Abstract
As the largest and the basal-most family of conifers, Pinaceae provides key insights into the evolutionary history of conifers. We present comparative chloroplast genomics and analysis of concatenated 49 chloroplast protein-coding genes common to 19 gymnosperms, including 15 species from 8 Pinaceous genera, to address the long-standing controversy about Pinaceae phylogeny. The complete cpDNAs of Cathaya argyrophylla and Cedrus deodara (Abitoideae) and draft cpDNAs of Larix decidua, Picea morrisonicola, and Pseudotsuga wilsoniana are reported. We found 21- and 42-kb inversions in congeneric species and different populations of Pinaceous species, which indicates that structural polymorphics may be common and ancient in Pinaceae. Our phylogenetic analyses reveal that Cedrus is clustered with Abies–Keteleeria rather than the basal-most genus of Pinaceae and that Cathaya is closer to Pinus than to Picea or Larix–Pseudotsuga. Topology and structural change tests and indel-distribution comparisons lend further evidence to our phylogenetic finding. Our molecular datings suggest that Pinaceae first evolved during Early Jurassic, and diversification of Pinaceous subfamilies and genera took place during Mid-Jurassic and Lower Cretaceous, respectively. Using different maximum-likelihood divergences as thresholds, we conclude that 2 (Abietoideae and Larix–Pseudotsuga–Piceae–Cathaya–Pinus), 4 (Cedrus, non-Cedrus Abietoideae, Larix–Pseudotsuga, and Piceae–Cathaya–Pinus), or 5 (Cedrus, non-Cedrus Abietoideae, Larix–Pseudotsuga, Picea, and Cathaya–Pinus) groups/subfamilies are more reasonable delimitations for Pinaceae. Specifically, our views on subfamilial classifications differ from previous studies in terms of the rank of Cedrus and with recognition of more than two subfamilies.
Keywords: chloroplast genome, Cedrus, Cathaya, Pesudotsuga, Pinaceae phylogeny, molecular dating
Introduction
Pinaceae (pine family) is the largest (more than 230 species), most economically important, and basal-most family of conifers (Hart 1987; Price et al. 1993; Chaw et al. 1995, 1997; Stefanovic et al. 1998; Gugerli et al. 2001); therefore, it can provide key insights into the evolutionary history of conifers. The Pinaceae are trees (2- to 100-m tall) that are mostly evergreen (except Larix and Pseudolarix; both being deciduous), resinous, and unisexual, with subopposite or whorled branches and spirally arranged linear (needle-like) leaves (Farjon 1990). Many of the species that are highly valuable for their timber include firs (Abies), cedars (Cedrus), larches (Larix), spruces (Picea), pines (Pinus), Douglas firs (Pseudotsuga), and hemlocks (Tsuga).
Pinaceae species often form the dominant component of boreal, coastal, and montane forests in the northern hemisphere (Farjon 1990; Liston et al. 2003). For instance, Pinus, the largest genus of the family, with more than 110 species, occupies an extended geographic range—North America, northern part of Asia, and Europe (Farjon 1990). Distributions of the Pinaceae genera are discontinuous, with major diversity centers in the mountains of southwest China, Mexico, and California (Farjon 1990). Fossil records indicate that Pinaceae ancestors appeared during late Triassic (∼220–208 Ma; Miller 1976) and widely spread over Asia and North America. However, in Europe, fossils only after Cretaceous are abundant (LePage and Basinger 1995; Liu and Basinger 2000; LePage 2003).
Twelve genera (i.e., Abies, Cathaya, Cedrus, Hesperopeuce, Keteleeria, Larix, Nothotsuga, Picea, Pinus, Pseudolarix, Pseudotsuga, and Tsuga) have been recognized in the family since the pioneering work of Van Tieghem (1891; supplementary table 1, Supplementary Material online). However, from nrITS studies, Hesperopeuce (only T. longibrateata) and Nothotsuga (only T. heterophylla) were retained in Tsuga rather than forming two separate genera (see review by Vining and Campbell 1997). A monophyletic origin of the Pinaceae genera was supported by many unique traits such as P-type plastids (i.e., plastids accumulating protein as a single product or in addition to starch; Behnke 1974), the 4-tiered proembryos (Dogra 1980), lack of flavonoids (Geiger and Quinn 1975), and an unusual indel at nucleotide position 195 of the nuclear 18S rRNA gene (Chaw et al. 1997).
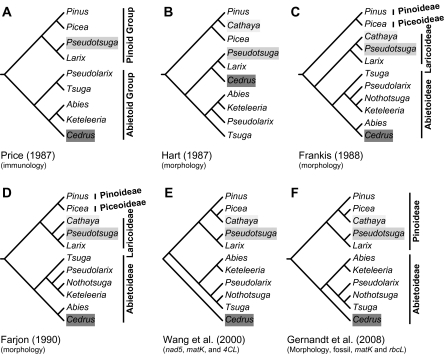
Six major competing views on the classification/phylogeny of Pinaceae genera and subfamilies (fig. 1; supplementary table 1, Supplementary Material online) have been proposed but debated. The major disputes are in the placements of Cathaya, Cedrus, Pseudolarix, and Pseudotsuga and the delimitation of subfamilies. Van Tieghem (1891) first divided Pinaceae genera into two groups (i.e., the Abietoid [=Abitoideae, including Abies, Cedrus, Keteleeria, Pseudolarix, and Tsuga] and Pinioid [Pinioideae, including Larix, Picea, Pinus, and Pseudotsuga] groups) on the basis of the location and number of resin canals. The two groups were adopted by Jeffrey (1905), Doyle (1945), and Price et al. (1987; Cathaya was not included; fig. 1A) from studies of wood anatomy, pollen morphology, and immunology of seed proteins, respectively. In contrast, Pinus was placed in its own subfamily, Pinioideae, by Vierhapper (1910) because of its unusually short shoots (needle fascicles) and distinctive thickened cone scales (see review by Price 1989). Vierhapper (1910), Pilger (1926), and a number of their followers (e.g., Florin 1931, 1963; Melchior and Werdermann 1954; Krüssmann 1985) divided the remaining genera into two subfamilies (supplementary table 1, Supplementary Material online) on the basis of “presence or absence of strongly condensed vegetative short shoots that bear the majority of the foliage leaves” (Price 1989). However, Price (1989) considered it highly artificial to divide the family on the basis of shoot dimorphism alone, with which other morphological traits show little concordance. Frankis (1988) and Farjon (1990) emphasized the importance of reproductive morphologies, such as cones, seeds, pollen types, and chromosome numbers and concurrently recognized four subfamilies in Pinaceae (supplementary table 1, Supplementary Material online) but disagreed with each other in the divergent course of the subfamilies and the evolutionary position of Cathaya (fig. 1). Wang et al. (2000), using three genes (nad5, matK, and 4CL) for phylogenetic analysis, proposed an eccentric view that Cedrus is the basal-most genus of Pinaceae. By inferring from chloroplast rbcL and matK genes and nonmolecular characters and integrating fossil and extant Pinaceous taxa, Gernandt et al. (2008) claimed that root placements varied for Pinaceae when different analysis methods were conducted.
FIG. 1.—
Six major competing views on the phylogeny of Pinaceous genera and subfamilies. All trees were redrawn and simplified from the cited references. The light, medium, and heavy gray backgrounds indicate the positions of Cathaya, Pseudotsuga, and Cedrus, respectively. Prior treatments without phylogenetic trees were not included. Modified trees were reconstructed using characters noted within the parentheses below cited studies. For subfamilial delimitations, refer to supplementary table 1 (Supplementary Material online) and text.
