Abstract
Population-level differences in the number of copies of genes resulting from gene duplication and loss have recently been recognized as an important source of variation in eukaryotes. However, except for a small number of cases, the phenotypic effects of this variation are unknown. Data from the Saccharomyces Genome Resequencing Project permit the study of duplication in genome sequences from a set of individuals within the same population. These sequences can be correlated with available information on the environments from which these yeast strains were isolated. We find that yeast show an abundance of duplicate genes that are lineage specific, leading to a large degree of variation in gene content between individual strains. There is a detectable bias for specific functions, indicating that selection is acting to preferentially retain certain duplicates. Most strikingly, we find that sets of over- and underrepresented duplicates correlate with the environment from which they were isolated. Together, these observations indicate that gene duplication can give rise to substantial phenotypic differences within populations that in turn can offer a shortcut to evolutionary adaptation.
Keywords: gene duplication, gene retention, evolutionary adaptation, genomics
Introduction
Gene duplication has long been recognized as an important source of new genes (Ohno 1970). Duplicated genes contribute to functional innovation, with the redundant duplicate often acquiring new functions that derive from an existing secondary function (Conant and Wolfe 2008). The importance of gene duplication is indicated by its frequency: for eukaryotes, it has been estimated that fully 50% of genes are expected to duplicate at least once in timescales of the order of 35–350 My (Lynch and Conery 2000). As opposed to de novo gene evolution, duplication is by far the more frequent mechanism used by evolution to generate novel genes.
Duplications can arise on a range of scales, from whole genomes to small motifs. Within the yeast Saccharomyces cerevisiae, a whole-genome duplication is thought to have occurred ≈100 Ma (Wolfe and Shields 1997; Kellis et al. 2004). Duplication of large segments of the genome is also common in yeasts, giving rise to new genes (Dujon et al. 2004; Koszul et al. 2004); in Candida glabrata, segmental duplications have given rise to entire new chromosomes (Polakova et al. 2009). The new genes generated by segmental duplications are found in tandem sets and are less diverged than those dispersed through the genome (Dujon et al. 2004). Many new genes are found in the subtelomeric regions (Horowitz et al. 1984). These regions are repeat rich (Pryde et al. 1997) and have high levels of recombination (Horowitz et al. 1984; Barton et al. 2008), leading to rapid turnover of genes and expansion of gene families (Louis 1995; Brown et al. 2010).
Duplicated genes can have one of a number of fates. Duplicates can be retained with the same function if increased dosage gives selective advantage (Spofford 1969; Otto and Whitton 2000; Hakes et al. 2007). If a duplicated gene is entirely redundant, it may no longer be subject to purifying selection. In this case, one member of the pair can degenerate and become a pseudogene (known as pseudogenization or nonfunctionalization). A third possibility is that sequences of duplicate pairs can diverge functionally, with one copy evolving a novel function (neofunctionalization) or, alternatively, the ancestral function being partitioned between the paralogs (subfunctionalization; Force et al. 1999; Lynch et al. 2001).
In addition to high rates of duplication, rates of gene loss are high (Kellis et al. 2004; Scannell et al. 2006). Specific subsets of genes are more likely to be retained, including those 1) with higher expression levels (Seoighe and Wolfe 1999); 2) involved in environmental responses (Kondrashov et al. 2002); 3) present in multiple evolutionary divergent lineages (Gu et al. 2002) 4) that are functionally constrained (Davis and Petrov 2004) and 5) derived from whole-genome duplication (Hakes et al. 2007). Any phenotypic changes associated with duplicate gene content may lead to selection for certain types of duplicates (Guan et al. 2007; Hakes et al. 2007). For these reasons, we hypothesize that the environment will alter the complement of duplicated genes retained in the genome.
Until recently, it has not been possible to correlate duplication events directly with environment within a population on a large scale, making the testing of this hypothesis difficult. However, the data from the Saccharomyces Genome Resequencing Project (Liti et al. 2009) gives us the first complete set of genomic data for a population where the natural environment for each of the strains is known. Strains of both S. cerevisiae and S. paradoxus selected for resequencing were isolated from a range of environments, including clinical isolates, laboratory isolates, strains used in fermentation and baking, and several isolated from the wild, including a number from oak bark.
