Abstract
Arabidopsis thaliana is the model plant and is grown worldwide by plant biologists seeking to dissect the molecular underpinning of plant growth and development. Gene copy number variation (CNV) is a common form of genome natural diversity that is currently poorly studied in plants and may have broad implications for model organism research, evolutionary biology, and crop science. Herein, comparative genomic hybridization (CGH) was used to identify and interrogate regions of gene CNV across the A. thaliana genome. A common temperature condition used for growth of A. thaliana in our laboratory and many around the globe is 22 °C. The current study sought to test whether A. thaliana, grown under different temperature (16 and 28 °C) and stress regimes (salicylic acid spray) for five generations, selecting for fecundity at each generation, displayed any differences in CNV relative to a plant lineage growing under normal conditions. Three siblings from each alternative temperature or stress lineage were also compared with the reference genome (22 °C) by CGH to determine repetitive and nonrepetitive CNVs. Findings document exceptional rates of CNV in the genome of A. thaliana over immediate family generational scales. A propensity for duplication and nonrepetitive CNVs was documented in 28 °C CGH, which was correlated with the greatest plant stress and infers a potential CNV–environmental interaction. A broad diversity of gene species were observed within CNVs, but transposable elements and biotic stress response genes were notably overrepresented as a proportion of total genes and genes initiating CNVs. Results support a model whereby segmental CNV and the genes encoded within these regions contribute to adaptive capacity of plants through natural genome variation.
Keywords: natural variation, genome duplication, gene copy number variation, comparative genomic hybridization, genome evolution, Arabidopsis
Introduction
Angiosperms rapidly radiated into diverse biomes via both vicariance, which drove adaptation to various ecological niches due to the slow breaking up of land masses and also by biotic and abiotic dispersal mechanisms, which was aided by the advanced seed dispersal mechanisms that were evolving (Lidgard and Crane 1988; Knapp et al. 2005). Consequently, the genome size of angiospermophyta have diversified markedly since their origin, at a rate beyond that of most other taxa (Gaut and Ross-Ibarra 2008). Genome size has been correlated with organism growth and ecology (Gregory 2002), and extremely large genomes may be limited both ecologically and evolutionarily (Lynch and Conery 2003; Morse et al. 2009). Forces of selection based on environmental conditions may be the major components that contribute to genome evolution and plasticity (Huynen and Bork 1998; Barrick et al. 2009), yet the relationship between genome reshuffling and natural selection remains poorly understood. Regions of genome plasticity and rates of genetic segment movement are not well characterized and may have important biological consequences, particularly in annual flowering plants where high natural diversity rates among siblings could provide adaptive advantage.
Recently, structural shifts in the entire genome by gene copy number variants (CNVs) have been identified as major genetic variables in the human genome (Sebat et al. 2004) accounting for human disease etiology (McCarroll and Altshuler 2007; Png et al. 2008) and phenotypic variability between individuals (Estivill and Armengol 2007). CNVs are microduplications and deletions, as opposed to whole-genome duplications (Sebat et al. 2004). Plants are the most prolific genome duplicators with examples such as oats (Avena sativa), which has a hexaploid genome structure indicating six polyploid copies of its genome (Linares et al. 1998). Despite the clear flexibility of the plant genome and the sequenced and well-annotated genome of Arabidopsis thaliana, high-resolution study of microduplications and microdeletions by CNV in plants lags behind its mammalian counterpart and may represent a valuable component of the genomic framework (Kliebenstein 2008) that delimits natural phenotypic variation. A study was released during preparation of this manuscript that assayed CNV in maize (Zea mays) (Springer et al. 2009) and showed its potential contribution to the heterosis of this crop during domestication.
The rationale for this study was that laboratories all around the world use the model plant, A. thaliana, of the same ecotype (in this case Columbia-0) to dissect the molecular underpinnings of plant growth and development. As an R strategist (MacArthur and Wilson 1967), A. thaliana produce a lot of seed, have a short life cycle, and thus lean to reproduction rather than stability. An overarching goal of the current study sought to test whether A. thaliana, grown under different temperature (16 and 28 °C) regimes and stress regimes (salicylic acid [SA] spray) for five generations and selected for fecundity at each generation, displayed any differences in gene CNV relative to a plant lineage growing under normal (22 °C) conditions. Three siblings from each evolved temperature or stress lineage were compared with the genome of a plant lineage grown under normal growth conditions by comparative genomic hybridization (CGH) to determine repetitive and nonrepetitive CNVs (fig. 1). Ultimately, it was hoped that this experiment would reveal whether CNVs appeared in a lineage-specific manner after force and selection were applied. Force and selection are the overarching rules of evolution and are also common themes of molecular genetics experimental design using A. thaliana.
FIG. 1.—
A schematic representation of the experimental design. Force and selection were made at five generational points stemming from a common ancestor. After which, three siblings were selected for interlineage comparison with a reference growth condition by CGH.
Materials and Methods
Experimental Design
In this article, five lineages were established from a single parent genotype, and these were grown for five generations under different environmental conditions. The treatments were temperature (16, 22, 28 °C) plus a SA line and a mock treatment line. Selection among these lineages was made between each of the five generations based on fecundity (seed production) whereby the plant with the greatest seed production was chosen for the next generation. Once evolved, three siblings of each lineage were chosen randomly and assayed by whole-genome CGH in an attempt to find regions of repetitive microdeletion and microduplication (fig. 1). The tiling array interval for the CGH chip corresponding to the level of resolution for deletion/duplication was 300 bp.