Cathaya Chun et Kuang (Chun and Kuang 1962), with a single species endemic to southern China, is the latest described genus in Pinaceae. Its affinity to other genera has been highly debated (see review by Wang et al. 1998). Florin (1963) placed it in the Abietoideae. By analysis of embryo development, Wang and Chen (1974) and Hart (1987) held that Cathaya is closely related to Pinus (fig. 1B). In contrast, by analysis of other vegetative organs, Hu and Wang (1984) and Frankis (1988) argued that the genus is more related to Pseudotsuga than to Larix (fig. 1C). On observing that Cathaya cones were produced on the leafy peduncles, Farjon (1990) claimed that Cathaya should be sister to the Laricoideae (previously including only Larix and Pseudotsuga (fig. 1D) [supplementary table 1, Supplementary Material online]). Recent phylogenetic analyses (Wang et al. 2000; Gernandt et al. 2008) recovered the Cathaya–Picea subclade and revealed that this subclade and Pinus form a clade but with low bootstrap support (fig. 1E and F). Associated with the controversial position of Cathaya, the phylogenetic position of Psuedotsuga has also been uncertain.
Pseudotsuga comprises about eight species ranging from Canada, United States, Mexico, and Japan to China (Farjon 1990). This genus, along with Larix and Cedrus, was first grouped as Laricinae (equivalent to the subfamily Laricoideae [supplementary table 1, Supplementary Material online]) by Melchior and Werdermann (1954), who emphasized that the three have both short and long shoots, monomorphic leaves, and strobili borne on the short shoots. Hart’s (1987) cladistic analysis substantiated this grouping. Later, Frankis (1988) substituted Cedrus with Cathaya (first described in 1962; refer to previous paragraph) in the Laricoideae and regarded Larix as a sister group to Cathaya–Pseudotsuga (fig. 1). Hart (1987) and Frankis (1988) also considered that their respective circumscribed Laricoideae is sister to Abietoideae rather than to the Pinus–Picea clade (fig. 1; supplementary table 1, Supplementary Material online) as posited by Price et al. (1987), whose view in turn was maintained by Farjon (1990), Wang et al. (2000), and Gernandt et al. (2008).
The cedar genus Cedrus, consisting of 4–5 species (Farjon 1990), is native to the mountains of the western Himalayan and Mediterranean regions. Cedrus is traditionally placed in the Abietoideae along with other four genera, Abies, Keteleeria, Pseudolarix, and Tsuga (supplementary table 1, Supplementary Material online). All of these five genera have erect and similar cone structures (Hu et al. 1989; Farjon 1990). Nevertheless, Cedrus was previously placed as sister to the Larix–Pesudotuga group (Hart 1987), the Abies–Keteleeria group (Price et al. 1987), or Abies (Frankis 1988; Farjon 1990). The earliest fossil record of Cedrus was documented in the Early Tertiary, ∼65 Ma (Miller 1976), which is much later than the record of a fossil cone species, Pinus belgica (135 Ma; Alvin 1960), and a fossil wood of the Pinus subg. Strobus (85 Ma; Meijer 2000). Hence, Wang et al. (2000) posited that Cedrus is the earliest divergent genus in Pinaceae, which appears to conflict with the fossil records. Liston et al. (2003) remarked that “the position of Cedrus remains problematic.”
In view of the aforementioned long-standing controversies surrounding traditional systematic/cladistics and contradictory molecular hypotheses for the evolution of Pinaceae, other lines of evidence are critically needed to better resolve the issues. To this end, we sequenced the chloroplast genomes (cpDNAs) of five key Pinaceae species (complete cpDNAs: Ca. argyrophylla and Ce. deodara; draft cpDNAs: Larix decidua, Picea morrisonicola, and Pseudotsuga wilsoniana) and performed cpDNA comparisons and phylogenetic analyses for our sampled data set, which includes 19 cpDNAs from 15 Pinaceous species and 4 reference species—a non-Pinaceae conifer (Cryptomeria japonica; Cupressaceae) (Hirao et al. 2008), Ginkgo biloba (Ginkgoaceae) (Jansen et al. 2007), and 2 cycad species (Jansen et al. 2007 and Wu et al. 2007). The 15 sampled Pinaceous species represent 8 of the 10 Pinaceous genera and all the 4 Pinaceous subfamilies. The cpDNA sequences are suggested to be useful candidates for resolving the plant phylogeny at deep levels of evolution because of their low rates of silent nucleotide substitutions and their structural characters, such as gene order/segment inversions, expansion/contraction of the inverted repeat (IR) regions, and loss/retention of genes (see review by Raubeson and Jansen 2005). For example, an inversion flanking the petN and ycf2 genes occurs in all cpDNAs of vascular plants except lycopods, which suggests that lycopsids are the basal-most lineage of vascular plants (Raubeson and Jansen 1992a); a common duplication of the trnH–rps19 gene cluster in IRs distinguishes monocots from dicots (Chang et al. 2006) and an intron loss in each of clpP and rps12 genes sustains the early split of the IR-lacking legumes (Jansen et al. 2008). Additionally, concatenating sequences from many genes may overcome the problem of multiple substitutions that results in loss of phylogenetic information between chloroplast lineages (Lockhart et al. 1999) and can reduce ‘‘sampling errors due to substitutional noise” (Sanderson and Doyle 2001).
However, important events in the phylogeny, such as gene duplications and gene/taxon diversifications, can be put on a timescale to address correct evolutionary history only with faithful estimations of divergence times (Kumar and Hedges 1998; Arbogast et al. 2002; Smith and Peterson 2002) and the availability of a reliable phylogenetic tree. Therefore, we also reestimated the divergence times of the Pinaceous subfamilies and genera by using the phylogenetic tree obtained in the present study and three reliable fossil records.
Materials and Methods
Amplification and Sequencing of Pinaceae cpDNAs
The plant materials of Ca. argyrophylla and Ce. deodara originated from Sichuan, China and India, respectively, were collected from Sanzhi, Taipei County, Taiwan. Larix decidua, P. morrisonicola, and P. wilsoniana were collected from Sitou Nature Education Area, Nantou County, Taiwan and were grown in the greenhouse at Academia Sinica. Young leaves were harvested, and genomic DNAs were extracted by use of a 2× CTAB protocol (Stewart and Rothwell 1993). The cpDNA fragments were amplified by long-range polymerase chain reaction (PCR) (TaKaRa LA Taq, Takara Bio Inc) with primers (supplementary table 2, Supplementary Material online) designed according to the conserved regions from published sequences. The entire cpDNA was amplified by approximately 12 partially overlapped PCR fragments (8–16 kb). Amplicons were purified and eluted by electrophoresis with low-melting agarose (SeaPlaque Agarose, LONZA) and subsequently used for hydroshearing, cloning, sequencing (ABI PRISM 3700, Applied Biosystems), and assembling. Final sequence lengths were more than 8× coverage of the cpDNAs.
Gene Annotation
The obtained cpDNA sequences of Pinaceous species were annotated by use of Dual Organellar GenoMe Annotator (Wyman et al. 2004). For genes with low sequence identity, manual annotation was performed. We first identified the positions of start and stop codons and then translated the genes into putative amino acids by standard/bacterial code.
Structural Comparison of CpDNAs
We used the program Mulan (Ovcharenko et al. 2005), available on the Web site at http://mulan.dcode.org/, to visualize gene order conservation (dot-plot analyses and dynamic conservation profiles) between the Pinaceae representatives Cryptomeria and Cycas taitungensis. Mulan comparative analyses involved threaded block alignment and identified evolutionarily conserved sequences at default value (>70% identity and >100 bp).
