Abstract
Many of the important changes in evolution are regulatory in nature. Sequenced bacterial genomes point to flexibility in regulatory circuits but we do not know how regulation is remodeled in evolving bacteria. Here, we study the regulatory changes that emerge in populations evolving under controlled conditions during experimental evolution of Escherichia coli in a phosphate-limited chemostat culture. Genomes were sequenced from five clones with different combinations of phenotypic properties that coexisted in a population after 37 days. Each of the distinct isolates contained a different mutation in 1 of 3 highly pleiotropic regulatory genes (hfq, spoT, or rpoS). The mutations resulted in dissimilar proteomic changes, consistent with the documented effects of hfq, spoT, and rpoS mutations. The different mutations do share a common benefit, however, in that the mutations each redirect cellular resources away from stress responses that are redundant in a constant selection environment. The hfq mutation lowers several individual stress responses as well the small RNA–dependent activation of rpoS translation and hence general stress resistance. The spoT mutation reduces ppGpp levels, decreasing the stringent response as well as rpoS expression. The mutations in and upstream of rpoS resulted in partial or complete loss of general stress resistance. Our observations suggest that the degeneracy at the core of bacterial stress regulation provides alternative solutions to a common evolutionary challenge. These results can explain phenotypic divergence in a constant environment and also how evolutionary jumps and adaptive radiations involve altered gene regulation.
Keywords: Escherichia coli genomics, experimental evolution, stress responses
Experimental evolution studies combined with genomics are beginning to reveal not only the mechanistic aspects of evolutionary change but also the underlying principles (Barrick et al. 2009). An inherent but poorly understood feature of evolution is the tendency to diversify. Divergence is evident from the richness of the biosphere and can also be studied in experimental populations of microbes (Rainey and Travisano 1998; Zhong et al. 2004; Barrett and Bell 2006; Maharjan et al. 2006; Kinnersley et al. 2009; Rozen et al. 2009). Ecological interactions have been proposed to contribute to population heterogeneity: examples include divergence in structured environments (Rainey and Travisano 1998), with alternative resources (Zhong et al. 2004; Barrett and Bell 2006), cross-feeding, and cheating between population members (Treves et al. 1998) and seasonal specialization and niche construction (Rozen et al. 2009).
Another source of co-persistence of diverged forms is from alternative adaptations intrinsic to the organism. An example is the coexistence of yeasts with two different modes of energy metabolism but equal competitive fitness (Gudelj et al. 2007). Parallel metabolic and transport adaptations were also proposed to explain diversity in Escherichia coli evolving under glucose limitation (Maharjan et al. 2006, 2007). Multiple alternative metabolic pathway solutions can also result in diversity with the same selection condition (Portnoy et al. 2008). Based on these indications, the hypothesis we test is that inherent degeneracy in cellular processes leads to divergent means of getting fit within the same environment without ecological partitioning. Biological degeneracy occurs at many levels (Edelman and Gally 2001); here, we demonstrate that alternative solutions to reorienting gene regulation can provide degenerate paths to fitness. Furthermore, we show that when mutations beneficially affect global regulatory genes, considerable phenotypic divergence can be rapidly achieved with a few alternative mutational steps, setting the scene for adaptive radiations.
Evolutionary jumps often involve altered patterns of gene regulation (Wittkopp et al. 2004; Hunter 2008) and regulatory gene changes correlate with adaptive radiations (Barrier et al. 2001). The annotation of transcriptional regulatory networks indicates that bacterial regulation is also diverse within and between bacterial species (Lozada-Chavez et al. 2006). Regulatory changes have been noted in evolving experimental populations: Altered global gene expression in yeast (Gresham et al. 2008) and E. coli (Philippe et al. 2007) are accompanied by altered epistatic interactions with other global regulators (Cooper et al. 2008). Still untested, however, is whether alternative, parallel paths to fitness also arise within the same population in a constant environment.