Growth Conditions and Multigenerational Study Conditions
All A. thaliana lines used in this study were of the Columbia (Col)-O ecotype. For growth of plants in the multigeneration study, plants were stratified for 3 days in 0.15% agar at 4 °C and then seeded directly into soil and grown in continuous light (200 mmol/m2/s) in an Adaptis A1000 environmental growth chamber (Conviron) set to continuous temperature of 16, 22, or 28 °C. Multigeneration studies utilized seed collected from a single plant (the Col-0 seed stocks were obtained from the laboratory of Chris Somerville). For measurements of germination rate and radical elongation extent experiments, seed were germinated in sterile conditions on plates. Plating conditions used surface-sterilized seed that were stratified for 3 days in 0.15% agar at 4 °C prior to plating and were grown in the darkness at the identical temperature regime described above. Plates contained autoclaved 0.5 × Murashige and Skoog (MS) mineral salts (Sigma) and 1% agar. SA treatments were applied with an aqueous mixture of 0.3 mM SA (Sigma Aldrich, catalog number S7401) with 0.025% Silwet-L77 surfactant. Plants were sprayed with a handheld mist sprayer, and approximately 200 ml of SA solution was applied to pots containing 4 individual plants every 14 days. The mock SA treatment contained the Silwet-L77 but no SA and was applied in the same manner as the SA treatment using a different mist sprayer.
Isolation of Plant Genomic DNA from Arabidopsis
Genomic DNA (gDNA) was extracted from leaves of Arabidopsis plants using the DNeasy Plant Kits (QIAGEN). A NANODROP spectrophotometer (Thermo Scientific) was used to check DNA quality. Samples had an A260/A280 ≥1.8 and A260/A230 ≥1.9 for optimal labeling yields. A total of 2 mg of gDNA per sample was used for labeling and hybridization of the CGH array.
Comparative Genome Hybridization and Data Mining
Microarray Design.
CGH measures DNA copy number differences between a test and reference genome. The Arabidopsis whole-genome CGH array was designed and performed as a full service fee-for-service product by Roche Nimblegen on request and is commercially available at Roche Nimblegen (http://www.nimblegen.com/). Initially, probe selection was constrained to have one match within the genome, which resulted in large gaps in coverage. The probe match requirements and selection were relaxed to allow up to five matches and were able to fill in the gaps. The average tiling interval was 300 bases and the median interval was 280 bases (supplementary table S4, Supplementary Material online). The format of CGH array was the 385K design format whereby the single array contains 385,000 probes.
Hybridization and CGH Analysis
DNA extraction and purification were performed according to Nimblegen criteria and samples sent to Roche Nimblegen for CGH sample preparation, labeling, and hybridization. Each gDNA sample comparison used for CGH was labeled using a NimbleGen Dual-Color DNA Labeling Kit. Pairs of samples intended for hybridization to the same array were labeled in parallel using Cy3-Random and Cy5-Random Nonamers from the same kit (Roche NimbleGen). The test samples were labeled with Cy3 (16, SA, and 28 °C treatment) and reference samples with Cy5 (22 °C treatment and Mock SA). Data were normalized by spatial correction of the raw data. Spatial correction reduced artifacts observed in CGH data from and resulted in minimal impact on overall noise and log2-ratio values in regions of CNV. Spatial correction was applied to correct position-dependent nonuniformity of signals across the array. Specifically, locally weighted polynomial regression (LOESS) was used to adjust signal intensities based on X,Y feature position. A qspline fit normalization (Workman et al. 2002) was applied to the data prior to segmentation analysis. By default, normalization is applied and compensated for inherent differences in signal between the two dyes. These statistical processes were performed by Roche Nimblegen as part of the full service product (Roche Nimblegen). Segmental analysis was performed using the SignalMAP software (Roche Nimblegen) to identify regions of CNV that were greater than 1 gene copy. Using the same software, all three biological replicates were overlaid to identify repetitive CNV events. These data were then overlaid with a GIFF annotation file that contains all genes in the Arabidopsis genome fitted to the segmental output in SignalMap (commercially available from Roche Nimblegen). Cross-referencing the annotation file provided the gene identities for each CNV. These were then cross-referenced with The Arabidopsis Information Resource mapping tool (www.arabidopsis.org) to provide putative annotations for each gene.
Results
Genome-Wide Distribution and Size of Repetitive CNVs
All five chromosomes in the A. thaliana genome contained repetitive gene CNV events (table 1). Repetitive gene CNV events were CNVs that were present in at least 2 out of the 3 sibling level CGH assays for each interlineage comparison (figs. 2–4). Chromosomes 1 and 2 were most prone to segmental duplication or deletion, based on the number of genes exposed to CNV (table 1). Size of CNV regions, which by definition are greater than 1 kb in size (The Copy Number Variation Project, Wellcome Trust Sanger Institute), ranged from 2 genes to 128 genes in length (3 to 300 kb). In the 16/22 CGH, there were 14 CNV events that contained a total of 400 gene CNVs that were all microdeletion events. SA spray/Mock CGH assay identified 13 CNV deletion events that contained 402 genes. By contrast, 28/22 CGH assay documented 11 CNV events, which comprised seven microduplications and four microdeletion events and a total of 292 genes were exposed to CNV (table 1). The contribution of CNV to genome variation among CNV events that are repetitive among siblings with a lineage showed that 1.4%, 1.1%, and 1.4% of genes in the genome were exposed to repetitive CNV in the 16/22, 28/22, and SA/Mock assays, respectively (27,549 genes according to EMBL:EBI INTEGR8, A. thaliana genome statistics). Overall, the 16/22 and SA/Mock treatments resulted in a loss of genome size, whereas the 28/22 resulted in a net gain of genome size (supplementary table S1, Supplementary Material online). The largest single CNV event was present in all interlineage comparisons and was a deletion event located on chromosome 2 between 3,244,499 and 3,526,499 bp (supplementary table S1, Supplementary Material online and figs. 2–4).
Table 1.
