Protein family splits over ascending protein similarity thresholds for six protein families that are universal at T25: (a) Translation elongation factor G, (b) 50S ribosomal protein L1, (c) 50S ribosomal protein L5, (d) 50S ribosomal protein L11, (e) 30S ribosomal protein S11, (f) valyl-tRNA synthetase. The splits are shown as colored boxes within columns. Currently recognized taxonomic groups are indicated in rows for comparison. For example, 50S ribosomal protein L5 (c) is universal at T25, whereas in T30 the protein family splits into an archaebacteria-specific family (blue) and a eubacteria-specific family (light purple).
Prokaryotic Genome Clusters
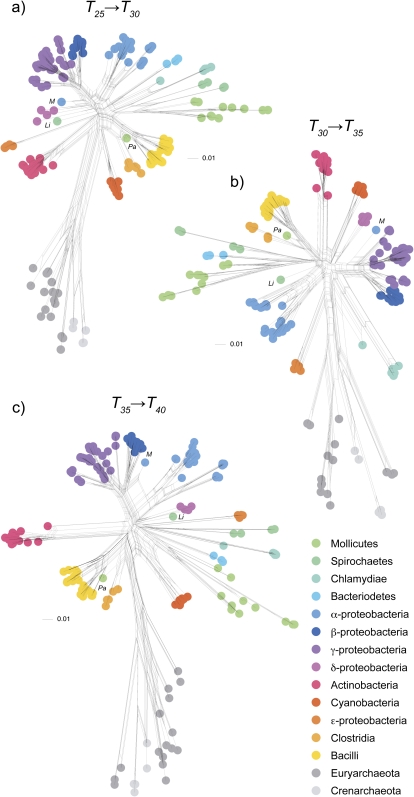
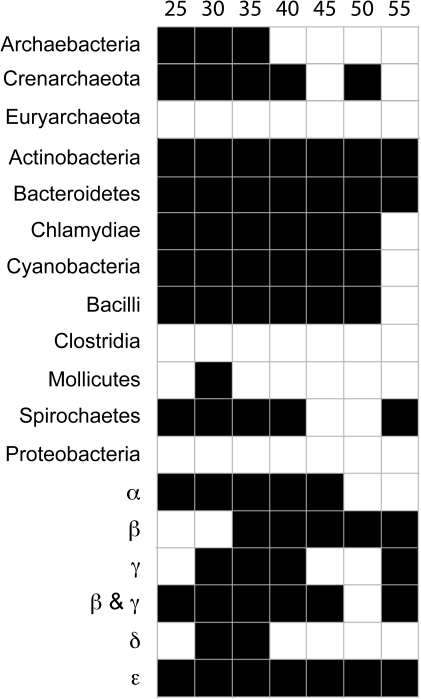
Comparison of networks obtained from different protein similarity thresholds shows that ancient genome splits contain more information about divergence of the major taxa than recent ones (fig. 4). This is because using higher protein similarity thresholds results in an increased proportion of taxon-specific families (supplementary table S1, Supplementary Material online), a shift of the split information to the tips of the network, and as a result, a collapse of the network into a star-like topology (supplementary fig. S1, Supplementary Material online). Overall, the protein family split networks tend to recover traditionally recognized prokaryotic groups at higher taxonomic levels (fig. 5). Splits of protein families in the lower thresholds, for example, the T25 → T30 splits and T35 → T40 splits, contain enough information to recover the divergence of the major prokaryotic groups, so that most of them are “monophyletic” in the sense of there being a split in the data that unites them to the exclusion of all other taxa irrespective of conflicting splits using figure 5. This is a somewhat liberal use of the word monophyletic in this context because it focuses on the criterion “is there any signal uniting them” as opposed to asking “does any signal divide them.” A network is a composite of multiple potentially conflicting signals, and the presence of a split separating out a clade suggests (in an unrooted sense) the presence of at least some phylogenetic evidence in favor of the clade being monophyletic for at least part of the genome. It is notable that only three higher groups examined here failed that monophyly criterion at all thresholds: the proteobacteria, the euryarchaeotes, and the clostridia (fig. 6). This is worth a brief consideration.