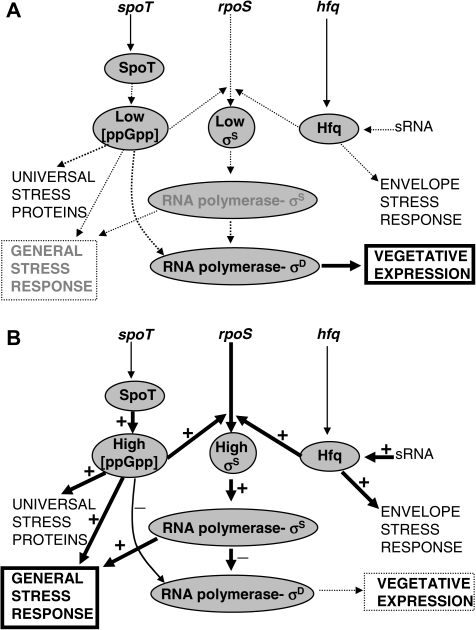
Divergence is evident in chemostats under controlled conditions (Zhong et al. 2004; Maharjan et al. 2006; Kinnersley et al. 2009). Here, we use limitation for inorganic phosphate (Pi) as the simplest possible nutritional selection condition, with glucose as the carbon source. The utilization of Pi by E. coli involves accumulation followed by incorporation into adenosine triphosphate (Torriani 1990). Nevertheless, continued Pi-limitation involves more than induction of the pho regulon (Torriani 1990) responsible for Pi scavenging because the reduced growth rate forced by limitation turns on global hunger and starvation responses (Ferenci 1999). Pi-limitation induces the general stress response regulated by RpoS or sigma factor σS (Bougdour et al. 2006), which controls expression of 10% of the genome (Weber et al. 2005). Pi-limitation also elevates the intracellular level of ppGpp (Spira et al. 1995), in turn inducing the stringent response that affects as many as 500 genes (Durfee et al. 2008). The reported experiments used a specific growth rate of 0.1 per hour, in which these responses are triggered (Ferenci 2007). RpoS and ppGpp are central to redirecting transcription between vegetative and stress/starvation states of E. coli and other bacteria, as summarized in figure 1. These nutrient limitation settings in global regulation are central to understanding the evolutionary selection conditions and set the scene for the experimental results below.
FIG. 1.—
Regulation in the induction of stress responses in Escherichia coli. In unstressed, vegetatively growing bacteria (A), RNA polymerase mainly initiates transcription by complexing with the sigma factor σD, and stress responses are uninduced. Under stressed conditions or when the growth rate is limited in chemostats (B), stress responses are activated. The rpoS gene encodes the RpoS protein, the sigma factor σS that interacts with core RNA polymerase to express several hundred genes involved in the general stress response (Hengge-Aronis 2002). Elevated levels of σS result in reduced transcription by σD and decreased expression of transporters, ribosomes, and metabolic functions (Ferenci 2005). spoT encodes a bifunctional enzyme controlling the level of the signal molecule ppGpp in the cell (Potrykus and Cashel 2008). Under slow growth and starvation conditions, SpoT is involved in elevating ppGpp levels, which in turn has a positive role in rpoS expression. ppGpp is also involved in modulating RNA polymerase activity at certain promoters (dotted arrow) and a role in induction of other proteins, for example, universal stress proteins (dashed arrow, [Magnusson et al. 2005]). Hfq in the presence of small RNAs is important in the translational control of rpoS expression as well as in other stress responses governed by other small RNAs, for example, the envelope stress response (dashed arrow, [Gottesman 2004]). Mutations in any of the spoT, rpoS, or hfq genes results in a shift of expression away from stress responses and toward vegetative functions transcribed by σD. The + and – in the figure denote activation or inhibition of steps, respectively. The thickness of the line denotes the contribution of the signals under the given conditions.
Phenotypic Differences between Coexisting Isolates from Pi-Limited Chemostats
A Pi-limited population growing at a dilution rate of 0.1 per hour (or ∼7-h doubling time) changed considerably over 37 days of continuous propagation. By day 37, the population exhibited considerable colony heterogeneity. Five isolates with different combinations of the traits shown in figure 2A were chosen for detailed analysis.
FIG. 2.—
Phenotypic and proteomic diversity in five Escherichia coli clones coevolved from the parental strain MC4100TF. (A) The phenotype of the five isolates from day 37 was characterized in five different ways (from top to bottom): colony morphology after growth on LB; staining with iodine for RpoS status; sensitivity to 1% methyl α-glucoside on glycerol plates; staining with X-P (5-bromo-4-chloro-3-indolylphosphate) for AP activity; and sensitivity to 3% SDS on L-agar plates. (B) The growth yield of isolates in Pi-limited chemostats at dilution rate 0.1 per hour (% change relative to ancestor) measured as a mean of four estimations is shown. (C) The number of protein changes under Pi-limitation detected by proteomics (more than 1.5-fold in three replicates relative to ancestor) are shown by the number next to each strain, which reflects the number of proteomic differences to ancestral levels. The numbers on lines indicate the number of changes shared by any pair of strains. (D) The number of C-sources metabolized was measured with 95 substrates in replicate 96-well Biolog plates as previously described (Maharjan et al. 2007). (E) Stress survival of isolates was compared with osmotic and oxidative stress by viable counts at increasing concentrations of NaCl and hydrogen peroxide.
To define the global extent of regulatory divergence, proteomic analysis of the five strains was undertaken (detailed results in supplementary table S1, Supplementary Material online). There were >30 protein changes relative to ancestor in each strain but only a minority were shared by isolates (fig. 2C). These results suggest divergent regulatory patterns under identical growth conditions. The genomic basis of divergence was investigated by complete high-coverage assembly of the five genomes and comparison with ancestor (Ferenci et al. 2009). Multiple mutations were found in all isolates (table 1): here, we focus mainly on the astonishing result that distinct global regulatory mutations were found in each of the five isolates.