Repetitive CNV That Occurred in At Least 2 Out of 3 Siblings within an Interlineage Comparison (28/22, 16/22, and SA/Mock)
| Event Number | Chromosome | Physical Location | CNV Type |
| 28/22 CGH | |||
| 1 | 1 | 9019499–9046499 | Duplication |
| 2 | 1 | 15085499–15151499 | Deletion |
| 3 | 1 | 15322499–15379499 | Deletion |
| 4 | 1 | 17248499–17266499 | Duplication |
| 5 | 2 | 1499–67499 | Duplication |
| 6 | 2 | 8971499–9139499 | Duplication |
| 7 | 2 | 13750499–13843499 | Duplication |
| 8 | 2 | 3241499–3508499 | Deletion |
| 9 | 4 | 1945499–1951499 | Duplication |
| 10 | 4 | 3193499–3259499 | Deletion |
| 11 | 5 | 3322499–3455499 | Duplication |
| Total genes | All | 292 | |
| Total TE | All | 32 | |
| 16/22 CGH | |||
| 1 | 1 | 8767499–8836499 | Deletion |
| 2 | 1 | 21751499–21850499 | Deletion |
| 3 | 1 | 27412499–27427499 | Deletion |
| 4 | 2 | 2593499–2674499 | Deletion |
| 5 | 2 | 3121499–3511499 | Deletion |
| 6 | 2 | 7604999–7610999 | Deletion |
| 7 | 2 | 12457499–12469499 | Deletion |
| 8 | 3 | 12667499–13147499 | Deletion |
| 9 | 3 | 16246499–16267499 | Deletion |
| 10 | 4 | 1699499–1741499 | Deletion |
| 11 | 4 | 5857499–5920499 | Deletion |
| 12 | 4 | 13621499–13636499 | Deletion |
| 13 | 5 | 11509499–11593499 | Deletion |
| 14 | 5 | 15250499–15262499 | Deletion |
| Total genes | All | 400 | |
| Total TE | All | 159 | |
| SA/M CGH | |||
| 1 | 1 | 8767499–8836499 | Deletion |
| 2 | 1 | 11482499–11524499 | Deletion |
| 3 | 1 | 21751499–21850499 | Deletion |
| 4 | 1 | 27412499–27427499 | Deletion |
| 5 | 2 | 2593499–2674499 | Deletion |
| 6 | 2 | 3121499–3514499 | Deletion |
| 7 | 2 | 12457499–12469499 | Deletion |
| 8 | 3 | 12667499–13141499 | Deletion |
| 9 | 3 | 16246499–16267499 | Deletion |
| 10 | 4 | 1699499–1741499 | Deletion |
| 11 | 4 | 5857499–5920499 | Deletion |
| 12 | 4 | 13621499–13636499 | Deletion |
| 13 | 5 | 11509499–11593499 | Deletion |
| Total genes | All | 402 | |
| Total TE | All | 159 | |
NOTE.—All data represent normalized CNV data, and the initiation and termination site of each CNV segment were documented as a physical location. Each event was defined as CNV event 1–11, 1–14, and 1–13 for 28/22, 16/22, and SA/Mock CGH, respectively. Chromosome number and whether the CNV was a deletion or duplication were documented for each interlineage comparison.
FIG. 2.—
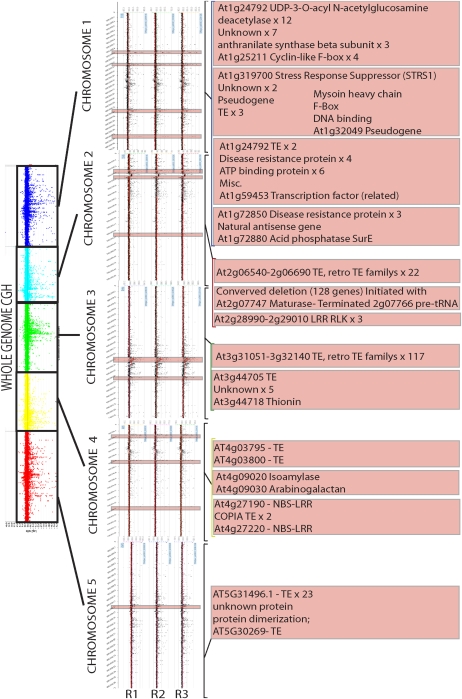
CGH analysis of CNV in the Arabidopsis genome in a plant lineage grown at 28 °C compared with a lineage grown at 22 °C for five generations. Both lineages were derived from a common ancestor plant. Right panel is an averaged rainbow view of the CGH analysis with the entire genome broken into color-coded panels. CNV events on each chromosome and each replicate are enlarged, itemized, and presented with functional annotation of the genes occurring in each event. Refer to supplementary table S1 (Supplementary Material online) for the exact annotation and metadata for each gene.
FIG. 3.—
CGH analysis of CNV in the Arabidopsis genome in a plant lineage grown at 16 °C compared with a lineage grown at 22 °C for five generations. Both lineages were derived from a common ancestor plant. Right panel is an averaged rainbow view of the CGH analysis. CNV events on each chromosome and each biological replicate are itemized and presented with functional annotation of the genes occurring in each event. Refer to supplementary table S1 (Supplementary Material online) for the exact annotation and metadata for each gene.
FIG. 4.—
CGH analysis of CNV in the Arabidopsis genome in a plant lineage grown under conditions whereby plants were exogenously sprayed with SA every 14 days compared with a lineage grown under conditions whereby plants were exogenously sprayed with a mock SA mixture for five generations at 22 °C. Both lineages were derived from a common ancestor plant. Right panel is an averaged rainbow view of the CGH analysis. CNV events on each chromosome and each biological replicate are itemized and presented with functional annotation of the genes occurring in each event. Refer to supplementary table S1 (Supplementary Material online) for the exact annotation and metadata for each gene.
Repetitive and Nonrepetitive CNV Events Assayed by Intersibling Comparison
Aimed at pinpointing the consistency and genome-wide distribution of CNV events within a single generation, three siblings from the fifth generation of each lineage were assayed via CGH (fig. 1), and the number of both repetitive and nonrepetitive CNV events counted (table 2). CNV events in siblings from the 16/22 and SA/Mock assays were highly uniform in their distribution among siblings, and no nonrepetitive CNVs were documented. By contrast, the number of CNV events among the three replicates varied substantially in the 28/22 assay (table 2). For instance, chromosome 3 contained no repetitive CNV events but among the siblings there were 6, 5, and 2 nonrepetitive CNV events. Moreover, the number of CNV events on chromosome 5 among siblings varied 2-fold from four in siblings 1 and 2 to eight events in sibling 3. The number of genes exposed to intersibling CNV variation was highest in the 28/22 CGH lineage with 0.38% of genes in the genome exposed to nonrepetitive CNV in an intersibling comparison.