Phylogenetic Analysis
We used 49 plastid protein-coding genes from 19 gymnosperms (supplementary table 3, Supplementary Material online) in the present study. Alignments were performed with the ClustalW method implemented in MEGA (version 4.0, Tamura et al. 2007; Kumar et al. 2008) with manual inspection. The aligned sequences were concatenated and then used for reconstructing the Pinaceae phylogeny. Li and Graur (1991) recommended that the use of more than one outgroup generally improves the estimate of tree topology. Both morphological and molecular studies of the conifers consistently supported that living conifers are monophyletic (Hart 1987; Raubeson and Jansen 1992b; Chaw et al. 1997), and Pinaceae is sister to the remaining conifer families as a whole (Hart 1987; Chaw et al. 1997; Stefanovic et al. 1998). Therefore, we included sequences from 1 Cupressaceae (C. japonica) (Hirao et al. 2008), 2 cycads (Cycas micronesica [Jansen et al. 2007] and C. taitungensis [Wu et al. 2007]), and 1 Ginkgo (G. biloba [Jansen et al. 2007]) to serve as outgroups. Maximum likelihood (ML) analyses, adopting the best-fit sequence evolution model selected by ModelTest (version 3.7; Posada and Buckley 2004) with the Akaike Information Criterion (AIC), were performed for the 49-gene combined data set. ML searches were conducted with GARLI (version 0.96b8, www.bio.utexas.edu/faculty/antisense/garli/Garli.html), which implements a genetic algorithm to perform rapid heuristic ML searches. PAUP* (Swofford 2003) was used to calculate the scores of ML trees from GARLI searches. One thousand bootstrap replicates were subsequently used to estimate ML branch support values. Bayesian phylogenetic analysis were performed using MrBayes (version 3.1.2; Ronquist and Huelsenbeck 2003) with sequence evolution model selected by ModelTest using AIC. The Markov chain Monte Carlo (MCMC) searches were started from a random tree and run for 2,000,000 generations, with topologies sampled every 100 generations. The values of -lnL reached a plateau before the first 2,000 trees in every analysis. The first 5,000 (corresponds to 25% of our samples) trees were discarded as burn-in (as suggested by the manual of MrBayes), and the remaining trees were used to construct the 50% majority-rule consensus tree and for inferring Bayesian posterior probabilities of nodal supports.
Testing Alternative Hypotheses
To assess the probability of alternative relationships among Cathaya, Cedrus, and four Pinaceous subfamilies, different hypothesized topologies were compared with the obtained unconstraint optimal phylogenies. Harmonic means (H) were obtained for unconstraint and constraint Bayesian phylogenetic analyses with use of MrBayes (version 3.1.2; Ronquist and Huelsenbeck 2003). The molecular models and MCMC searches for the constraint analyses were the same as those for the unconstraint analyses in the phylogenetic analyses. Twice the deviation of H between constraint and unconstraint analyses was used for consulting the Bayes factor criteria of significance (Bayes factor = 2δH; Kass and Raftery 1995). AU tests were performed with use of CONSEL (version 0.1i; Shimodaira and Hasegawa 2001). Alternative topologies (including the best ML tree) were tested, holding all other relationships constant to those found in the best GARLI ML tree. Likelihood values for these topologies were estimated by PAUP* under the general time reversible (GTR) + I + Γ model.
Molecular Dating
A likelihood ratio test of nucleotide substitution rate constancy across lineages indicated that our data rejected a constant molecular clock model (P = 4.06 × 10−20). Divergence times were therefore estimated under a relaxed molecular clock model by a penalized likelihood method (Sanderson 2002) implemented in r8s (Sanderson 2003). The smoothing parameter (λ) was determined by cross-validation. The ML topology for the 49-gene combined data set was used for the estimation. Deviations of divergence times were estimated by a nonparametric bootstrapping method (Baldwin and Sanderson 1998; Sanderson and Doyle 2001). Bootstrapping results were used for repeating the dating procedure 100 times, generating 100 topologically identical trees by use of SEQBOOT in PHYLIP (Felsenstein 2005).
Results and Discussion
Evolution of CpDNAs in Pinaceae
Genomic Structures of Ca. argyrophylla and Ce. deodara.
The complete cpDNAs of Ca. argyrophylla and Ce. deodara (DNA Data Bank of Japan [DDBJ] accession numbers AB547400 and AB480043, respectively) are circular molecules of 107,122 and 119,298 bp (supplementary fig. 1, Supplementary Material online), respectively. As compared with the four reference species (i.e., two Cycas spp., G. biloba, and Cr. japonica—a conifer), the two studied species have a pair of extremely reduced IRs (429 and 236 bp, respectively) and a common loss of all 11 ndh genes, similar to the elucidated cpDNAs of Keteleeria davidiana and Pinus (table 1). However, the corresponding IR region in cpDNA of Cryptomeria has even more reduced to 114 bp and retains only the gene, trnI. The sizes of the large single copy (LSC) and small single copy (SSC) are 64,197 and 42,067 bp, respectively, for Cathaya and 65,052 and 53,775 bp for Cedrus, respectively. Of note, our Ce. deodara is 1,226 bp longer than the published one (Parks et al. 2009), and the size difference is due to length variations in their noncoding regions. The LSC regions of Pinaceous genera are ∼25 kb shorter, on average, than that of Cycas (table 1), whereas the SSC regions of Pinaceae are at least ∼20 kb longer than that of Cycas because of the degradation of Pinaceae IRB and integration of the large ancestral IR fragment into SSC.
Table 1.
Comparisons of CpDNA Features among Cycas, Cryptomeria, and Two Pinaceae Subfamilies
| Features | Cycadaceae |
Cupressaceae |
Abitoideae |
Pinoideae |
||||
| Cycas taitungensis | Cryptomeria japonica | Cedrus deodara | Keteleeria davidiana | Cathaya argyrophylla | Pinus thunbergii | P. koraiensis | ||
| Size (bp) | 163,403 | 131,810 | 119,299 | 117,720 | 107,122 | 119,707 | 117,190 | |
| LSC length | 90,216 | NA | 65,052 | 64,648 | 64,197 | 65,696 | 64,563 | |
| SSC length | 23,039 | NA | 53,775 | 52,538 | 42,067 | 53,021 | 51,717 | |
| IR length | 25,074 | NA | 236 | 267 | 429 | 495 | 455 | |
| % AT content | 60.5 | 64.7 | 60.9 | 61.4 | 61.2 | 61.5 | 61.2 | |
| % Coding genes | 57.2 | 60.8 | 56.4 | 57.7 | 58.7 | 56.7 | 57.7 | |
| Total | 133 | 118 | 114 | 113 | 106 | 115 | 113 | |
| Number of protein-coding genes | 87 | 82 | 75 | 75 | 70 | 75 | 73 | |
| Number of duplicated genes | 15 | 2 | 6 | 5 | 3 | 6 | 5 | |
| Number of tRNA genes | 38 | 32 | 35 | 34 | 32 | 36 | 36 | |
| Number of rRNA genes | 8 | 4 | 4 | 4 | 4 | 4 | 4 | |
| Number of genes with introns | 20 | 17 | 14 | 14 | 13 | 14 | 14 | |
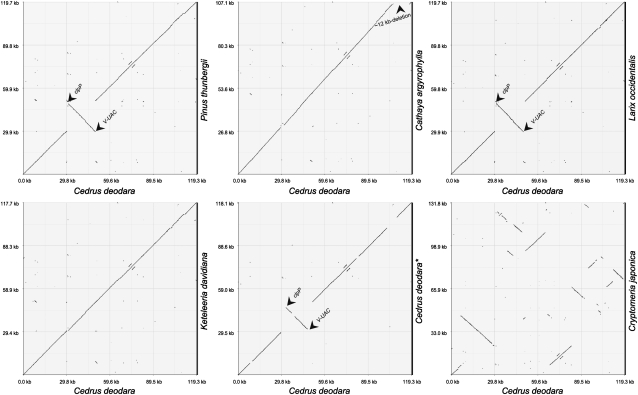
The small size and low gene content in Cathaya cpDNA are due to a ∼12 kb-deletion in its SSC region (fig. 2, supplementary fig. 1, Supplementary Material online), which corresponds to the region with five genes—ycf2, trnL-CAA, rps7, 3′-rps12, and trnV-GAC —in Cedrus cpDNA. Moreover, in Cathaya, its trnT-GGU (in SSC), psaM, and ycf12 (in LSC) are single rather than duplicated as in other elucidated Pinaceae cpDNAs, and its SSC region has a unique pseudogene, ψpsbB, located between trnE-UUC and trnY-GUA (supplementary fig. 1, Supplementary Material online).