Table 1.
DNA Changes in the Evolved Genomes
| Strain | Region/Gene | Product | Genome Position | Type | Nucleotide | Codon | Amino acid Change |
| BW4218 | ig (insC-isrC) | NA | 1961811 | SNP | A -> G | NA | NA |
| ig (rpoS leader) | Transcript involved in translational control of rpoS | 2751440 | SNP | C -> A | NA | NA | |
| valS | Valyl-tRNA synthetase | 4418389 | SNP | C -> T | GCG -> GTG | R736L | |
| BW4223 | hfq | RNA-binding protein | 4337201 | SNP | G -> T | CAG -> CAT | Q52H |
| lysZ | tRNA (lys) | 683353–683559 | Deletion | NA | NA | NA | |
| BW4227 | ig (insC-isrC) | 1961811 | SNP | A -> G | NA | NA | |
| rpoS | RNA polymerase (σS) | 2750861 | Indel | C -> * | Frameshift | NA | |
| valS | Valyl-tRNA synthetase | 4418389 | SNP | C -> T | GCG -> GTG | R736L | |
| BW4236 | ig (insC-isrC) | NA | 1961811 | SNP | A -> G | NA | NA |
| spoT | Bifunctional (p)ppGpp synthetase II | 3710130 | SNP | A -> T | AAC -> TAC | N459Y | |
| valS | Valyl-tRNA synthetase | 4418389 | SNP | C -> T | GCG -> GTG | R736L | |
| BW4239 | menD | Bifunctional 2-oxoglutarate decarboxylase | 2261978 | SNP | G -> T | CGC -> CTC | A370E |
| gabD | Succinate-semialdehyde dehydrogenase I | 2675241 | SNP | G -> A | ATG -> ATA | M45I | |
| rpoS | RNA polymerase (σS) | 2751015 | SNP | T -> C | TTG -> TCG | N124S | |
| trkH | Potassium transporter | 3921075 | SNP | T -> A | CTG -> CAG | L80Q | |
| ig (purH-rrsE) | NA | 4095306 | SNP | C -> T | NA | NA | |
| trkG | Potassium transporter subunit | 1314085 | Insertion (IS2) | NA | NA | NA |
NOTE.—ig, intergene; *, absent at given position; NA, not applicable; SNP, single nucleotide polymorphism. Genes involved in global regulation are in bold. Genome positions are based on genome sequence of Escherichia coli K-12 strain BW2952 (Ferenci et al. 2009).
We found 2–6 mutations in the strains shown in table 1, with BW4239 containing 6 but the others 2–3 mutations. In total, there were 13 mutations, and most were single nucleotide polymorphisms. No mutator mutations were found in the sequenced genomes. Three of the clones shared two mutations (in valS and intergene between insC-isrC) not obviously contributing to fitness; presumably, these mutations arose in the population before the different regulatory mutations.
Fitness through hfq, rpoS, and spoT Mutations
A single hfq mutation (Q52H substitution in the RNA chaperone Hfq) was found in BW4223. Hfq is essential for the regulatory effects of dozens of small RNAs involved in regulation in E. coli (Gottesman 2004), including stress responses through control of RpoS translation (fig. 1). The phenotypic effects described below indicate that Q52H affects several regulatory roles.
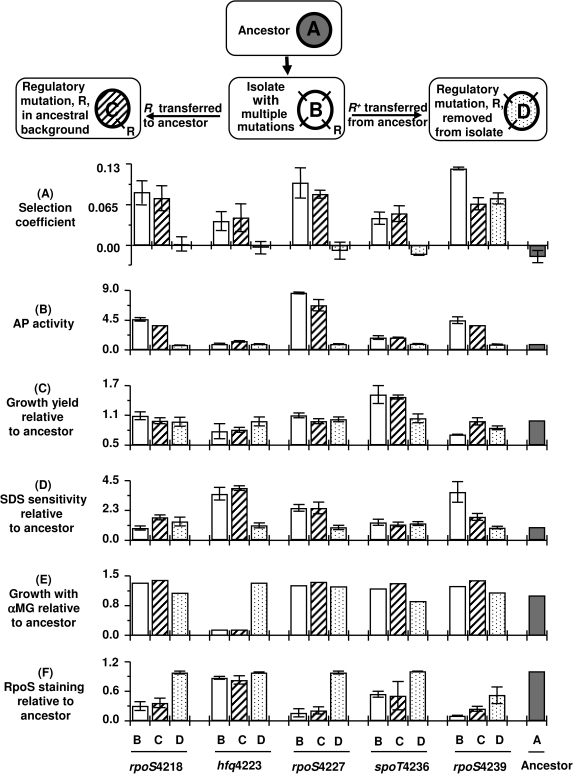
This mutation is sufficient to confer fitness under Pi-limitation when the Q52H allele is transferred to ancestor, as shown in figure 3A. A remarkably diverse set of effects were caused by hfq in BW4223 but explicable from the known pleiotropy (Tsui et al. 1994) and diverse roles of Hfq in regulation: alterations in glucose uptake (Vanderpool 2007) leading to sensitivity to methyl-α−glucoside, decreased RpoS levels (Hengge-Aronis 2002) and increased outer membrane permeability (causing elevated detergent sensitivity) (Valentin-Hansen et al. 2007) are all present (figs. 2A and 3D). The decrease in RpoS was 18% but significantly different to wild type (t-test, P = 0.0017). All these properties were evident when the regulatory mutation was transferred to a clean ancestral background without the other mutations in the isolate (fig. 3A–F). Thus, the hfq mutation was largely responsible not only for fitness but also the multiple property changes of BW4223, and indeed, the isolate in which the hfq mutation was replaced by the wild-type allele had ancestral properties as shown in figure 3A–F. This suggests that the lysZ mutation in BW4223 does not significantly contribute to the evolved properties.