Table 2.
CNV Events Per Chromosome Per Sibling Were Analyzed to Establish a Combined Number for Repetitive and Nonrepetitive CNV Events Per Chromosome in a Lineage
| CNV Events |
|||
| Sibling 1 | Sibling 2 | Sibling 3 | |
| 28/22 | |||
| Chromosome-1 | 4 | 4 | 5 |
| Chromosome-2 | 4 | 4 | 3 |
| Chromosome-3 | 6 | 5 | 2 |
| Chromosome-4 | 5 | 5 | 6 |
| Chromosome-5 | 4 | 4 | 8 |
| 16/22 | |||
| Chromosome-1 | 3 | 3 | 3 |
| Chromosome-2 | 4 | 4 | 4 |
| Chromosome-3 | 2 | 2 | 2 |
| Chromosome-4 | 3 | 3 | 3 |
| Chromosome-5 | 2 | 2 | 1 |
| SA/M | |||
| Chromosome-1 | 3 | 3 | 3 |
| Chromosome-2 | 4 | 4 | 4 |
| Chromosome-3 | 2 | 2 | 2 |
| Chromosome-4 | 3 | 3 | 3 |
| Chromosome-5 | 2 | 1 | 1 |
NOTE.—CNV events that were not common among 2 out 3 biological replicates were considered outlier events within this study.
Border Initiation–Termination Sites for Repetitive CNV Events among Siblings
To explore the physical location whereby CNV events began and ended, the CGH probes corresponding to CNVs were mapped against the genome and compared. In the 28/22 CGH assay, 8 out of the 11 CNV events displayed variable initiation or termination sites of the CNV segment among siblings. Alternatively, the 16/22 CGH assay had variable border regions in 3 out of 14 CNV (table 3). SA/Mock also had 3 variable border CNV regions among siblings out of 13 CNV events. Therefore, in addition to displaying greater duplication rate and nonrepetitive CNV events, the 28/22 assay displayed far greater variability of border regions than 16/22 or SA/Mock (table 3). Among duplication events in the 28/22, 5 out of 7 displayed variable initiation–termination sites among siblings, whereas out of 17 different deletion events documented in all lineages, only 5 had variable initiation–termination sites (table 3).
Table 3.
Initiation and Termination Sites for Repetitive CNV Events in Each CGH Interlineage Comparison Was Assessed for Each Sibling within a Lineage
| Sibling 1 | Sibling 2 | Sibling 3 | |
| Physical Location of Initiation and Termination of CNV Event |
|||
| 28/22 | |||
| Event 1 | 8935499–9079499 | 8905499–9232499 | 9025499–9076499 |
| Event 2 | 15091499–15139499 | 15091499–15151499 | 15085499–15151499 |
| Event 3 | 15325499–15376499 | 15322499–15379499 | 15322499–15379499 |
| Event 4 | 17251499–17269499 | 17248499–17266499 | 17258999–17261999 |
| Event 5 | 1–67499 | 1–67499 | 1–67499 |
| Event 6 | 3241499–3508499 | 3241499–3508499 | 3241499–3511499 |
| Event 7 | 8971499–9139499 | 8938499–9154499 | 8947499–9016499 |
| Event 8 | 13750499–13843499 | 13753499–13939499 | NA |
| Event 9 | 1945499–1951499 | 1942499–1951499 | 1945499–1951499 |
| Event 10 | 3193499–3259499 | 3196499–3262499 | 3196499–3262499 |
| Event 11 | 3322499–3445499 | 3316499–3445999 | NA |
| 16/22 | |||
| Event 1 | 8767499–8836499 | 8767499–8836499 | 8767499–8836499 |
| Event 2 | 21751499–21850499 | 21751499–21850499 | 21751499–21850499 |
| Event 3 | 27412499–27427499 | 27412499–27427499 | 27412499–27427499 |
| Event 4 | 2593499–2674499 | 2593499–2674499 | 2593499–2674499 |
| Event 5 | 3121499–3511499 | 3121499–3511499 | 3121499–3235499 |
| Event 6 | 7604999–7610999 | 7604999–7610999 | 7604999–7610999 |
| Event 7 | 12457499–12469499 | 12457499–12469499 | 12457499–12469499 |
| Event 8 | 12667499–13141499 | 12667499–13141499 | 12667499–13186499 |
| Event 9 | 16246499–16267499 | 16246499–16267499 | 16246499–16267499 |
| Event 10 | 1699499–1741499 | 1699499–1741499 | 1699499–1741499 |
| Event 11 | 5857499–5920499 | 5857499–5920499 | 5857499–5920499 |
| Event 12 | 13621499–13636499 | 13621499–13636499 | 13621499–13636499 |
| Event 13 | 11509499–11593499 | 11509499–11593499 | 11509499–11626499 |
| Event 14 | 15250499–15262499 | 15250499–15262499 | 15250499–15262499 |
| SA/M | |||
| Event 1 | 8767499–8836499 | 8767499–8836499 | 8767499–8836499 |
| Event 2 | 11482499–11524499 | 11482499–11524499 | 11482499–11524499 |
| Event 3 | 21751499–21850499 | 21751499–21850499 | 21751499–21850499 |
| Event 4 | 27412499–27427499 | 27412499–27427499 | 27412499–27427499 |
| Event 5 | 2593499–2674499 | 2593499–2674499 | 2593499–2674499 |
| Event 6 | 3121499–3511499 | 3121499–3514499 | 3121499–3514499 |
| Event 7 | 12457499–12469499 | 12457499–12469499 | 12457499–12469499 |
| Event 8 | 12667499–13141499 | 12667499–13141499 | 12667499–13141499 |
| Event 9 | 16246499–16267499 | 16246499–16267499 | 16246499–16267499 |
| Event 10 | 1699499–1741499 | 1699499–1741499 | 1636499–1741499 |
| Event 11 | 5857499–5920499 | 5857499–5920499 | 5857499–5920499 |
| Event 12 | 13621499–13636499 | 13621499–13636499 | 13621499–13636499 |
| Event 13 | 11509499–11593499 | 11509499–11593499 | 11509499–11626499 |
NOTE.—Repetitive CNV events were defined as occurring in 2 out of 3 biological replicates or siblings among lineage-specific CGH comparisons. The current table documents the physical location where each overlapping CNV event was initiated and terminated on in order to determine conservation among siblings. Refer to table 1 for the corresponding chromosome for each event.