FIG. 4.—
Protein family splits networks for the lowest three protein similarity cutoffs. Networks for higher protein similarity cutoffs are presented in supplementary figure S1 (Supplementary Material online).
FIG. 5.—
Midpoint root location in the T25 → T30 protein family splits network.
FIG. 6.—
Detectable monophyly of groups under different similarity cutoffs, monophyly here meaning the presence of a split uniting the group irrespective of the presence of conflicting splits. Black square indicates that the respective group is monophyletic in that sense under the given cutoff.
In general, the lack of monophyly for groups in the present analysis is most easily attributed to patchy patterns of gene sharing across groups, for example, as afforded by LGT during evolution. That the proteobacteria are not monophyletic in our analyses is largely attributable to their frequency in the sample size and their general tendency to harbor large and diverse genomes with abundant LGT (Lang and Beatty 2007; Dagan et al. 2008). More curious is the lack of monophyly for the clostridia, which contains many acetogens (Pierce et al. 2008; Ljungdahl 2009) and the euryarchaeotes, where the methanogens reside (Thauer et al. 2008). Acetogens and methanogens are strict anaerobes and inhabit environments that have existed since there was first life on earth (Martin et al. 2008), they both gain their energy from the reduction of CO2 with H2, they both harbor forms that can generate their chemiosmotic ion gradients without the participation of cytochromes (Müller 2003) or quinones (Thauer et al. 2008; Biegel et al. 2009). The lack of monophyly might relate to the large amounts of gene exchange across higher taxa involving these groups, for example, as in the hundreds of clostridial genes found in Thermotogales (Zhaxybayeva et al. 2009), or the dozens (Chistoserdova et al. 1998) to hundreds (Deppenmeier et al. 2002) to thousands of genes (Ng et al. 2000) that have been exchanged between some euryarchaeotes and eubacteria. Another possible interpretation is that if LGT is as prevalent in the environment and over geological time as some are claiming (Doolittle and Bapteste 2007), then the oldest prokarytic groups will have had the greatest opportunity to exchange genes with other groups hence, eroding their monophyly be the measure of whole genome comparison used here. In that sense, and with the corresponding caveats, the lack of monophyly for the clostridia and euryarchaeotes could reflect their antiquity relative to the other groups sampled here.
In the three most ancient split networks (fig. 4), archaebacteria are monophyletic but within this kingdom the euryarchaeotes are paraphyletic, consistent with the findings of other recent studies (Fukami-Kobayashi et al. 2007; Cox et al. 2008; Puigbò et al. 2009). Only three species out of the 191 genomes do not branch with their traditionally assigned taxonomic group within the splits networks (for details, see supplementary table S2, Supplementary Material online).
The Root of Prokaryotes
The concept of rooting is familiar in the realm of phylogenetic trees but has so far not been developed in the context of phylogenetic networks. The simplest form of rooting entails finding the two most distance species and placing the root on their midpoint, but it also entails a global rate constancy assumption (Farris 1972). Midpoint rooting for a network must, however, take into account multiple paths between pairs of taxa. Here, the midpoints are calculated for all equally shortest paths between the two most distant species and then all midpoints are “averaged” into a new root location within the network (see Materials and Methods). The two most distant species in the T25 → T30 network are Thermoplasma acidophilum (Euryarchaeota) and Mycoplasma pneumoniae (Tenericutes). Averaging the midpoint among all shortest paths results in a root location on the split between archaebacteria and eubacteria (fig. 5).