FIG. 3.—
Fitness and phenotypes in strains with altered regulatory genes. We compared the five evolved isolates (Type B in the schematic) and two classes of derivative strains manipulated for each regulatory allele by cotransduction with linked markers (see Materials and Methods). In one class, each mutated regulatory gene (R) was transferred into a clean ancestral background (Type C). In the other class, the regulatory mutation present in each isolate was replaced by the ancestral regulatory gene (Type D) while retaining the other evolved mutations. (A) Competitive fitness was measured for Type B, C, and D strains in Pi-limited chemostats. Competitions were all against a reference strain BW3454 containing a metC::Tn10 countable marker; this strain is marginally fitter than ancestor. Competing strains were mixed 50:50 after 16 h individual acclimatization in chemostats. Selection coefficients (Dykhuizen and Hartl 1983) were calculated from changes in population proportions. The mean and standard deviations were obtained from 3 to 5 replicates. (B) The alkaline phosphatase was measured using ONP-Pi as substrate after growth in media containing 30 μM KH2PO4. (C) The growth yield of strains was measured from the absorbance in L-broth after overnight growth to steady state and shown relative to ancestor. (D) Sensitivity to 3% SDS was measured by growth in microtitre plates, with densities divided by that of the ancestor shown. (E) Sensitivity to 1% methyl α-glucoside (α-MG) was measured in patches on glycerol plates. Patches were scanned densitometrically, and density relative to ancestor is shown. (F) The RpoS level was estimated by glycogen staining as in figure 2 and scanning densities relative to that of ancestor.
Three different rpoS mutations were found by genomic sequencing of BW4218, BW4227, and BW4239: two intragenic and causing a null phenotype and one upstream of rpoS in the transcribed sequence before the rpoS gene (table 1). The upstream mutation is in a region involved in mRNA secondary structure formation that is involved in the translational control of rpoS expression (Hengge-Aronis 2002). The mutation may prevent the activation of the RNA transcript (removal of the secondary structure) and hence decrease rpoS translation.
All three mutations provide a strong fitness benefit under Pi-limitation and in BW4218 or BW4227, the rpoS mutation was the dominant fitness determinant in the isolate (fig. 3A). The established benefit of rpoS mutations at slow growth rates is in the relief of repression of vegetative genes through competing sigma factors (fig. 1, [Ferenci 2005]). In BW4239, other mutations besides rpoS contributed to fitness, and replacement of the rpoS4239 mutation with the wild-type allele only partly reduced the selection coefficient of the isolate. The nature of the benefit conferred by the other mutations in BW4239 is not yet known but will be investigated in the future.
Another evolved regulatory difference in the same population was a spoT mutation in BW4236 (table 1). The positive selection coefficient of BW4236 was almost entirely due to the spoT mutation (fig. 3A). SpoT is a bifunctional enzyme that can either degrade or synthesize ppGpp, a stress alarmone in E. coli (Gentry and Cashel 1996). Under starvation conditions, the elevation of ppGpp is due to SpoT involvement. The most direct effect of the N459Y substitution was to reduce ppGpp levels relative to ancestor (from 0.092 ± 0.004 to 0.052 ± 0.001 relative units). Unsurprisingly, the spoT mutation in BW4236 also changed multiple properties, including reduced RpoS consistent with the ppGpp–RpoS relationship (Spira et al. 2008) and a markedly increased growth yield (fig. 2B). The other mutations in BW4236 did not contribute to fitness or phenotypes (fig. 3A–F).
The N459Y substitution has not been previously described in SpoT, but mutations close to this site were found in the Lenski experimental populations increasing fitness in glucose culture (Philippe et al. 2007). In contrast to our results, they did not find decreased ppGpp, and the benefit was in the increased growth yield, which we also observe in batch culture (fig. 3C). Under Pi-limitation, the RpoS reduction is also a likely benefit as discussed above.