An additional question was whether a nonrandom pattern of initiation gene species existed among CNV events. Five of the initiation genes among the 11 repetitive CNV events in the 28/22 assay were transposable elements (TEs) (supplementary table S1, Supplementary Material online). Others included a secretory membrane protein, other RNA, maturase, nodulin, protein kinase, and Grl5. In the 16/22 assay, initiation genes among the 14 replicated CNV regions included a UDP-3-O-acyl N-acetylglucosamine deacetylase family protein/F-box protein, mRNA splicing, and a protease inhibitor, maturase, five TEs, two unknown proteins, three disease resistance loci including an leucine-rich repeat (LRR) protein kinase, disease resistance protein (nucleotide-binding site [NBS]–LRR), and a resistance locus o (MLO) protein. In the SA/Mock assay, 13 CNVs were apparent and the initiation genes included a UDP-3-O-acyl N-acetylglucosamine deacetylase family protein/F-box protein, maturase, isoamylase, six TEs, three disease resistance loci including an LRR protein kinase, disease resistance protein (NBS-LRR), and a stress responsive suppressor (supplementary table S1, Supplementary Material online). Hence, TEs were the most representative initiating gene in CNV events occurring 42% of the time across all repetitive CNVs (supplementary table S1, Supplementary Material online) but it is important to note that this number was similar to the ratio of TEs among total genes in CNV events.
Variant Gene Identity and Function
16/22 °C
TEs, retro-TEs, and transposases are well documented to represent the largest contributor to genome size and flexibility (Ma et al. 2004). TEs comprised approximately 40% of all genes among repetitive CNV events the 16/22 assay. The most abundant classes were COPIA, GYPSY, CACTA, and non-LTR TEs. Additional TEs identified in the CGH analysis were MUTATOR TE, hAT-like TE, HELICASE TE, Sadhu noncoding TE, and retroelement pol polyproteins. CNVs that were comprised primarily of TEs were located on chromosome 3, 4, and 5 in the 16/22 assay (supplementary table S1, Supplementary Material online). The size of these segments varied, with the largest containing 117 genes and second largest contained 25 genes occurring on chromosome 3 (12,667,227–13,139,009 bp) and chromosome 5 (11,510,033–11,610,717 bp), respectively. In addition to TEs, there were defined transcripts that were highly represented in certain deletion event. Chromosome 1 had three main deletion events, the first was comprised of 36 genes, 6 of which were natural antisense genes and 12 of which were annotated as UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase genes. The second deletion on chromosome 1 contained 25 genes, and there were six adenosine triphosphate-binding protein and four disease resistance (CC-NBS-LRR class) genes. The third deletion event was 6 genes long and contained 3 disease resistance protein (CC-NBS-LRR class) genes, 2 acid phosphatase survival (SurE) genes, and an antisense gene. Hence, it was documented that disease resistance genes were prone to CNV in these experimental conditions (supplementary table S1, Supplementary Material online).
Chromosome 2 also had three deletion events, the first of which was comprised primarily of TEs, the second was conserved across all treatments, and the third deletion event contained a string of 3 LRR-receptor like kinases (RLK) (supplementary table S1, Supplementary Material online). Of particular interest was the second and highly conserved event among all experimental lineages. The CNV event was 128 genes in length and began with a maturase and ended with a pre-tRNA gene. Unlike other CNVs documented in this study, it comprised an overrepresentation of pre-tRNAs (16% of genes in CNV segment). In additional to pre-tRNAs, there were 62 genes of unknown function (48% of genes in CNV segment), 16 TEs (12.5% of genes in CNV segment), and several genes annotated as cytochromes (3%), adenosine triphosphatase (3%), or genes associated with primary metabolism (3%) (supplementary table S1, Supplementary Material online).
Chromosome 3 had two deletion events, the first of which was a large block of 117 genes that were largely TEs. The second region was 7 genes in length, and 5 were unknown genes, 1 a TE, and 1 a thionin gene. Chromosome 4 contained three small deletion events, the first contained an unknown gene and a Methyl-CpG–binding domain containing gene. The second contained an mRNA splicing, theoredoxin, pre-tRNA, and a DJ-1 gene. The third was comprised solely of TEs. A single CNV occurred on chromosome 5 and was largely composed of TEs. Despite the most abundant genes located within CNV deletion events being TEs, the identification of CNV regions that contained only NBS-LRR genes was unexpected (supplementary table S1, Supplementary Material online).
SA/Mock
Repetitive CNV events in the SA/Mock lineages were identical to those described above for 16/22 with the exception three CNVs. One additional deletion event occurred on chromosome 1 between 11,503,499 and 11,524,499 bp (At1g31993–At1g32045) and comprised stress response suppressor STRS1, three TEs, an F-BOX protein, a MYOSIN heavy chain related protein, two unknown, and two pseudogenes (supplementary table S1, Supplementary Material online). Two deletion events that occurred in the 16/22 lineage where absent in the SA/Mock lineage, and these occurred on chromosome 2 between 7,604,999 and 7,610,999 and chromosome 5 between 15,250,499 and 15,262,499 bp (supplementary table S1, Supplementary Material online).