In order to test the robustness of the root placement between archaebacteria and eubacteria, we applied a jackknife resampling approach to our network rooting procedure. In this approach, the rooting procedure is iterated, whereby in each iteration the most distant species from the previous iteration are excluded from the network until the result location of the root changes. Here, we repeated the rooting procedure until the root was no longer located between archaebacteria and eubacteria. The robustness of the root location is thus dependent on the number of iterations. The original placement of the root is between T. acidophilum and M. pneumoniae. After excluding those two species from the network, we find that the root is placed between Sulfolobus acidocaldarius DSM 639 and Mycoplasma genitalium. Excluding the most distant pair in each step results in smaller distances as the iterations proceed (supplementary table S3, Supplementary Material online). After applying the exclusion and rerooting procedure iteratively for 20 times, we still find the root on the split separating archaebacteria (Methanosarcina acetivorans) from eubacteria (Mycobacterium bovis). Further exclusion of M. acetivorans as a member of the euryarchaeota group results in a network devoid of archaebacteria, rerooting of which places the root on a split between Actinobacteria and the remaining eubacteria.
The split networks reconstructed for increasing Ti also show that the split found in the rooted network is also the most ancient split among prokaryotes because it is the strongest split at the lowest amino acid identity thresholds and weakens when higher thresholds (more closely related proteins only) are queried (fig. 4, supplementary fig. S1, Supplementary Material online). However, just as with rooting trees, this approach to rooting the network can be sensitive to rate variation because split weight can be affected by variation in the rate of sequence change among groups. Hence, it was important to test for lineage- or genome-specific rate variation, which we did for 191 genomes using eight ancient paralogs that were duplicated in the common ancestor of genomes sampled here.
Comparison of Evolutionary Rates among Lineages
Ancient paralogs are protein pairs that were duplicated prior to the divergence of eubacteria and archaebacteria (fig. 2a). Here, we use eight such paralogs in order to compare evolutionary rates among prokaryotic lineages (Kollman and Doolittle 2000): 1) adenosine triphosphate (ATP) synthase α (atpA) and β (atpB) subunits, 2) carbamoyl-phosphate synthase small (carA) and large (carB) subunits, 3) SRP proteins (ftsY, ffh), 4) isoleucyl-tRNA synthetase (ileS) and valyl-tRNA synthetase (valS), 5) aspartate carbamoyltransferase (pyrB) and ornithine carbamoyltransferase (argF), 6) threonyl-tRNA synthetase (thrS) and seryl-tRNA synthetase (serS), 7) translation elongation factors EF-G (fusA) and EF-Tu (tufA), and 8) tyrosyl-tRNA synthetase (tyrS) and tryptophanyl-tRNA synthetase (trpS). For all possible species pairs that represent two different higher taxa (called here lineages for convenience) shown in supplementary table S4 (Supplementary Material online), we investigated the corresponding ancient paralog quartet. Of course, LGT of ancient paralogs can generate topologies other than that expected by vertical inheritance alone (Zhaxybayeva et al. 2005). We therefore tested each quartet for a vertical topology (fig. 2b) using the SH test. Quartets of vertical topology (tvert) were then tested for a global clock model (rglobal) using the maximum-likelihood ratio test (Yang 2007). In the cases where rglobal was rejected, the quartet was tested for lineage-specific rates (fig. 2c). Quartets of vertical topology that accepted the lineage-specific rates model (rlineage) permit identification of lineage-specific rate increases, that is, which of the two genomes is undergoing more rapid sequence change.
Thus, orthologs of the eight ancient paralogs were identified in all genomes and were used to assemble 115,750 sequence quartet alignments. Alignment quality was tested using the HoT procedure (Landan and Graur 2007). Employing a conservative cutoff for alignment reliability of identical sum-of-pairs score >80% resulted in 56,297 alignments for which we reconstructed maximum-likelihood trees; the remaining 59,453 alignments were excluded because about half (49 ± 16%) of the site patterns (columns) in the alignment were irreproducible in the simplest alignment comparison (N-terminal vs. C-terminal seeding). The proportion of tvert trees within consistent alignments is very high, ranging between 90% of ATP synthase quartets and 100% of the carbamoyl-phosphate synthetase and translation elongation factor EF-Tu and EF-G quartets (supplementary table S5, Supplementary Material online). In total, of the 56,297 reproducible alignments, 55,765 (99%) gave a tvert quartet result. This high proportion of vertical topologies—for the paralogous two taxon case—suggests that LGT of these genes between the higher level taxonomic groups sampled here is quite rare, whereby this result does not address the frequency of transfer of these genes among closely related lineages. Using a maximum-likelihood ratio test, we were able to accept a global clock model for most (75%) of the tvert quartets. Furthermore, 58% (5,611) of the quartets comparing archaebacterial and eubacterial lineages passed the global clock model test. Hence, in most cases, there is no significant difference in evolutionary rates between the different lineages for the proteins we tested (Novichkov et al. 2004).