Phosphate Transport and pho Regulation in Evolved Isolates
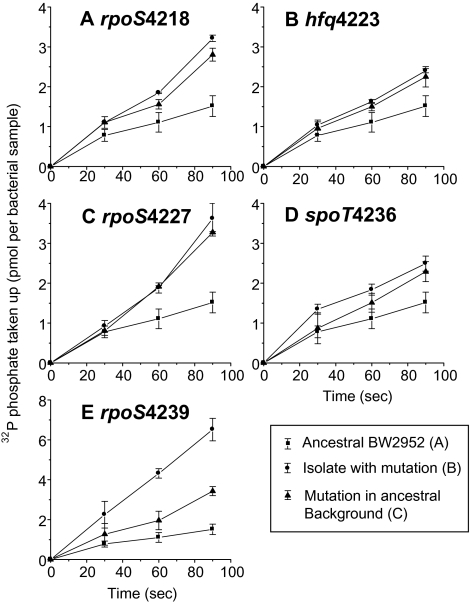
Previous studies showed that pho genes are negatively regulated by RpoS (Taschner et al. 2004), so relief of this repression is a likely benefit either through rpoS mutations or perhaps indirectly through hfq and spoT mutations. As shown in figure 4, the isolates and strains with the regulatory mutations all exhibited somewhat elevated transport rates when measured with 32Pi. However, the increase relative to ancestor was highest in BW4239, in which fitness (fig. 3A) and transport (fig. 4E) are both boosted by mutations other than the rpoS mutation. The regulatory mutations in isolation all increase transport, in line with a common reduction in negative regulation by RpoS. The hfq mutation has the lowest positive effect (fig. 4B), and this is mirrored by the minor increase in phoA gene expression shown in figure 3B. These small increases in transport-related functions are in contrast to glucose-limited populations, in which evolved transport rates are boosted up to 8-fold (Maharjan et al. 2006, 2007).
FIG. 4.—
The effect of regulatory mutations on transport rates measured with 32Pi. Pi-limited chemostat cultures were cultured for 30 h to establish limitation and then assayed for transport rates when measured with a low concentration (1 μM) 32Pi. In each panel, the ancestral BW2952 strain (▪) is compared with the chemostat isolate containing the mutation shown (•) as well as the type C strain from figure 3 in which the regulatory mutation is in a clean ancestral background (□). Each assayed sample contained 5 × 107 bacterial cells.
A Shared Fitness Trade-off through hfq, spoT, and rpoS Mutations
A shared consequence of the regulatory mutations was the reduction in RpoS levels, as shown in figure 2A and figure 3F. In confirmation of the staining data, even in the non-rpoS mutants BW4223 and BW4236, the RpoS protein levels dropped by 21% and 32%, respectively when measured by quantitative immunoblotting (Spira et al. 2008). In this respect, the rpoS, spoT, and hfq mutations were convergent in resetting the stress protection and nutritional competence (SPANC) balance, the trade-off between SPANC determined by RpoS (fig. 1, [Ferenci 2005]). Each mutation changed the SPANC balance toward better nutrition not only under Pi-limitation but also in general; all the regulatory mutations permitted more carbon sources to be metabolized, as tested with 95 Biolog substrates (fig. 2D). So the effect of the mutations was evident in unselected traits like acetate utilization. Conversely, the common cost of the adaptations was decreased stress resistance, as seen with high osmolarity or peroxide stress (fig. 2E). Altogether, these results demonstrate that the strategy of improving nutritional capability has degenerate mutational solutions. The changes to Pi uptake observed in figure 4 are also part of the global regulatory change toward better nutrition through reduced negative control by RpoS.
Conclusions
These results have broad implications for evolutionary diversification in general and regulatory divergence in particular.
Our results demonstrate that there is sufficient internal regulatory flexibility in a living organism to simultaneously undertake divergent adaptive pathways in the same environment. The importance of regulatory mutations was supported by our observation that each of the five distinct regulatory mutations was integral to the fitness of the isolates under Pi-limitation (fig. 3A). Hence, reordering of the regulatory network at the core of bacterial transcriptional control shown in figure 1 was essential in adapting to Pi-limitation. Even if the different regulatory adaptations do not persist to fixation, the transitory coexistence of the different types allows parallel exploration of the mutational possibilities for further evolution in the different backgrounds.