28/22 °C
Distinct from both 16/22 and SA/Mock assays, repetitive microduplications among siblings were prominent in the 28/22 assays. No CNV events occurred on chromosome 3 and single CNV events occurred on chromosome 4 and 5 that contained a total of eight genes. Moreover, only a single deletion event was common to either of the other CGH experiments. Hence, 97% of genes exposed to CNV in 28/22 occurred on chromosome 1 and 2. On chromosome 1, two deletion (four genes—all TEs) and two duplication (17 genes) CNV segments were present. The first duplication event was initiated by a MUTATOR transposon (At1g25784) followed by a Ulp protease, which was homologous to the SUMO protease. Of the remaining ten genes, five were unknown and a cytochrome B561, extensin, esterase, haloacid dealogenase, and phosphorylase were present (supplementary table S1, Supplementary Material online). The second microduplicated segment contained two TEs, an other RNA gene, DREB ERF/AP2, and genes encoding proteins of unknown function.
Chromosome 2 contained 265 of the 290 CNV genes, in total 91% of all genes among four CNV segments. The first event occurred at the initiation of the chromosome and contained several rRNAs, unknown proteins, and a suite of TEs (supplementary table S1, Supplementary Material online). The second event was a deletion that was common to all treatments and contained 128 genes. This event was comprised of a large number of genes annotated as pseudogenes many of which were pseudogenes associated with primary metabolism, pre-tRNAs, and unknown proteins. The third CNV event on chromosome 2 was a large duplication. This CNV comprised only two TEs out of 89 genes. Genes within this intriguing segment included a fructose biphosphate aldolase, mannose-6-phosphate reductase, triosephosphate isomerase, lipoic acid synthase, diacylglycerol kinase, and peptidyl-prolyl cis-trans isomerase (supplementary table S1, Supplementary Material online). Moreover, regulatory genes such as bZIP transcription factors, auxin response genes, and MCM10 (involved in the initiation of DNA replication) were duplicated. Several genes in this segment were annotated as light or temperature response genes, such as the chloroplast-localized THYLAKOID FORMATION 1 gene involved in vesicle-mediated formation of thylakoid membranes. Genes associated with circadium rhythm were also present in this segment such as FIONA1 (a central oscillator-associated component, two copies were duplicated) and XAP5, which is involved in light regulation of the circadian clock and photomorphogenesis. Other genes such as SYT1 affect temperature tolerance. The fourth duplicated CNV segment that occurred on chromosome 2 contained a diverse range of gene annotations. Regulatory genes such as 2 AXR1 genes (auxin homeostasis), two 3'-5' exonuclease-nucleic acid–binding proteins, 2 F-BOX, MEKK, calcium-binding EF hand family protein, 2 R2R3 factor genes, and RCD1 were duplicated. This duplicated region also had cell wall-associated genes, such as four cellulose synthase like genes, a galactosyltransferase family protein, and a hydroxyproline-rich glycoprotein.
Chromosome 3 contained no repetitive CNV events among siblings that could be considered lineage stable CNVs. Chromosome 4 contained two small CNV events. The first was a deletion event, which contained a tandem duplicate of a NODULIN-like gene and six genes of unknown function. The second event on chromosome 4 was a duplication comprised of a tandem duplicate GYPSY TE (supplementary table S1, Supplementary Material online). A single repetitive CNV event occurred on chromosome 5 and was initiated by a protein kinase, followed by a gene encoding a pathogenesis-related protein, C2H2 ring zinc finger domain containing gene, C3HC4 ring zinc finger containing gene, gene of unknown function, and an LRR-RLK terminated the CNV event.
Regions of Tandem-Duplicated Genes Were Common in CNV Events
Genes subject to tandem duplication were highly abundant and comprised 52% of all genes in the 28/22 assay (supplementary table S1, Supplementary Material online). Furthermore, the 16/22 and SA/Mock also had 52% tandem duplication rate among genes exposed to CNV. Considering that tandem-duplicated genes are highly abundant in Arabidopsis and represent approximately 15% of the genome (Vision et al. 2000; Zhang et al. 2002; Blanc et al. 2003), it was noteworthy that a substantial overrepresentation of tandem duplicates occurred among CNV segments (supplementary table S1, Supplementary Material online). Patterns of past gene duplication defined as back to back copies of genes that share nucleotide sequence similarity and retail the same functional annotation occurred frequently within regions of CNV (supplementary table S1, Supplementary Material online; regions of similarity are colored). In both SA/Mock and 16/22 CGH comparisons, the first deletion on chromosome 1 was a good example of where 12 out of 36 genes in this CNV were encoded by UDP-3-O-acyl N-acetylglucosamine deacetylase family protein/F-BOX protein. Moreover, a pattern of gene duplication appeared in this CNV and involved a Cyclin-like F-BOX, anthranilate synthase beta subunit and a potential natural antisense gene (supplementary table S1, Supplementary Material online). TEs were frequently associated with tandem duplication within CNV events (supplementary table S1, Supplementary Material online). Furthermore, CNV event seven (table 1) in the 16/22 and SA/Mock assays revealed that three LRR-RLKs were deleted and comprised the entire CNV event. Other examples of entire CNV events comprising duplications were demonstrated by events two and three on chromosome 1 in the 28/22 CGH, in this case both gene duplicates were TEs (CACTA and GYPSY).