FIG. 2.—
Comparison of cpDNA structures among Pinaceae representatives, Cryptomeria japonica (Cupressaceae), and 2 Cycas spp. (Cycadaceae). Dot-plot analyses of the cpDNAs of two Cedrus species (Parks et al. 2009 and this study), Cathaya, Larix, Pinus, and Keteleeria, and between the cpDNAs of Cedrus and Cryptomeria. Note that the cpDNA of Cathaya has a unique ∼12-kb deletion and that the cpDNAs of Cedrus, Larix, and Pinus have an inversion of 21 kb (from clpP to trnV-UAC; arrows). The gene order of Cryptomeria cpDNA differs greatly from those of Pinaceae cpDNAs.
A ψycf2 (∼200 bp) is generally present in the elucidated cpDNAs of Pinaceae except Cathaya. Wu et al. (2007), in their 2-step model, used this pseudogene to reconstruct the evolutionary history of IR-lost cpDNAs in Pinus. However, in Cathaya, another ycf2 residue (here designated ψycf2′) is located downstream of the ∼12-kb deletion and lies adjacent to the IRA (supplementary fig. 1, Supplementary Material online). An alignment of the trnH-GUG and ψycf2′ and their intergenic spacers of Cathaya and other available Pinaceous representatives revealed that ψycf2′ is highly homologous (identities >80%) to the 5′ regions of ycf2 (supplementary fig. 2, Supplementary Material online) in other Pinaceae, whereas the ψycf2 sequence annotated by Wu et al. (2007) is an internal residual sequence of ycf2.
The cpDNA of Cedrus contains 114 genes (75 protein-coding, 35 tRNA, and 4 rRNA genes), similar to those of K. davidiana, Pinus koraiensis, and P. thunbergii, whereas the cpDNA of Cathaya contains only 106 genes (including 70 protein-coding, 32 tRNA, and 4 rRNA genes) (table 1). The AT content of the only sequenced non-Pinaceae conifer cpDNA, Cr. japonica, is slightly higher (by ∼3% and 4%) than those of Pinaceae and Cycas cpDNAs (table 1). Moreover, the AT contents of the first, second, and third codon positions in the concatenated 49 common protein-coding genes are ∼1.4%, 2.0%, and 3.2% higher, respectively, in Cryptomeria than in Pinaceae, which suggests that Cryptomeria cpDNA has a biased usage of the AT-rich codons.
Our Two Reported Pinaceous CpDNAs Are Reliable.
The long-range PCR strategy was employed to completely cover a cpDNA without pure chloroplast extraction (Goremykin et al. 2003). Except for P. thunbergii (Wakasugi et al. 1994), the rest of the published Pinaceae cpDNAs were obtained by long PCR amplifications (Cronn et al. 2008; Parks et al. 2009;Wu et al. 2009; this study). The long PCR amplifications rely highly on PCR performance. We have designed many conserved primer pairs by aligning sequences from the published cpDNAs of seed plants. We increased the PCR performance to specifically yield a single band over 8 kb per PCR run. Longer amplicons (∼10 vs. ∼3.6 kb) and fewer segments (12 vs. 35 segments) per cpDNA than that used in previous studies (Cronn et al. 2008; Parks et al. 2009) greatly reduced the time required for PCR and for amplicon verifications. The reliability of the present two cpDNA sequences was evident in two aspects: 1) the results of annotation did not reveal many unexpected pseudogenes, so the amplified sequences were from cpDNAs rather than nuclear or mitochondrial DNAs and 2) underrepresented gaps could be closed by a single amplicon yielded from contig-specific primers.
Structural Rearrangement in the Pinaceae CpDNAs.
Our comparative analysis revealed that in terms of cpDNA organization, Pinaceae and Cycas are more similar to each other than to Cryptomeria, and the former two are unparallel to the latter (fig. 2; supplementary fig. 3, Supplementary Material online). These data suggest that Pinaceae is the basal-most family (see cited references in Introduction). Previously, the cpDNA of Pseudotsuga menziesii was reported to have a 42-kb inversion relative to Pinus radiata and nonconiferous plants (Strauss et al. 1988). Tsumura et al. (2000) also found that 5 and 2 species of Japanese Abies and Tsuga, respectively, have the same 42-kb cpDNA inversion polymorphism, and the authors defined the inversion as being between two short IRs (trnS-psaM-trnG and ψtrnG-psaM-trnS). Milligan et al. (1989) noted that the rearranged cpDNAs typical of those in several IR-lost legumes may be caused by the presence of numerous dispersed repeated sequences that facilitate recombination and rearrangement. Therefore, Tsumura et al. (2000) concluded that “probably this polymorphism has been maintained within populations and species in both genera because [the] mutation rate of the 42-kb inversion is high.” The 42-kb inversion is absent from Cathya and Ce. deodora but present in P. wilsoniana (Lin CP, Wu CS, Hsu CY, Chaw SM, unpublished data). Moreover, similar to the IR-lost legume cpDNAs, the inversions are associated with a short IR.
On comparing the cpDNA organizations between P. thunbergii and Japanese Abies and Tsuga, Tsumura et al. (2000) also uncovered a 21-kb inversion (between ycf12-trnT and trnE-trnG). We further detected its presence in the elucidated cpDNAs of Pinus spp. (Wakasugi et al. 1994; Noh et al. 2003; Cronn et al. 2008), Picea sitchensi (Cronn et al. 2008), Abies firma, Ce. deodora, and Larix occidentalis (Parks et al. 2009) but its absence in Keteleeria (Wu et al. 2009), Cathaya, and Ce. deodora (this study) (fig. 2; supplementary fig. 3, Supplementary Material online). Therefore, the 21-kb inversion is polymorphic among congeneric species and intraspecific populations (e.g., Ce. deodora). More intensive cpDNA samplings from all the Pinaceae genera and comprehensive comparisons of the repeated sequence types may help clarify the spectrum, mechanism, and evolution of these two large inversions in Pinaceae.
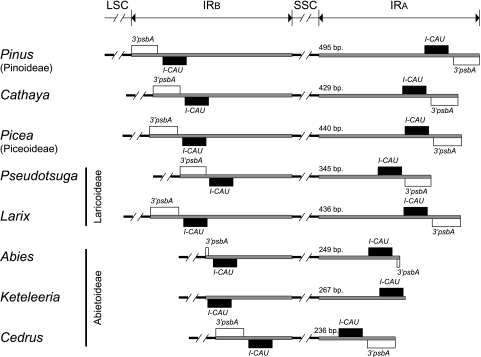
The Reduced IRs of Abietoideae Are Further Reduced.
In the cpDNAs of the 15 elucidated Pinaceae (except Keteleeria), the reduced IRs contain only the gene trnI-CAU and a 3′ fragment of psbA. The lengths of IRs vary from 236 to 495 bp (fig. 3). To investigate and comprehend the IR dynamics and evolution in the Pinaceae cpDNAs, we also determined the IR lengths in A. firma (Abietoideae), L. decidua (Laricoideae), P. morrisonicola (Piceoideae), and P. wilsoniana (Laricoideae). Figure 3 shows that IRs are shorter in the sampled Abietoideae than in other subfamilies. Remarkably, Abies and Keteleeria appear to have the IRs further shortened from the IR-LSC junction, whereas the reduced IRs of Cedrus are further reduced from the IR-SSC junction (fig. 3), which implies that Abies and Keteleeria are closer to each other than to Cedrus.
FIG. 3.—
Comparison of length dynamics of IRs among representative cpDNAs of Pinaceae species. Farjon’s (1990) subfamilies were adopted, with Cathaya excluded from the Laricoideae. Eight representative genera of the four subfamilies are presented, and IR regions are scaled. Note that the lengths of IRs are much shorter in Abietoideae than in other subfamilies. See text for further explanation.