We performed this test specifically to address the empirical validity of repeated assertions that the archaebacteria are an evolutionarily young group of organisms—only 850 million (Cavalier-Smith 2006a, 2009, 2010a, 2010b) or 1 billion (de Duve 2007) years of age—whose distinctness at the molecular level is attributable to some unspecified mutational mechanism of increased sequence change, quantum evolution (Cavalier-Smith 2010b), within the genome of archaebacteria in general or the archaebacterial common ancestor. Our results clearly indicate that there is no such lineage-specific effect for the archaebacteria (supplementary table S4, Supplementary Material online), although lineage-specific effects can be detected for other groups.
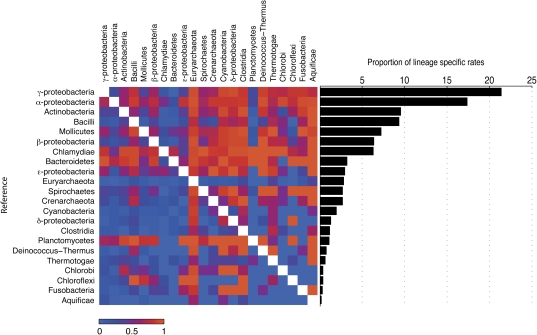
About one fifth (19%) of the total tvert quartets uncover significant lineage-specific rate increases (rlineage; supplementary table S4, Supplementary Material online); in these cases, both paralogs from the same lineage have the same degree of increased rate. Using these 10,728 rlineage quartets, we can compare the rates among lineages and rank lineages into slow- versus fast-evolving categories. The fastest lineages in this ranking are the γ-Proteobacteria, the α-Proteobacteria, the Actinobacteria, and the Bacilli (fig. 7). The splits of these four lineages within the splits networks are furthermore distinct across most protein similarity thresholds (fig. 3), suggesting a slight bias in the eubacterial clustering due to infraeubacterial evolutionary rate variation. But the two archaebacterial classes, euryarchaeota and crenarchaeota, are found to have at best an average rate in the lineage comparisons. They are slower than most eubacterial classes in the pairwise comparison (fig. 7), with only 4% (euryarchaeotes) and 10% (crenarchaeotes) of the tvert quartets suggesting a higher rate in the respective archaebacterial class. Hence, the weight of the rooted split between archaebacteria and eubacteria cannot be attributed to faster archaebacterial evolutionary rates. Furthermore, the argument that archaebacteria are only 850–1,000 MY old (Cavalier-Smith 2006a; de Duve 2007) is rejected because its corollary that their molecular distinctness can be explained away by assuming an increased archaebacterial evolutionary rate is shown here to be untrue. Our findings are, however, fully consistent with the view that the archaebacteria are a very ancient lineage of organisms, at least as ancient as the eubacteria (Stetter 2006; Thauer 2007), a view that is furthermore consistent with isotope data for the antiquity of archaebacterial metabolism.
FIG. 7.—
Rate comparisons of rlineage quartets. (a) A color-coded matrix showing the proportion of rlineage quartets in which the reference taxon (left) evolves in higher evolutionary rate than the compared taxon (top) in a 100% (red) to 0% (blue) scale. (b) Proportion of rlineage with elevated rates in the reference species from the total rlineage quartets in which the taxon is represented.