Plant Growth among Lineages
Plants grown at 16, 22, and 28 °C displayed different rates of growth measured from seedling to mature phase. The lifecycle of A. thaliana at 22 °C was 6 weeks in constant light conditions, whereas plants grown at 16 °C required between 8 and 9 weeks to complete their lifecycle. Growth form of plants grown at 16 versus 22 °C was not markedly different, apart from a greater degree of anthocyanin accumulation in leaves, siliques, and stems of the 16 °C grown plants (data not presented, previously documented by Leyva et al. 1995). Plants grown at 28 °C displayed low germination rates (supplementary table S2, Supplementary Material online) consistent with previous reports (Kurek et al. 2007). Although initial growth rates were not significantly different to 22 °C as measured for hypocotyl elongation rates (supplementary table S3, Supplementary Material online), the plants became dwarfed and accumulated 40–50% less biomass than plants grown at 22 °C, and seed set was reduced under high temperature treatment (data not presented). These phenotypes were not detailed since the impact of high temperature on photosynthesis, biomass production and seed production have been previously documented (e.g., Feller et al. 1998; Salvucci and Crafts-Brandner 2004; Kurek et al. 2007). Low temperature regimes (16 °C) have also been shown to have no significant impact on germination rates but some effect on flowering time (Nordborg and Bergelson 1999). Plants grown under conditions whereby SA was applied by exogenous misting every 14 days also had an impact on plant growth, which has been previously documented (Nawrath and Métraux 1999) and therefore not represented. Plants were selected for fecundity at each generation, and seed from a single individual propagated in the proceeding generation (fig. 1). Seed from plants evolved at 22, 16, and 28 °C for five generations were grown on sterile, half strength MS agar plates for five days in the darkness to induce etiolation at each of the selection temperatures. The aim of this experiment was to determine whether germination and/or elongation rates were different in plants acclimated to 16 °C when grown at 28 °C relative to plants acclimated at 28 °C. Results showed that greater germination rates occurred in plants acclimated to 28 °C than those acclimated at 22 °C and 16 °C when growth at 28 °C after five generations (supplementary table S3, Supplementary Material online). Plants that were grown at 16 °C or 22 °C did not differ with respect to their germination rate (supplementary table S3, Supplementary Material online), hypocotyl or root elongation capacity.
Discussion
Life scientists, particularly those studying model organisms often assume isogenic properties of their “wild-type” organism. However, these same experimental biologists are often posing constant selection forces on model organisms through simple environmental changes between or within laboratories, mutagenesis, and/or seeking phenotypes of interest. Hence, we as scientists are playing a role in natural selection with unknown consequences because natural variation and subsequent phenotypic selection are the driving force behind evolution (McClintock 1984). Natural variation among a population of individuals is likely to arise from a complex interplay of genetic variability, such as single nucleotide polymorphisms (Borevitz et al. 2007; Clark et al. 2007; Nordborg and Weigel 2008) and gene CNV (Sebat et al. 2004; Conrad et al. 2006; Mileyko et al. 2008; Springer et al. 2009). CNV has already been shown to be a genomic polymorphism of enormous importance to human biology (Sebat et al. 2004) and yet is poorly studied in plants. The overarching goal of this experiment was to take initial steps to determine whether CNV could be detected among individuals in a population of A. thaliana over immediate family generation scale and secondly, if CNV did occur, what were the genomic features of CNV events. It was shown herein that the genome of A. thaliana displayed unexpectedly high rates of physical change by CNV among closely related lineages, affecting not tens but hundreds of genes between individuals separated by only five generations. Because force and selection were applied to individual lineages and multiple siblings within a lineage sustained the same CNV polymorphisms, results presented herein document an unprecedented level of genome divergence by CNV.
As mentioned above, findings of this experiment showed that CNV events were consistent among siblings in interlineage CGH assays. Therefore, these assays infer stability of CNVs into the lineage. The instance where this premise did not always hold true was in the case of the highly stressed lineage of plants evolved at 28 °C. Here, CNV instability was apparent, and numerous nonrepetitive CNVs were documented between siblings (approximately 0.38% of all 27,549 genes in the A. thaliana genome). Association between stress and genome flexibility can occur in A. thaliana by increased rates of recombination under stress conditions (Lucht et al. 2002). This theory has been long established as a mechanism by which an organism may improve its evolutionary advantage (McClintock 1984). Because both allelic recombination (Abu Bakar et al. 2009) and nonallelic recombination (Cheeseman et al. 2009) have been proposed to generate CNV, a plausible explanation for the high rates of nonrepetitive CNV between siblings in the 28/22 assays is that an environment–genome variation interaction exists via CNV. A mechanism by which stress is sensed and transferred to stimulate CNV (via recombination?) remains unclear. Documenting the genes that are highly abundant in CNV, particularly in plants, may have important rational basis to explain steady-state changes in the genome.
It was found that approximately 52% of CNVs were comprised of tandem-duplicated genes (supplementary table S1, Supplementary Material online) suggesting that these regions of the genome are prone to CNV. Past research has suggested that tandem duplicates can display divergence in expression in A. thaliana (Ganko et al. 2007). Divergence of preserved duplicates has the potential to lead to differential gene expression based on tissue or cellular compartmentalization (Innan and Kondrashov 2010). Variant deletion or duplication of already tandem-duplicated genes, as documented here, may be a means to attempt to specialize or adapt the genome while shielding against deleterious single gene copy polymorphisms. Such a postulate would be consistent with Hanada et al. (2009), who suggest that duplicate genes contribute to genome robustness and protect the plant from severe phenotypes. Another form of common genome diversification is mediated by TEs (Bennetzen 2000). Cataloging the identity of genes that occurred in the repetitive CNV segments among siblings (figs. 2–4; supplementary table S1, Supplementary Material online) showed that TEs were highly abundant components of both duplicated and deleted regions (supplementary table S1, Supplementary Material online) consistent with other studies (Bennetzen 2000; Feschotte et al. 2002). A major discrepancy in this pattern was that while TEs comprised 40% of all genes represented in repetitive CNV events among siblings in the 16/22 lineage CGH, they comprised only 10% of genes in repetitive CNV events among siblings in the 28/22 lineage. These data suggested that variants in plants that were successively being selected with a strong environmental force had reduced frequency of TEs in CNV events. The biological consequence of this disparity was unclear, but it is tempting to speculate that an association between selection and functional gene species would be correlated with adaptation. However, this fails to explain why fewer TEs were associated with stable CNVs in the 28/22 lineage because this would require a mechanism that was capable of preferential CNV selection.