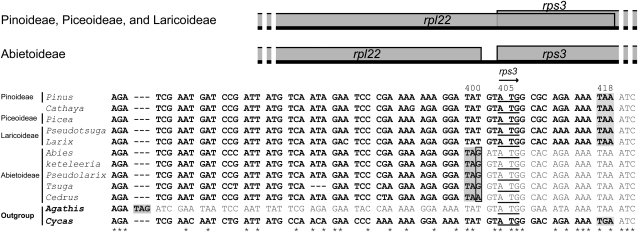
A Point Mutation Caused An Earlier Stop in the Coding Regions of Abietoideae rpl22.
We discovered that the 3′ region of rpl22 contains a six-codon difference among some elucidated Pinaceae cpDNAs. To gain a general picture of this gene evolution among the ten Pinaceous genera, we also sequenced this region from the remaining two genera, Tsuga (T. chinensis; DDBJ accession number AB547462) and Pseudolarix (P. kaempferi; DDBJ accession number AB547461). Cycas taitungensis (GenBank accession number NC_009618) and Agathis dammara (DDBJ accession number AB547460) were used as outgroups because this region of Cryptomeria is unalignable with those of Pinaceae. The length of rpl22 was shorter in the Abietoideae than in other Pinaceae species (fig. 4). As compared with the outgroup sequences, those of rpl22 of Abietoideae have a common point mutation (from T to G or A) at nucleotide position 402, which leads to an earlier stop of the gene. However, the 3′ ends of rpl22 in Larix, Pseudotsuga, Cathaya, Pinus, and Picea retain the Cycas feature of overlap with the gene rps3.
FIG. 4.—
Comparison of length dynamics of rpl22 among representative species of Pinaceae. Upper: a linear representation of two neighboring genes, rpl22 and 5′rps3. Note that coding sequences of the two genes overlap in Pinioideae, Cathya, Piceoideae, and Laricoideae. Lower: nucleotide sequence alignment of the 3′rpl22 and 5′rps3 region. The sequences of Cycas taitungensis (GenBank accession number NC_009618) and Agathis dammara (DDBJ accession number AB547460) were used as outgroups. The arrow indicates the transcription direction. Nucleotide sequences of rpl22 are in bold; stop codons are in shadow, and observed point mutations are boxed. The start codons of rps3 are underlined. Nucleotide positions are counted from the first codon position of Cycas rpl22. An asterisk at the bottom of the sequence alignment indicates conserved nucleotides.
Phylogenetic Analyses
CpDNA Data.
The compiled data set contained 49 concatenated protein-coding genes from 19 completely or partially elucidated cpDNAs of gymnosperms. Two Cycas species and Ginkgo were designated as outgroups, and Cr. japonica was an internal check. Excluding gaps and ambiguous sites, the final alignment was 29,691 bp, among which 8,141 bp are variable and 4,680 bp parsimony informative. Bayesian inference (BI) and single ML trees were obtained under the best-fit model (GTR + I + Γ) from the AIC implemented in ModelTest 3.7 (Posada and Buckley 2004).
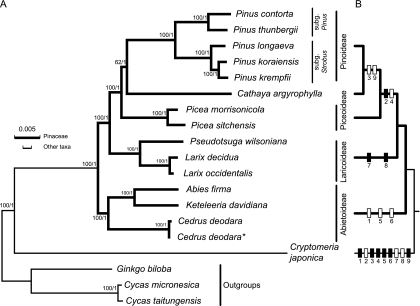
Cedrus Is Sister to Abies–Keteleeria Clade.
Figure 5A shows the two phylogenetic trees, reconstructed by two independent methods (ML and BI), with identical topologies. Crypotmeria was consistently revealed as an outgroup to the monophyletic Pinaceae genera and Abietoideae as the basal-most subfamily to the other three, with strong bootstrap support. Within the Abietoideae, Cedrus is clearly a sister group to the two sampled genera, Abies and Keteleeria. With Cedrus forced to be the outgroup of the other seven sampled Pinaceous genera, the constraint and optimal topologies showed statistically significant difference by the AU test and Bayes factor analysis (supplementary fig. 4, Supplementary Material online), which implies that Cedrus is not an outgroup to the rest of the Pinaceous genera. In the aligned rpl22 and rps3 gene cluster (fig. 4), all the five sampled Abietoideae genera have identical nonsense mutations at nucleotide position 402, so their rpl22 and rps3 are commonly separated by two nucleotides. Therefore, our cpDNA data strongly indicate that Cedrus and the other two representative genera of Abietoideae comprise a monophyletic group, and Cedrus is not the basal-most genus of Pinaceae. These results confirm the placement of Cedrus in Abietoideae by Price et al. (1987) and Gernandt et al. (2008) but contradict the view that the genus is a sister group to Larix–Pseudotsuga (Hart 1987), Abies (Frankis 1988; Farjon 1990), or the rest of the Pinaceae genera (Wang et al. 2000) (fig. 1).
FIG. 5.—
Chloroplast phylogenomics of Pinaceae genera. (A) A ML tree inferred from analysis of a data set containing 49 concatenated protein-coding genes in 19 cpDNA taxa by use of the GTR + I + Γ model. Only the ML tree is shown because the generated BI tree has identical topologies. Cycas and Ginkgo were used as the outgroups, and Cryptomeria was used as an internal check. The thick and thin scale bars at the upper left corner denote the respective branch lengths (substitutions per site) of Pinaceae and other taxa. Subfamilial names at the right were adopted from Fajon’s (1990) classification with modification. The two values at nodes represent the percentage of bootstrap supports (ML tree)/posterior probabilities (BI tree). (B) A simplified tree shows the distribution of nine informative indels in six introns (for the intron names and the indel locations, see supplementary fig. 5, Supplementary Material online) for respective subfamilies and the genus Cathaya. Insertions and deletions are indicated by solid and blank bars, respectively. See text for explanation.
Larix–Pseudotsuga Is a Distinct Clade and Clustered with Picea–Cathaya–Pinus.
The tree topology in figure 5A clearly suggests that the first split of Pinaceae occurs between Abietoideae and the rest of the sampled five genera, followed by Larix–Pseudotsuga clade (Laricoideae) and a clade containing Picea, Cathaya, and Pinus. This close sisterhood between Larix and Pseudotsuga has been previously noted on the basis of their resemblance in seed proteins (Prager et al. 1976; Price et al. 1987) and common possession of derived characters such as nonsaccate pollen, an extremely modified micropylar apparatus during pollination, fiber–sclerids in the bark, and similar asymmetric karyotypes (see review by Price 1989). Therefore, our cpDNA data and the aforementioned studies reject the view that the Larix–Pseudotsuga clade is a sister group to Cedrus (Hart 1987) or to Cathaya (Frankis 1988; Farjon 1990).
Cathaya Is Likely a Sister to Pinus.
Figure 5A depicts that Cathaya is embedded in a highly supported large clade containing Pinus (Pinoideae) and Picea (Piceoideae) and is a sister group to Pinus but only with moderate support. Although the AU test (P = 0.233) and Bayes factor analysis [2ln (BF) = 8.42] showed a nonsignificant difference between the unconstrained Cathaya–Pinus and constrained Cathaya–Picea topologies (supplementary fig. 4, Supplementary Material online), a number of other characters substantiating the sisterhood relationship between Cathya and Pinus have been observed before but have often been neglected. These characters are pollen morphology, the embryogeny and structure of mature embryos (Wang and Chen 1974; Hu et al. 1976), phytochemical data (He et al. 1981), and the ovule structure, as well as development of female gametophytes (Chen et al. 1995).
A sister relationship between Cathaya and Pseudotsuga (Frankis 1988) or between Cathaya and the Larix–Pseudotsuga clade (Farjon 1990) have never been supported in DNA-based studies (Wang et al. 2000; Gernandt et al. 2008) (fig. 1). Moreover, Cathaya was also claimed to be sister to Picea in previous studies using molecular markers (Wang et al. 2000; Gernandt et al. 2008), but the bootstrap supports were week. Here, our phylogenetic trees clearly indicate that Cathaya and Pinus form a clade with a strong support (PP = 1) in the BI tree and a moderate support (BP = 62%) in the ML tree (fig. 5A). These results agree well with the study based on reproductive characters mentioned above.