Life at the Root
The debate about the position of the root in the tree of life has focused mainly on its position and to some extent on the biology of the first organisms. The issues of microbial lifestyle (autotrophy vs. heterotrophy: Lane et al. 2010) and cellularity, that is, the transition from replicating molecules in inorganic compartments to genetically specified replicating cells (Martin and Russell 2003; Koonin and Martin 2005; Branciamore et al. 2009) have received attention of late. However, by far the most heavily debated aspect of life at the root concerns temperature.
The view of thermophilic origins attracted much attention following the suggestions by Karl Stetter (Stetter et al. 1990) and Pace (1991) that prokaryotes inhabiting many of the extreme kinds of environments we see today are, to some extent, inhabiting environments that existed in a fully “modern” form on early earth: anoxic volcanic settings and hydrothermal vents, both which are often quite hot (>80 °C). In trees rooted between the prokaryotic domains, the hyperthermophiles branched first, suggesting that maybe the first organisms were hyperthermophilic archaea and bacteria (Stetter et al. 1990; Pace 1991). That view spawned the counterhypothesis of thermoreduction (Forterre 1995, 1996), which posits that the hyperthermophilic origins scenario is wrong by virtue of a misplaced root. In that view, the eukaryotes are seen as the ancestral form of life on earth, prokaryotes having evolved from them via reductive evolution. Although thermoreduction in the original sense can now be excluded because all eukaryotes either have or had mitochondria (Cox et al. 2008; van der Giezen 2009), meaning that eukaryotes as we know them cannot be ancestral to prokaryotes, the issue of temperature at life’s root remains current.
Recently, gene trees have been used to infer the temperature of early earth environments based on statistical arguments (Gaucher et al. 2003, 2008). Boussau et al. (2008), for example, suggested that the first organisms (the common ancestor of archaeabacteria and eubacteria in their view) arose and lived at about room temperature (∼20 °C) based on the estimated GC content of inferred ancestral sequences in maximum-likelihood trees. Is such a low temperature for life at the root realistic? Amend and McCollom (2009) recently calculated that in geochemically promising environments for the origin of life, the Gibbs energy of reaction (ΔGr) toward the synthesis of total prokaryotic cell mass was unfavorable (+500 Joules per gram of cells) at 25 °C but exergonic at 50, 75, and 100 °C, with values of –1,016, –873, and –628 Joules per gram of cells, respectively, dropping sharply again at 125 °C (Amend and McCollom 2009). Clearly, the synthesis of the first cells must have entailed a fundamentally exergonic reaction, as life cannot have arisen against the laws of thermodynamics. If thermodynamics are favorable in the range of 50–100 °C but not at 25 °C, then this can be taken as a constraint for phylogenetic models rather than a variable for estimation, when it comes to considering temperature at the root.
Part of the rational against the view of thermophilic origins was once founded in the circumstance that nucleoside triphosphates are very unstable at temperatures around 100 °C (Forterre 1996), for which reason such temperatures were deemed to be incompatible with the notion of an RNA world. However, Constanzo et al. (2009) recently reported that RNA chains dozens to over 100 nucleotides in length arise spontaneously, in hot (>80 °C) water, and without catalysts yet not from nucleoside triphosphates rather from the ribonucleoside 3′,5′ cyclic monophosphates at concentrations around 1 mM. Temperatures around 85 °C yielded rapid polymerization, below 60 °C the reaction rates dropped sharply (Constanzo et al. 2009). Thus, from the thermodynamic and chemical perspective, life at the root might be more likely in the range of 50–100 °C than at values approaching room temperature. That view is consistent with the recent discovery of a novel bifunctional fructose-1,6-bisphosphate aldolase/phosphatase from thermophilic eubacteria and archaebacteria that provides comparative biochemical evidence in favor of chemolithoautotrophic origins (Say and Fuchs 2010).