An important element of this study was to ask whether nonrandom patterns appeared in the whole-genome distribution of CNV events. Regional CNV clusters at the whole-genome scale were most prevalent on chromosome 2. For instance, 265 out of 290 genes exposed to repetitive CNVs among siblings were located on chromosome 2 from the 28/22 CGH assay. Also, the largest CNV event occurred on chromosome 2 and was a conserved deletion among all treatments of 128 genes (table 1). Patterns of CNV were not predominantly associated with centromeres and could not be physically associated with any region of genome. Without assaying hundreds rather than three interlineage comparisons, which is economically challenging, it is not feasible to draw any conclusions as to the distribution patterns of CNVs at the genome scale in A. thaliana at the current time. Moreover, because selection was based on fecundity and not phenotypic abnormality, it was not an aim of this experiment to delineate between phenotype and CNV. Taking a broad view of the genomic landscape over interlineage selection with a strong forcing function such as temperature (a reality under current climate change scenarios, Tester and Langridge 2010), a systematic view of polymorphisms in interlineage evolution with at least 20+ individuals compared back with a common ancestor genome is needed to acquire a more elaborate picture of genome evolution.
Conclusions
The current study establishes several important paradigms related to CNV in plants for consideration in plant genome evolution and natural diversity: 1) A. thaliana lineages separated by five generations from a common genotype displayed a substantial rate of retained CNVs among siblings (up to 402 genes); 2) CNVs are comprised of a diverse range of gene species but where overrepresented by TEs, biotic stress response genes, and one conserved CNV was heavily comprised of pre-tRNAs (chromosome 2 position 3,121,499–3,511,499 bp); 3) environmental stress was correlated with duplication CNV events and a higher rate of nonrepetitive CNVs among siblings; and 4) experimental biologists using the model plant A. thaliana are dealing with an organism that undergoes unexpectedly high rates of CNV. To broadly conclude, the experiment described herein aimed to take some first steps to understand how CNV contributes to interlineage natural variation in A. thaliana and found that it represented a significant genomic factor underlying natural diversity.
Supplementary Material
Supplementary tables S1–S3 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
Sue Olson, Kyle Munn, and Doug Durso of Roche Nimblegen are kindly acknowledged for array design and technical assistance. I wish to thank an anonymous reviewer who greatly provided insightful comments. Randy Collins (technical support), Darby Harris, Pamela Compton (comments) (University of Kentucky), and Doug Cook (comments) (UC Davis) are also acknowledged. Funding was derived from The University of Kentucky Research Foundation and National Science Foundation grants NSF-IOS 0922947 and NSF-EFRI: 0937657.
References
- Abu Bakar S, Hollox EJ, Armour JAL. Allelic recombination between distinct genomic locations generates copy number diversity in human β-defensins. Proc Natl Acad Sci U S A. 2009;106:853–858. doi: 10.1073/pnas.0809073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I, et al. Gene copy number variation throughout the Plasmodium falciparum genome. BMC Genomics. 2009;10:353. doi: 10.1186/1471-2164-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Estivill X, Armengol L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3(10):e190. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller U, Crafts-Brandner SJ, Salvucci ME. Moderately high temperatures inhibit ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase-mediated activation of Rubisco. Plant Physiol. 1998;116:539–546. doi: 10.1104/pp.116.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant retrotransposons, where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol. 2007;24:2298–2309. doi: 10.1093/molbev/msm158. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Ross-Ibarra J. Selection on major components of angiosperm genomes. Science. 2008;320:484–486. doi: 10.1126/science.1153586. [DOI] [PubMed] [Google Scholar]
- Gregory LM. Genome size and development complexity. Genetica. 2002;115:131–146. doi: 10.1023/a:1016032400147. [DOI] [PubMed] [Google Scholar]
- Hanada K, et al. Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol Evol. 2009;1:409–414. doi: 10.1093/gbe/evp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci U S A. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One. 2008;3(3):e1838. doi: 10.1371/journal.pone.0001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M, et al. Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech) PLoS Biol. 2005;3:e14. doi: 10.1371/journal.pbio.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, et al. Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell. 2007;19:3230–3241. doi: 10.1105/tpc.107.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Jarillo JA, Salinas J, Martinez-Zapter JM. Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995;108:39–46. doi: 10.1104/pp.108.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidgard S, Crane PR. Quantitative analyses of the early angiosperm radiation. Nature. 1988;331:344–346. [Google Scholar]
- Linares C, Ferrer E, Fominaya A. Discrimination of the closely related A and D genomes of the hexaploid Avena sativa L. Proc Natl Acad Sci U S A. 1998;95:12450–12455. doi: 10.1073/pnas.95.21.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht JM, et al. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO. The theory of island biogeography. Princeton (NJ): Princeton University Press; 1967. [Google Scholar]
- McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Morse AM, et al. Evolution of genome size and complexity in pinus. PLoS One. 2009;4(2):e4332. doi: 10.1371/journal.pone.0004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileyko Y, Joh RI, Weitz JS. Small-scale copy number variation and large-scale changes in gene expression. Proc Natl Acad Sci U S A. 2008;105:16659–16664. doi: 10.1073/pnas.0806239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Bergelson J. The effect of seed and rosette cold treatment on germination and flowering time in some Arabidopsis thaliana (Brassicaceae) ecotypes. Am J Bot. 1999;86:470–475. [PubMed] [Google Scholar]
- Nordborg M, Weigel D. Next generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- Png C, et al. Copy number variation (CNV) that involves a promoter region and homo-oligomerization domain of the major secretory intestinal mucin MUC2 gene, is associated with Inflammatory Bowel Diseases (IBD) Gastroenterol. 2008;134:A-460. [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ. Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiol. 2004;134:1460–1470. doi: 10.1104/pp.103.038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Springer NM, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5(11):e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327:818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Workman C, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:0048.1–0048.16. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]