Distribution of Intron–Indels in Pinaceae Lineages in the Phylogenetic Context.
Because no informative indels were detected in the protein-coding genes, we examined the 14 intron-containing genes that are common to the Pinaceae cpDNAs (table 1) (supplementary table 5, Supplementary Material online). Notably, Cathaya cpDNA has uniquely lost the only intron within the 3′rps12, and Cryptomeria cpDNA has 17 intron-containing genes because it retains three additional ones (ndhA, ndhB, and rps16; Hirao et al. 2008). To evaluate the existence of informative indels that can be used for inferring relationships within Pinaceae lineages, the nucleotide sequences of all 14 introns were aligned, with those of Cryptomeria used as the outgroup. A total of 9 indels, including 6 deletions (2 of 3, 1 of 4, 1 of 5, 1 of 6, and 1 of 18 nt) and 3 insertions (2 of 4 and 1 of 5 nt) were detected in the 6 intron-containing genes: trnA-GUC, trnG-UCC, trnI-GAU, atpF, rpl2, and rpl16 (supplementary fig. 5, Supplementary Material online). Distributions of these indels on the cpDNA phylogeny were then plotted onto the cpDNA phylogenetic trees of Pinaceae (fig. 5B).
Foremost, monophyly of the three sampled Abietoideae genera is supported by their shared three indels (fig. 5B, indels 1, 5, and 6) in the introns of atpF, trnG-UCC, and trnI-GAU, respectively (supplementary fig. 5, Supplementary Material online). However, a unique 4- and a distinct 5-nt insertion (fig. 5B, indels 8 and 7) in the introns of trnI-GAU and rpl2, respectively, are exclusively present in the Larix–Pseudotsuga subclade but not Cathaya (supplementary fig. 5, Supplementary Material online), which indicates the close affinity between Larix and Pseudotsuga but their remoteness from Cathaya. Monophyly of the Cathaya–Pinus–Picea subclade is strongly substantiated by a specific 4-nt insertion and an 18-nt deletion in the introns of trnA-UGC and trnG-UCC, respectively (fig. 5B, indels 2 and 4; supplementary fig. 5, Supplementary Material online). A sisterhood relationship between Cathya and Pinus is evidenced by their two common multinucleotide deletions, one in the trnG-UCC (a 6-nt indel) and the other in rpl16 introns (a 3-nt indel) (fig. 5B, indels 3 and 9; supplementary fig. 5, Supplementary Material online).
Cryptomeria Has Accelerated Nucleotide Substitution Rates and the Pinus–Cathaya Clade Has Significantly Faster Rates than Do Other Pinaceous Genera
Our likelihood ratio test of the constancy of nucleotide substitution rate across lineages indicates that the present cpDNA data set rejects a constant molecular clock model (P = 4.06 × 10−20), and our phylogenetic trees (fig. 5A) show that Cryptomeria has an extremely longer branch than do the Pinaceae genera. Comparisons of the ML pairwise distances among Cryptomeria, Pinus, and Cycas (with Ginkgo used as the outgroup) revealed that Cryptomeria exhibits exceptional accelerated rates in most protein-coding genes (supplementary fig. 6, Supplementary Material online), especially the infA, petL, ribosomal-protein (rpl and rps), and RNA polymerase (rpo) gene families. We also used Tajima’s relative rate test (Tajima 1993) to compare the nucleotide substitution rates among Pinaceous genera using generic representatives that have median evolutionary rates (supplementary table 4, Supplementary Material online). Abietoideae and Picea species were similar in having relatively slower rates, but their rates differ from those of other Pinaceae, whereas Cathaya has a distinctively faster substitution rate than other subfamilies have (P < 0.05). Therefore, we used a relaxed molecular clock model for the molecular dating analysis described in the following section.
Phylogeographic Implications Based on Genomic Dating
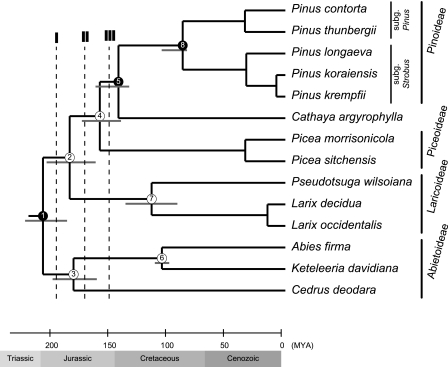
A correct phylogeny is a prerequisite for molecular dating. Hence, the ML tree in figure 5A was used to reestimate the divergence times for major splitting events of Pinaceae lineages. We used three reliable fossil records as calibration points: the emergence of Pinus (dated 135 Ma; Alvin 1960), the oldest Pinaceae-type cone (dated 225 Ma; Miller 1999), and subg. Strobus (dated 85 Ma; Meijer 2000). Combinations of different calibration points yielded six estimates of nodal ages (table 2). Only minor differences were obtained among nodal ages estimated from these three calibration dates but using the 135 Ma nodal age of Pinus resulted in slightly younger estimates for all nodes. By averaging the six estimates of nodal ages, Abietoideae appeared to branch off during Jurassic, ∼209.5 Ma, and Larix–Pseudotsuga split from Picea–Cathaya–Pinus ∼186.5 Ma. Subsequently, Picea separated from the Cathaya–Pinus subclade ∼160.4 Ma and then Cathaya and Pinus deviated from each other ∼144.5 Ma. Remarkably, Cedrus diverged from other Abietoideae genera ∼183.1 Ma, which is almost concurrent with the divergence time of the Larix–Pseudotsuga subclade from the Picea–Cathaya–Pinus subclade and suggests that Cedrus is ancient. Our phylogenomic analyses also provide novel implications for the historical biogeography of Pinaceae genera—namely, the origin of the ancestral Pinaceae was during Early Jurassic in Laurasia, followed by radiations into two lineages (i.e., Abietoideae and the rest of the five genera, including Larix, Pseudotsuga, Picea, Cathaya, and Pinus, during Mid-Jurassic; fig. 6); Cathaya and Keteleeria, specifically endemic to southern China and Taiwan, emerged during Early Cretaceous (144–100 Ma; fig. 6, node 5 and 6), when the first flowering plants were known to exist and began to diversify and spread (Soltis PS and Soltis DE 2004); and the extant two Pinus subgenera (Strobus and Pinus) completely diverged before Late Cretaceous (fig. 6, node 8). Our nodal age estimates are highly compatible with those obtained from the Pseudolarix–Tsuga calibration (Gernandt et al. 2008).
Table 2.
Ages of Pinaceae Nodes (Ma) Inferred from the Phylogenetic Tree in figure 4 Using the Penalized Likelihood Analyses
| Node | Age ± standard error (Ma) |
|||||
| RCa 1 | RC 2 | RC 3 | RUCa 1 | RUC 2 | RUC 3 | |
| Pinaceae root | 225.0b | 225.0b | 225.0b | 201.3 ± 0.7 | 192.2 ± 0.5 | 188.7 ± 0.5 |
| Larix–Pinus–Picea | 199.4 ± 0.6 | 206.4 ± 0.8 | 198.0 ± 0.8 | 184.2 ± 0.8 | 166.7 ± 0.5 | 164.0 ± 0.3 |
| Abietoideae | 188.0 ± 1.1 | 198.5 ± 0.9 | 201.2 ± 0.6 | 183.2 ± 0.6 | 164.8 ± 0.4 | 163.0 ± 0.6 |
| Cathaya–Pinus–Picea | 173.8 ± 0.9 | 175.9 ± 1.4 | 159.6 ± 1.4 | 168.5 ± 0.7 | 142.1 ± 0.2 | 142.4 ± 0.2 |
| Cathaya–Pinus | 164.1 ± 0.9 | 135.0c | 135.0c | 161.6 ± 0.7 | 135.0c | 135.0c |
| Abies–Keteleeria | 110.0 ± 1.1 | 108.4 ± 1.6 | 112.8 ± 1.4 | 104.8 ± 1.5 | 103.8 ± 0.5 | 100.4 ± 0.6 |
| Larix–Pseudotsuga | 123.4 ± 1.1 | 138.2 ± 2.3 | 127.2 ± 1.5 | 117.3 ± 1.6 | 93.4 ± 0.5 | 94.1 ± 0.5 |
| subg. Pinus + subg. Strobus | 85.0d | 106.9 ± 0.5 | 85.0d | 85.0d | 85.5 ± 0.0 | 85.0d |
“RC” and “RUC” represent root constrained and unconstrained, respectively.