Conclusions
Recent studies on the position of the root of prokaryotic life have suggested that it lies within anoxygenic photosynthetic eubacteria (Cavalier-Smith 2006b) or within the eubacteria between the actinobacteria and the firmicutes (Lake et al. 2009). In such eubacterial root scenarios, the archaebacteria are seen as derived from specific groups of the eubacteria, in which case an elevated rate must be invoked for the archaebacteria in order to account for their molecular divergence. We have shown that no indication of such an archeabacterial rate elevation exists in available genome sequence data. Our analyses indicate that the deepest divide in the living world is that between archaebacteria and eubacteria, as earlier studies indicated (Gogarten et al. 1989; Iwabe et al. 1989) and as is compatible with much recent genome data (Koonin 2009). Like supertree approaches (Pisani et al. 2007), our method takes the signal of all genes—including those that have undergone LGT—into account rather than demanding that gene families harboring LGT events first be identified and purged from the data. In contrast to supertree and supermatrix methods, however, our procedure is independent of individual phylogenetic trees and utilizes an approach entailing phylogenetic networks to the study of evolutionary genome comparisons.
Supplementary Material
Supplementary figure S1 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft and the European Research Council for financial support. D.B. was supported by the Alexander von Humboldt Foundation and by a Marsden grant through the Royal Society of New Zealand.
References
- Amend JP, McCollom TM. Energetics of biomolecule synthesis on early Earth. In: Zaikowski L, Friedrich JM, Seidel SR, editors. Chemical evolution II: from the origins of life to modern society. Washington, DC: American Chemical Society; 2009. pp. 63–94. [Google Scholar]
- Bapteste E, Philippe H. The potential value of indels as phylogenetic markers: position of trichomonads as a case study. Mol Biol Evol. 2002;19:972–977. doi: 10.1093/oxfordjournals.molbev.a004156. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Hedges SB. A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol. 2009;26:335–343. doi: 10.1093/molbev/msn247. [DOI] [PubMed] [Google Scholar]
- Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr Opin Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- Biegel E, Schmidt S, Muller V. Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii. Environ Microbiol. 2009;11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. Parallel adaptations to high temperatures in the Archaean eon. Nature. 2008;456:942–945. doi: 10.1038/nature07393. [DOI] [PubMed] [Google Scholar]
- Branciamore S, Gallori E, Szathmary E, Czaran T. The origin of life: chemical evolution of a metabolic system in a mineral honeycomb? J Mol Evol. 2009;69:458–469. doi: 10.1007/s00239-009-9278-6. [DOI] [PubMed] [Google Scholar]
- Brasier MD, Mcloughlin N, Green O, Wacey D. A fresh look at the fossil evidence for early archaean cellular life. Philos Trans R Soc Lond B Biol Sci. 2006;361:887–902. doi: 10.1098/rstb.2006.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-transfer-RNA synthetase gene duplications. Proc Natl Acad Sci U S A. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Butterfield NJ. Bangiomorpha pubescens n. Gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- Canfield DE. Biogeochemistry—gas with an ancient history. Nature. 2006;440:426–427. doi: 10.1038/440426a. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell evolution and earth history: stasis and revolution. Philos Trans R Soc Lond B Biol Sci. 2006a;361:969–1006. doi: 10.1098/rstb.2006.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006b;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Predation and eukaryote cell origins: a coevolutionary perspective. Int J Biochem Cell Biol. 2009;41:307–322. doi: 10.1016/j.biocel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Deep phylogeny, ancestral groups and the four ages of life. Philos Trans R Soc Lond B Biol Sci. 2010a;365:111–132. doi: 10.1098/rstb.2009.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol Direct. 2010b;5:7. doi: 10.1186/1745-6150-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci U S A. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H, et al. Molecular organization of selected prokaryotic S-layer proteins. Can J Microbiol. 2005;51:731–743. doi: 10.1139/w05-093. [DOI] [PubMed] [Google Scholar]