The data indicate that the stress/nutrition trade-off is a driver of diversification. This is consistent with results on natural isolates of the species E. coli as well as with E. coli populations in infections (Levert M, Zamfir O, Clermont O, Bouvet O, Lespinats S, Hipeaux MC, Branger C, Picard B, Saint-Ruf C, Norel F, Balliau T, Zivy M, Le Nagard H, Cruvellier S, Chane-Woon-Ming B, Nilsson S, Gudelj I, Phan K, Ferenci T, Tenaillon O, Denamur E, unpublished data; Spira B, Galbiati HF, Betteridge T, Phan K, Ferenci T, unpublished data). Hence, changing of the SPANC balance in members of the species extends to nonlaboratory situations. Our ancestral strain has endogenous levels of ppGpp and RpoS toward the higher end for natural isolates (King et al. 2004), which accentuates the cost of stress resistance. Nevertheless, RpoS/ppGpp levels are still higher in several naturally stress-resistant isolates and in some pathogenic lineages of E. coli, and these carry an even greater nutritional cost than in our ancestor. Trade-offs are generally important in life-history traits such as between fecundity and survival (Stearns 1992), so the regulatory rebalancing we describe has broader implications in explaining the role of trade-offs in evolution.
The indications are that biological degeneracy is important in evolution not simply to add complexity (Edelman and Gally 2001) but also to increase evolvability (Lenski et al. 2006). We find that degeneracy contributes to adaptation: the possibility of three alternative adaptations contributes to the capacity to evolve and indeed an increased capacity for risk hedging. In the specific examples studied here, the different rpoS, hfq, and spoT mutations may have similar benefits under Pi-limitation but very different fitness profiles in other environments. For example, as shown in figure 2, osmotically stressful environments counterselect more against the rpoS null mutations but membrane-damaging conditions such as detergents are more detrimental for hfq strains. In effect, functional and regulatory degeneracies provide an increased scope for gaining fitness, just as duplicated genes allow increased evolutionary options.
A conclusion from this study is that the emergence of alternative adaptations does not require distinct niches, confirming conclusions from other studies (Maharjan et al. 2006, 2007; Gudelj et al. 2007; Portnoy et al. 2008). Furthermore, our results suggest that sympatric divergence (in the biogeographical sense, in the same environment, see [Fitzpatrick et al. 2008]) may be the result of internal subspecialization of degenerate regulatory capabilities. Our experiments provide a case of sympatric divergence where there is no question of allopatry, whereas in natural populations it is extremely difficult to exclude allopatry through fine-grained subdivision of the habitat (Fitzpatrick et al. 2008). More speculatively, our results indicate how sympatric divergence can arise in a population biology sense (where interbreeding is reduced in the same environment [Fitzpatrick et al. 2008]). If highly global regulatory specialization is common, then this itself could cause reduced compatibility for interbreeding. The high frequency of major regulatory changes in evolution (Wittkopp et al. 2004; Hunter 2008) may provide the basis for sympatric speciation.
Materials and Methods
Growth Media and Strains
Bacteria were cultured in T-salts minimal medium supplemented with glucose (0.2% w/v) plus 30 μM KH2PO4 for chemostats or 1 mM KH2PO4 for batch culture (Spira et al. 1995). Bacteria for phenotypic tests were grown or minimal medium A or L-broth (as described by Miller [1972]). All growth was at 37 oC. For long-term chemostats, MC4100TF was grown overnight in T-salts and inoculated into an 80-ml chemostat containing T-salts, 0.2% glucose and 30 μM KH2PO4 as described (Spira and Ferenci 2008). The bacterial concentration in the chemostat was stable through 37 days, between 1.5 and 2.5 × 108 bacteria/ml.
The strains used in this study are described in table 2. The different alleles of rpoS, hfq, and spoT were transferred from evolved strains into ancestral strain or from ancestral strain into evolved strains by P1 transduction as described in Miller (1972). For the transfer of the spoT mutation, zib563::Tn10 was used as the linked selection marker. For the transfer of rpoS and hfq from evolved isolates into ancestral strain or from ancestral into evolved isolates, we first constructed cysD::amp and purA::tet strains using the protocol described in Yu and Court (1998). The proximity of cysD::Amp locus to rpoS and purA::tet locus to hfq allowed cotransduction (>90% cotransduction in both cases). The transductants were tested for alleles by sequencing.
Table 2.