Age-fixed node, an oldest Pinaceae-type cone, 225 Ma (Miller 1999).
Age-fixed node, the oldest fossil of Pinus, 135 Ma (Alvin 1960).
Age-fixed node, a wood fossil of subg. Strobus, 85 Ma (Meijer 2000).
FIG. 6.—
A chronogram illustrating divergence times of Pinaceae genera. Branch lengths of the tree are averages from all calibration strategies (table 2). Nodes fixed with fossil ages are shown in black circles. Maximum and minimum estimated ages are denoted by gray lines below nodes. The three dot lines, I, II, and III, are used as thresholds for subfamily delimitations.
Interestingly, diversification of Pinaceae genera was synchronized with the formation of continents, which began to take on their modern forms during the Cretaceous. A subsequent dispersal via the Bering land bridge between formerly isolated Asian and American continents during the Tertiary period might be responsible for the contemporary pan-north Hemisphere distribution of most of the Pinaceae genera. However, the existence of three endemic Pinaceae genera (Cathaya, Keteleeria, and Pseudolarix [not sampled in this study]) in southern China may suggest a southern China origin of the Pinaceae or a more heterogeneous habitat in that region, which provides distinct niches for evolution of these endemic genera.
Implication of Subfamilial Classifications
Price (1989) argued that recognition of two subfamilies (i.e., Abietoideae and Pinioideae, including Larix–Pseudotsuga, Picea, Cathya, and Pinus), corresponding to Van Tieghem’s (1891) two groups or three groups (i.e., Abietoideae, Laricoideae, and the monogeneric Pinioideae), seems to be the most reasonable alternatives and natural. However, Frankis (1988) and Farjon (1990) recognized four subfamilies—Abietoideae, Laricoideae (including Larix, Cathaya, and Pseudotsuga) and two monotypic subfamilies, Piceoideae and Pinoideae—on the basis of reproductive morphologies and chromosome numbers. Similar to Price (1989), Liston et al. (2003) preferred a more broadly circumscribed Pinoideae. The divergence pattern in our cpDNA phylogenetic tree (fig. 6) clearly suggests an unquestionable division of two subfamilies in Pinaceae (i.e., Abietoideae and the rest of the 5 genera [line I]). With the ML divergence between Picea and Pinus used as a threshold (line II), four groups (or subfamilies) should be recognized —Cedrus, non-Cedrus Abietoideae, Larix–Pseudotsuga, and Piceae–Cathaya–Pinus. If Picea is considered as comprising its own monogeneric subfamily (line III), then in Pinaceae five groups/subfamilies are proposed, and Cathaya should be grouped with Pinus. Most importantly, our views on the subfamilial classifications differ from those of previous studies in the ranking of Cedrus if more than two subfamilies are recognized. In other words, we consider Cedrus as an ancient and highly distinctive genus that could be considered as forming its own subfamily.
Conclusions
Structural comparisons of the organization of cpDNAs among eight sampled Pinaceous genera revealed that two large inversions (21 and 42 kb) frequently exist in congeneric species and intraspecific populations. Interestingly, distributions of these inversions have never been reported in other families of seed plants. More comprehensive samplings and comparisons of the repeated sequence types may help clarify the spectrum, mechanism, and evolution of these two inversions in Pinaceae. Our cpDNA-scale analyses greatly improve the resolutions of Pinaceae phylogeny and clearly place Cedrus within the sampled Abietoideae. These results are further corroborated by evidence from indel distributions in introns, reduction of IRs, an earlier stop of rpl22, and statistical topology tests. Therefore, the cpDNA data reject the Cedrus-basal hypothesis (Wang et al. 2000). In good agreement with previous embryonic comparative results (Wang and Chen 1974), our phylogenetic trees and indel distributions strongly suggest that Larix and Pseudotsuga form a monophytic clade, and Cathaya is closer to Pinus than to Picea or the Larix–Pseudotsuga group. Our age estimates indicate that the Late Mesozoic (or Cretaceous) and Laurasia were the respective time and space that the Pinaceae ancestor started diverging into the extant genera. The divergence time of Cedrus from the rest of Abietoideae is almost concurrent with that of the Larix–Pseudotsuga from Picea–Cathaya–Pinus clades. We conclude that two subfamilies (i.e., Abietoideae and Pinioideae, including Larix, Pseudotsuga, Picea, Cathaya, and Pinus) or, alternatively, five subfamilies (i.e., Cedrus, the rest of Abietoideae, Laricoideae, Picea, and Cathya–Pinus) appear to be the most reasonable for the subdivision of Pinaceae.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
This work was supported by research grants from the National Science Council, Taiwan (NSC972621B001003MY3) and the Biodiversity Research Center, Academia Sinica (to S.M.C.). We thank Yi-Ming Chen for the materials of Cathaya and Cedrus and Shu-Mei Liu, Shu-Jen Chou, and Mei-Jane Fang for the help with DNA shearing and sequencing. We are thankful to the two anonymous reviewers for their critical reading and valuable suggestions.