- Constanzo G, Pino S, Ciciriello F, Di Mauro E. Generation of long RNA chains in water. J Biol Chem. 2009;284:33206–33216. doi: 10.1074/jbc.M109.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U S A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci U S A. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U. The unique biochemistry of methanogenesis. Prog Nucleic Acid Res Mol Biol. 2002;71:223–283. doi: 10.1016/s0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- Di Giulio M. The evidence that the tree of life is not rooted within the Archaea is unreliable: a reply to Skophammer et al. 2007. Gene. 2007;394:105–106. doi: 10.1016/j.gene.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Dimarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dress AW, Huson DH. Constructing splits graphs. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:109–115. doi: 10.1109/TCBB.2004.27. [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Engelhardt H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J Struct Biol. 2007;160:115–124. doi: 10.1016/j.jsb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS. Estimating phylogenetic trees from distance matrices. Am Nat. 1972;106:645–668. [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Fischer WW. Biogeochemistry—life before the rise of oxygen. Nature. 2008;455:1051–1052. doi: 10.1038/4551051a. [DOI] [PubMed] [Google Scholar]
- Forterre P. Thermoreduction: a hypothesis for the origin of prokaryotes. C R Acad Sci III. 1995;318:415–422. [PubMed] [Google Scholar]
- Forterre P. A hot topic: the origin of hyperthermophiles. Cell. 1996;85:789–792. doi: 10.1016/s0092-8674(00)81262-3. [DOI] [PubMed] [Google Scholar]
- Frols S, White MF, Schleper C. Reactions to UV damage in the model archaeon sulfolobus solfataricus. Biochem Soc Trans. 2009;37:36–41. doi: 10.1042/BST0370036. [DOI] [PubMed] [Google Scholar]
- Fujihashi M, et al. Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol. 2007;365:903–910. doi: 10.1016/j.jmb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Minezaki Y, Tateno Y, Nishikawa K. A tree of life based on protein domain organizations. Mol Biol Evol. 2007;24:1181–1189. doi: 10.1093/molbev/msm034. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, et al. Evolution of the vacuolar H+-ATPase—implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DE, Overbeek R, Olsen GJ, Woese CR. An archaeal genomic signature. Proc Natl Acad Sci U S A. 2000;97:3304–3308. doi: 10.1073/pnas.050564797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassineau NV, Abell P, Appel PWU, Lowry D, Nisbet EG. Early life signatures in sulfur and carbon isotopes from Isua, Barberton, Wabigoon (Steep Rock), and Belingwe greenstone belts (3.8 to 2.7 Ga) Geol Soc Am Mem. 2006;198:33–52. [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Lorenzini E. Phylogeny and molecular signatures (conserved proteins and indels) that are specific for the Bacteroidetes and Chlorobi species. BMC Evol Biol. 2007;7:71. doi: 10.1186/1471-2148-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaux EJ, Knoll AH, Walter MR. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- Knoll AH, Javaux EJ, Hewitt D, Cohen P. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Doolittle RF. Determining the relative rates of change for prokaryotic and eukaryotic proteins with anciently duplicated paralogs. J Mol Evol. 2000;51:173–181. doi: 10.1007/s002390010078. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Herbold CW, Rivera MC, Servin JA, Skophammer RG. Rooting the tree of life using nonubiquitous genes. Mol Biol Evol. 2007;24:130–136. doi: 10.1093/molbev/msl140. [DOI] [PubMed] [Google Scholar]
- Lake JA, Servin JA, Herbold CW, Skophammer RG. Evidence for a new root of the tree of life. Syst Biol. 2008;57:835–843. doi: 10.1080/10635150802555933. [DOI] [PubMed] [Google Scholar]
- Lake JA, Skophammer RG, Herbold CW, Servin JA. Genome beginnings: rooting the tree of life. Philos Trans R Soc Lond B Biol Sci. 2009;364:2177–2185. doi: 10.1098/rstb.2009.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G, Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- Lane N, Allen JF, Martin W. How did LUCA make a living? Chemiosmosis and the origin of life. Bioessays. 2010;32:271–280. doi: 10.1002/bies.200900131. [DOI] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Ljungdahl LG. A life with acetogens, thermophiles, and cellulolytic anaerobes. Annu Rev Microbiol. 2009;63:1–25. doi: 10.1146/annurev.micro.091208.073617. [DOI] [PubMed] [Google Scholar]