Strains Used in the Study
| Strains | Relevant Genotype | Reference or Origin |
| MC4100TF | F-araD139 D(argF-lac)U169 rspL150 deoCl relA1 thiA ptsF25 flb5301 rbsR | Spira et al. (2008) |
| BW4218 | Chemostat evolved isolate | This study |
| BW4223 | Chemostat evolved isolate | This study |
| BW4227 | Chemostat evolved isolate | This study |
| BW4236 | Chemostat evolved isolate | This study |
| BW3454 | MC4100TF metC162::Tn10 | Notley-McRobb and Ferenci (1999) |
| BW4239 | Chemostat evolved isolate | This study |
| BW5151 | DY330 purA:: Tn10 | This study |
| BW5153 | MC4100TF purA:: Tn10 | This study |
| BW5166 | MC4100 hfq4223 | This study |
| BW6006 | BW4223 hfq4100 | This study |
| BW5197 | BW4236 spoT4100TF | This study |
| BW5199 | MC4100 spoT4236 | This study |
| BW5200 | MC4100TF zib563::Tn10 | Spira et al. (2008) |
| BW6007 | DY330 cysD::amp | This study |
| BW6008 | MC4100TF cysD::amp | This study |
| BW6009 | BW4218 cysD::amp | This study |
| BW6010 | BW4227 cysD::amp | This study |
| BW6011 | BW4239 cysD::amp | This study |
| BW6012 | MC4100 rpoS4218 | This study |
| BW6013 | MC4100 rpoS4227 | This study |
| BW6014 | MC4100 rpoS4239 | This study |
| BW6015 | BW4218 rpoS4100 | This study |
| BW6016 | BW4227 rpoS4100 | This study |
| BW6017 | BW4239 rpoS4100 | This study |
| DY330 | W3110 ΔlacU169 gal490 λcl857 Δ(cro-bioA) | Yu et al. (2000) |
Detection of rpoS Status
The level of RpoS was assessed by staining glycogen with iodine; the intensity of the brown color varies according to the level of σS in the cell (Notley-McRobb et al. 2002). For quantitation, photographs were scanned densitometrically across 2-μl spotted patches on L-agar using Image J software and densities related to ancestor values. For blots, bacterial cultures were grown overnight in LB medium at 37 oC. Proteins from 2 × 109 cells were resolved by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis in a 12.5% gel, and RpoS in blots detected with diluted monoclonal anti-RpoS antibodies (NeoClone). The Super Signal West Pico kit (Pierce) was used to detect the RpoS bands as recommended by the manufacturer. The signal intensities on autoradiograms were scanned and computed using the Image J software. At least three replicate cultures were used and tested for statistical significance.
Alkaline Phosphatase Assay
p-nitrophenyl-phosphate (p-NPP) was used as substrate as described (Spira et al. 1995), and AP activity units are defined as the increase in absorbance at 410 nm/min. Optical cell density at 600/nm.
SDS Susceptibility Assay
Sensitivity to SDS was assayed from overnight cultures grown in L-broth by spotting of cultures (2 μl) onto L-agar plates containing 3% (w/v) SDS. Liquid cultures containing 3% SDS were followed by measuring absorbance of 6-fold replicates of strains in L-broth in microtitre plates.
Methyl α-glucoside (α-MG)
To assay sensitivity to α-MG, culture (2 μl) was spotted onto minimal medium A 0.2% glycerol agar plate with or without 1% α-MG. For quantitation, photographs were scanned densitometrically across the growth patches using Image J software and densities related to ancestor values.
Growth Yields
The yields were determined by measuring optical density of the cultures at 600 nm.
Biolog Assay
The catabolism of the starting strain and the chemostat isolates with 95 substrates were determined using the commercially available Biolog GN2 (Biolog) as previously described (King et al. 2004).
Stress Resistance Assays
Bacteria from overnight cultures in L-broth were washed twice and diluted in 0.9% NaCl to a density of ∼4 × 103 cells/ml. For oxidative stress, freshly diluted H2O2 was added to 1 ml culture to final concentrations of 1, 2, 3, 4, and 5 mM and held at room temperature for 30 min. For osmolarity, suspensions of 4 × 103 cells/ml were incubated in 1, 2, 3, 4, 5 M NaCl for 1 h at room temperature.
Pi Uptake Assay
For transport assays, 500 μl bacteria from 30-h-old Pi-limited chemostat cultures were mixed with 5 μl of 100 μM KH2PO4 and 10 μl of 1μCi32P/μl (MP Biomedicals). Samples (100 μl) taken at time points were filtered through pore size 0.45-μm filters, washed immediately with 5-ml washing solution (T-salt plus 100 μM KH2PO4). The uptake rates were determined by measuring the scintillation of 32P in the 5 × 107 cells on the filters.
ppGpp Assay
Cells growing exponentially in T-salts/glucose were supplemented with and 0.25 mM 32P-KH2PO4 (100 μCi/ml) at an OD600 = 0.2. Samples were harvested after 70, 80, and 90 min. The labeled samples were analyzed as in Spira et al. (2008).
Fitness Experiments in Chemostats
For fitness comparisons, a tetracycline-resistant derivative of MC4100TF carrying a metC::Tn10 insertion was used, and medium was supplemented with 4 μg/ml methionine. Chemostat competitions were as previously described (Maharjan et al. 2006) and the selection coefficients based on the equations in Dykhuizen and Hartl (1983).
Proteomics and genomics details and strategies are described in the supplementary tables S1 and S2 (Supplementary Material online) and associated legends.
Supplementary Material
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Alex Uren and Anna Yeung for characterizing isolates in chemostats. R.P.M. and C.M. were supported by Australian Research Council grants to T.F. and P.R.R. B.S. was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP—Brazil). L.W. and L.F. were partly supported by the National Natural Science Foundation of China (NSFC) Key Programs Grant 30530010, the Chinese National Science Fund for Distinguished Young Scholars (30788001), NSFC General Program Grants 30870070, 30870078, and 30771175, National 863 Program of China Grants 2006AA020703 and 2006AA06Z409, National 973 Program of China Grant 2009CB522603, and National Key Programs for Infectious Diseases of China 2008ZX1004-002, 2008ZX1004-009, and 2009ZX10004-108.