References
- Alvin K. Further conifers of the Pinaceae from the Wealden formation of Belgium. Inst R Sci Nat Belg Mém. 1960;146:1–39. [Google Scholar]
- Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Syst. 2002;33:707–740. [Google Scholar]
- Baldwin B, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance. Proc Natl Acad Sci U S A. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke HD. Sieve element plastids of Gymnospermae: their ultrastructure in relation to systematics. Plant Syst Evol. 1974;123:1–12. [Google Scholar]
- Chaw SM, Sung HM, Long H, Zharkikh A, Li WH. The phylogenetic positions of the conifer genera Amentotaxus, Phyllocladus, and Nageia inferred from 18S rRNA sequences. J Mol Evol. 1995;41:224–230. doi: 10.1007/BF00170676. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Zharkikh A, Sung HM, Leu TC, Li WH. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Mol Biol Evol. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- Chang CC, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chen ZK, Zhang JH, Zhou F. The ovule structure and development of female gametophyte in Cathata (Pinaceae) Cathaya. 1995;7:165–176. [Google Scholar]
- Chun WY, Kuang KZ. De genere Cathaya Chun et Kaung. Acta Bot Sin. 1962;10:245–246. [In Chinese with English abstract] [Google Scholar]
- Cronn R, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra PD. Embryogeny of gymnosperms and taxonomy—an assessment. In: Nair PKK, editor. Glimpses in plant research. Vol. 5. New Delhi (India): Vikas Publishing House; 1980. pp. 114–128. [Google Scholar]
- Doyle JJ. Developmental lines in pollination mechanisms in the Coniferales. Sci Proc Roy Dublin Soc. 1945;24:43–62. [Google Scholar]
- Farjon A. Pinaceae. 1990. Konigstein (Germany): Koeltz Scientific Book. [Google Scholar]
- Felsenstein J. Distributed by the author. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle (WA): Department of Genome Sciences, University of Washington. [Google Scholar]
- Florin R. Untersuchungen zur Stammesegeschichte der Coniferales und Cordaitales. Kgl Svensk Vetensk Akad Handl. 1931;10:3–588. [Google Scholar]
- Florin R. The distribution of conifer and taxad genera in time and space. Acta Horti Berg. 1963;20:121–312. [Google Scholar]
- Frankis MP. Generic inter-relationships in Pinaceae. Notes R Bot Gard Edinb. 1988;45:527–548. [Google Scholar]
- Geiger H, Quinn C. Biflavonoids. In: Harborne JB, editor. The flavonoids. London: Chapman and Hall; 1975. pp. 692–742. [Google Scholar]
- Gernandt DS, et al. Use of simultaneous analyses to guide fossil-based calibrations of Pinaceae phylogeny. Int J Plant Sci. 2008;169:1086–1099. [Google Scholar]
- Goremykin V, Hirsch-Ernst KI, Wo S, Hellwig FH. The chloroplast genome of the ‘‘basal’’ angiosperm Calycanthus fertilis—structural and phylogenetic analyses. Plant Syst Evol. 2003;242:119–135. [Google Scholar]
- Gugerli F, et al. The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol Phylogenet Evol. 2001;21:167–175. doi: 10.1006/mpev.2001.1004. [DOI] [PubMed] [Google Scholar]
- Hart JA. A cladistic analysis of conifers: preliminary results. J Arn Arb. 1987;68:269–307. [Google Scholar]
- He GF, Ma ZW, Yin WF, Cheng ML. On serratene components in relation to the systematic position of Cathaya (Pinaceae) Acta Phytotaxon Sin. 1981;19:440–443. [In Chinese with English abstract] [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YS, Napp-Zinn K, Winne D. Comparative anatomy of seed-scales of female cones of Pinaceae. Bot Jahrb Syst. 1989;111(1):63–85. [Google Scholar]
- Hu YS, Wang FH. Anatomical studies of Cathaya (Pinaceae) Am J Bot. 1984;71:727–735. [Google Scholar]
- Hu YS, Wang FH, Chang YC. On the comparative morphology and systematic position of Cathaya (Pinaceae) Acta Phytotaxon Sin. 1976;14:73–78. [In Chinese with English abstract] [Google Scholar]
- Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Mol Phylogenet Evol. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey EC. The comparative anatomy and phylogeny of the Coniferales. Part 2. The Abietineae. Mem Boston Soc Nat Hist. 1905;6:l–37. [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Krüssmann G. Manual of Cultivated Conifers. Portland (OR): Timber Press; 1985. p. 361. [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- LePage BA. The evolution, biogeography and palaeoecology of the Pinaceae based on fossil and extant representatives. Acta Hortic. 2003;615:29–52. [Google Scholar]
- LePage BA, Basinger JF. Evolutionary history of the genus Pseudolarix Gordon (Pinaceae) Int J Plant Sci. 1995;156:910–950. [Google Scholar]
- Li WH, Graur D. Fundamentals of molecular evolution. Sunderland (MA): Sinauer Associates; 1991. [Google Scholar]
- Liston A, Gernandt DS, Vining TF, Campbell CS, Piñero D. Molecular phylogeny of Pinaceae and Pinus. Acta Hortic. 2003;615:107–114. [Google Scholar]
- Liu YS, Basinger JF. Fossil Cathaya (Pinaceae) pollen from the Canadian high arctic. Int J Plant Sci. 2000;161:829–847. [Google Scholar]
- Lockhart PJ, Howe CJ, Barbrook AC, Larkum AWD, Penny D. Spectral analysis, systematic bias, and the evolution of chloroplasts. Mol Biol Evol. 1999;16:573–576. [Google Scholar]
- Meijer JJF. Fossil woods from the Late Cretaceous Aachen formation. Rev Palaeobot Palynol. 2000;112:297–336. doi: 10.1016/s0034-6667(00)00007-5. [DOI] [PubMed] [Google Scholar]
- Melchior H, Werdermann E. A. Englers Syllabus der PflanzenfamilienI. Allg Teil Bakterien bis Gymnospermen. Vol. 12. Berlin (Germany): 1954. [Google Scholar]
- Miller CN. Early evolution in the Pinaceae. Rev Palaeobot Palynol. 1976;21:101–117. [Google Scholar]
- Miller CN. Implications of fossil conifers for the phylogenetic relationships of living families. Bot Rev. 1999;65:239–277. [Google Scholar]
- Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structure reorganization in subclover chloroplast DNA. Mol Evol Biol. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- Noh EW, et al. Complete nucleotide sequence of Pinus koraiensis. 2003 Direct Submission to GenBank, Accession No. NC_004677. [Google Scholar]
- Ovcharenko GL, et al. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilger R. Coniferae. In: Engler A, Prantl K, editors. Die natũrlichen Pflanzenfamilien. Leipzig (Germany): Englmann; 1926. Vol.13. p. 121–166. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Prager EM, Fowler DP, Wilson AC. Rates of evolution in conifers (Pinaceae) Evolution. 1976;30:637–649. doi: 10.1111/j.1558-5646.1976.tb00945.x. [DOI] [PubMed] [Google Scholar]
- Price RA. The genera of Pinaceae in the southeastern United States. J Arnold Arbor Harv Univ. 1989;70:247–305. [Google Scholar]
- Price RA, Olsen-Stojkovich J, Lowenstein JM. Relationships among the genera of Pinaceae: an immunological comparison. Syst Bot. 1987;12:91–97. [Google Scholar]
- Price RA, et al. Familial relationships of the conifers from rbcL sequence data. Am J Bot. 1993;80:172. [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992a;255:1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. A rare chloroplast DNA structural mutation is shared by all conifers. Biochem Syst Ecol. 1992b;20:17–24. [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry RI, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Wallingford (UK): CABI; 2005. pp. 45–68. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Doyle JA. Sources of error and confidence intervals in estimating the age of angiosperms from rbcL and 18S rDNA data. Am J Bot. 2001;88:1499–1516. [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Smith AB, Peterson KJ. Dating the time of origin of major clades: molecular clocks and the fossil record. Annu Rev Earth Planet Sci. 2002;30:65–88. [Google Scholar]
- Soltis PS, Soltis DE. The origin and diversification of angiosperms. Am J Bot. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Jager M, Deutsch J, Broutin J, Masselot M. Phylogenetic relationships of conifers inferred from partial 28S rRNA gene sequences. Am J Bot. 1998;85:688–697. [PubMed] [Google Scholar]
- Stewart WN, Rothwell GW. Paleobotany and the evolution of plants. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Strauss SH, Palmer JD, Howe GT, Doerksen AH. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc Natl Acad Sci U S A. 1988;85:3898–3902. doi: 10.1073/pnas.85.11.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland (MA): Sinauer; 2003. [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsumura Y, Suyama Y, Yoshimura K. Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol Biol Evol. 2000;17:1302–1312. doi: 10.1093/oxfordjournals.molbev.a026414. [DOI] [PubMed] [Google Scholar]
- Van Tieghem P. Structure et affinites des Abies et des genres les plus voisins. Bull Soc Bot Fr. 1891;38:406–415. [Google Scholar]
- Vierhapper F. Entwurf eines neuen Systemes der Coniferen. Abh KK Zool-Bot Ges Wien. 1910;5(4):1–56. [Google Scholar]
- Vining TF, Campbell CS. Phylogenetic signal in sequence repeats within nuclear ribosomal DNA internal transcribed spacer 1 in Tsuga. Am J Bot. 1997;84(Suppl):241. [Google Scholar]
- Wakasugi T, et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci U S A. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FH, Chen TK. The embryogeny of Cathaya (Pinaceae) Acta Bot Sin. 1974;16:64–69. [In Chinese with English abstract] [Google Scholar]
- Wang XQ, Han Y, Hong DY. A molecular systematic study of Cathaya, a relic genus of the Pinaceae in China. Plant Syst Evol. 1998;213:165–172. [Google Scholar]
- Wang XQ, Tank DC, Sang T. Phylogeny and divergence times in Pinaceae: evidence from three genomes. Mol Biol Evol. 2000;17:773–781. doi: 10.1093/oxfordjournals.molbev.a026356. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 2009;52:115–124. doi: 10.1016/j.ympev.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Liu SM, Chaw SM. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]