- Martin HH, König H. Beta-lactamases are absent from archaea (archaebacteria) Microb Drug Resist. 1996;2:269–272. doi: 10.1089/mdr.1996.2.269. [DOI] [PubMed] [Google Scholar]
- Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6:805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- Martin W, Russell M. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:59–85. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, Cotton JA, Pisani D. The prokaryotic tree of life: past, present… and future? Trends Ecol Evol. 2008;23:276–281. doi: 10.1016/j.tree.2008.01.008. [DOI] [PubMed] [Google Scholar]
- McInerney JO, Pisani D. Genetics—paradigm for life. Science. 2007;318:1390–1391. doi: 10.1126/science.1151657. [DOI] [PubMed] [Google Scholar]
- Müller V. Energy conservation in acetogenic bacteria. Appl Environ Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WV, et al. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet E. Palaeobiology: the realms of Archaean life. Nature. 2000;405:625–626. doi: 10.1038/35015187. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, et al. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. Origin of life: facing up to the physical setting. Cell. 1991;65:531–533. doi: 10.1016/0092-8674(91)90082-a. [DOI] [PubMed] [Google Scholar]
- Pierce E, et al. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum) Environ Microbiol. 2008;10:2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, Mcinerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Wolf YI, Koonin EV. Search for a 'Tree of Life' in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, et al. Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res. 2003;13:1800–1809. doi: 10.1101/gr.1474603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen B. Filamentous microfossils in a 3,235-million-year-old volcanogenic massive sulphide deposit. Nature. 2000;405:676–679. doi: 10.1038/35015063. [DOI] [PubMed] [Google Scholar]
- Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature. 2008;455:1101–1104. doi: 10.1038/nature07381. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Say RF, Fuchs G. Fructose-1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature. 2010;464:1077–1081. doi: 10.1038/nature08884. [DOI] [PubMed] [Google Scholar]
- Shen Y, Buick R, Canfield DE. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature. 2001;410:77–81. doi: 10.1038/35065071. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Skophammer RG, Herbold CW, Rivera MC, Servin JA, Lake JA. Evidence that the root of the tree of life is not within the Archaea. Mol Biol Evol. 2006;23:1648–1651. doi: 10.1093/molbev/msl046. [DOI] [PubMed] [Google Scholar]
- Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. H2-rich fluids from serpentinization: geochemical and biotic implications. Proc Natl Acad Sci U S A. 2004;101:12818–12823. doi: 10.1073/pnas.0405289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Stetter KO. Hyperthermophiles in the history of life. Philos Trans R Soc Lond B Biol Sci. 2006;361:1837–1842. doi: 10.1098/rstb.2006.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter KO, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124. [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thauer RK. A fifth pathway of carbon fixation. Science. 2007;318:1732–1733. doi: 10.1126/science.1152209. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nature Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440:516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- Ventura GT, et al. Molecular evidence of Late Archean archaea and the presence of a subsurface hydrothermal biosphere. Proc Natl Acad Sci U S A. 2007;104:14260–14265. doi: 10.1073/pnas.0610903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JT, Chen J, Mat WK, Ng SK, Xue H. Polyphasic evidence delineating the root of life and roots of biological domains. Gene. 2007;403:39–52. doi: 10.1016/j.gene.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Yang Z. Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol. 2000;17:1081–1090. doi: 10.1093/oxfordjournals.molbev.a026389. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Lapierre P, Gogarten JP. Ancient gene duplications and the root(s) of the tree of life. Protoplasma. 2005;227:53–64. doi: 10.1007/s00709-005-0135-1. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci U S A. 2009;106:5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]