References
- Barrett RDH, Bell G. The dynamics of diversification in evolving Pseudomonas populations. Evolution. 2006;60:484–490. [PubMed] [Google Scholar]
- Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1274. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Barrier M, Robichaux RH, Purugganan MD. Accelerated regulatory gene evolution in an adaptive radiation. Proc Natl Acad Sci U S A. 2001;98:10208–10213. doi: 10.1073/pnas.181257698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Wickner S, Gottesman S. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 2006;20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Remold SK, Lenski RE, Schneider D. Expression profiles reveal parallel evolution of epistatic interactions involving the CRP regulon in Escherichia coli. PLoS Genet. 2008;4:e35. doi: 10.1371/journal.pgen.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Hartl DE. Selection in chemostats. Microbiol Rev. 1983;47:150–168. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proc Natl Acad Sci U S A. 2001;98:13763–13768. doi: 10.1073/pnas.231499798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Regulation by nutrient limitation. Curr Opin Microbiol. 1999;2:208–213. doi: 10.1016/S1369-5274(99)80036-8. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv Microb Physiol. 2007;53:169–229. doi: 10.1016/S0065-2911(07)53003-1. [DOI] [PubMed] [Google Scholar]
- Ferenci T, et al. Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J Bacteriol. 2009;191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick BM, Fordyce JA, Gavrilets S. What, if anything, is sympatric speciation? J Evol Biol. 2008;21:1452–1459. doi: 10.1111/j.1420-9101.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudelj I, Beardmore RE, Arkin SS, Maclean RC. Constraints on microbial metabolism drive evolutionary diversification in homogeneous environments. J Evol Biol. 2007;20:1882–1889. doi: 10.1111/j.1420-9101.2007.01376.x. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. The great leap forward. Major evolutionary jumps might be caused by changes in gene regulation rather than the emergence of new genes. EMBO Rep. 2008;9:608–611. doi: 10.1038/embor.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T, Ishihama A, Kori A, Ferenci T. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol. 2004;186:5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley MA, Holben WE, Rosenzweig F. E Unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. 2009 doi: 10.1371/journal.pgen.1000713. PLoS Genet. 5:e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Barrick JE, Ofria C. Balancing robustness and evolvability. 2006 doi: 10.1371/journal.pbio.0040428. art. no. e428. PLoS Biol. 4:2190–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Maharjan R, Seeto S, Ferenci T. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J Bacteriol. 2007;189:2350–2358. doi: 10.1128/JB.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan R, Seeto S, Notley-McRobb L, Ferenci T. Clonal adaptive radiation in a constant environment. Science. 2006;313:514–517. doi: 10.1126/science.1129865. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Notley-McRobb L, Ferenci T. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ Microbiol. 1999;1:33–43. doi: 10.1046/j.1462-2920.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- Portnoy VA, Herrgard MJ, Palsson BO. Aerobic fermentation of D-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain. Appl Environ Microbiol. 2008;74:7561–7569. doi: 10.1128/AEM.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett. 2009;12:34–44. doi: 10.1111/j.1461-0248.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- Spira B, Ferenci T. Alkaline phosphatase as a reporter of sS levels and rpoS polymorphisms in different E. coli strains. Arch Microbiol. 2008;189:43–47. doi: 10.1007/s00203-007-0291-0. [DOI] [PubMed] [Google Scholar]
- Spira B, Hu X, Ferenci T. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology. 2008;154:2887–2895. doi: 10.1099/mic.0.2008/018457-0. [DOI] [PubMed] [Google Scholar]
- Spira B, Silberstein N, Yagil E. Guanosine 3',5'-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SM. The evolution of life histories. Oxford: Oxford University Press; 1992. [Google Scholar]
- Taschner NP, Yagil E, Spira B. A differential effect of sigma(S) on the expression of the Pi regulon genes of Escherichia coli. Microbiology. 2004;150:2985–2992. doi: 10.1099/mic.0.27124-0. [DOI] [PubMed] [Google Scholar]
- Torriani A. From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays. 1990;12:371–376. doi: 10.1002/bies.950120804. [DOI] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr Opin Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigma(S)-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Yu DG, Court DL. A new system to place single copies of genes, sites and lacZ fusions on the Escherichia coli chromosome. Gene. 1998;223:77–81. doi: 10.1016/s0378-1119(98)00163-2. [DOI] [PubMed] [Google Scholar]
- Yu DG, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Khodursky A, Dykhuizen DE, Dean AM. Evolutionary genomics of ecological specialization. Proc Natl Acad Sci U S A. 2004;101:11719–11724. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]