Analyzing these population sequence data, we find that there are large numbers of duplicates in both S. cerevisiae and S. paradoxus, including many that occur in only a single lineage. These related gene sets show substantial evolutionary divergence, with those found in a single lineage displaying a marked increase in sequence evolution. We also observe clear evidence of selection and adaptation to the environment directly linked to gene duplication. In particular, we show that clustering on the basis of the function of genes present in duplicate can be used to reconstruct clusters of environments from which these yeast strains were isolated. Moreover, there are larger differences in duplicate gene complement between S. cerevisiae strains than S. paradoxus, which corresponds to the wider range of environments from which they were isolated. We conclude that duplicates are a key source of variation, enabling genome-wide adaptation to the environment of yeast populations.
Materials and Methods
Identifying Genes
Genome sequence data for S. cerevisiae and S. paradoxus assembled by parallel contig assembly program (PCAP) (Huang et al. 2003) were downloaded from the Sanger FTP site. Two different assemblies are provided by Liti et al. (2009): those assembled by the parallel-alignment assembling (PALAS) method and those directly assembled with PCAP. The PCAP assembled data were chosen above the PALAS assembled data because the PCAP assemblies maintain the inherent variation between strains, whereas the PALAS assemblies lose some variation by using reference genomes as templates for assembly (Liti et al. 2009). AUGUSTUS (Stanke and Waack 2003), an ab initio gene prediction program, was used to generate coding and protein sequences from the assembled sequence data. AUGUSTUS was run using the following parameters: training data S. cerevisiae strain S288c, gene prediction on both strands, and predict gene using a complete gene model. The predicted genes were annotated with the known S. cerevisiae open reading frames (ORFs) using BlastN (Altschul et al. 1990) with an E value threshold of 1 × 10−8.
Duplicate Gene Analysis
Duplicates were identified using GenomeHistory (Conant and Wagner 2002) with the following parameters: Blast E value threshold 1 × 10−8, minimum ORF translation length 100 nt, and minimum aligned residues 100 nt. An identity threshold of 40% was used to decrease the occurrence of potential false-positive paralogy assignments (Hakes et al. 2007). Potential sets of lineage-specific duplicates (LSDs) were identified from the duplicate and genomic sequence data. An LSD must be identified as a duplicate pair in one strain and be absent in all others. In addition, if any other strain contains both genes of the LSD pair, regardless of their annotation as a duplicate pair, the LSD is treated as a false positive and removed. Ka and Ks values (number of nonsynonymous and synonymous substitutions per site, respectively) were taken from GenomeHistory. For this analysis, one anomalous duplicate pair was identified and removed from the data set. The pair in S. paradoxus strain A4 had Ks and Ka values of 1,039.62 and 45.59, respectively, which were extremely high compared with the species averages (Ks = 5.73, Ka = 0.32).
Detecting Asymmetric Divergence in Duplicate Pairs
Asymmetrically diverging pairs were identified by aligning coding sequences of the duplicate pair with that of its nonduplicated Kluyveromyces waltii ortholog as determined by Kellis et al. (2004). The alignment was performed using MUSCLE (Edgar 2004) and a maximum likelihood phylogenetic tree inferred using PhyML, with the Hasegawa–Kishino–Yano 85 substitution model (Guindon and Gascuel 2003). The K. waltii sequence was used as an outgroup. The ratio of the S. cerevisiae branch lengths was determined for each duplicate pair in order to identify sequences that have asymmetrically diverged. To control for missing data, this analysis was limited to sequences whose length is at least 65% of the reference sequence. For S. cerevisiae, we analyzed 8,584 duplicate pairs and 7,451 duplicate pairs for S. paradoxus.
Genomic Distribution of Duplicate Genes
Each of the 16 S. cerevisiae and S. paradoxus chromosomes were split into 70-kb bins. Duplicates were assigned to these bins using the reference strains S288c (S. cerevisiae) and CBS432 (S. paradoxus) with Blast-like alignment tool (Kent 2002). To generate a random distribution of genes, 1,000 Monte Carlo simulations were used. In each simulation, duplicate status was randomly assigned to the sequenced genes for each strain. Only sequenced genes could be assigned duplicate status to control for the missing data in some strains. The number of assigned duplicates was equal to the number of identified duplicates for each strain. This generates a random distribution of duplicate genes controlling for bias in the position of genes on each chromosome and the incomplete genome sequences of some strains. After 1,000 simulations, the average number of assigned duplicates in each bin was calculated and tallied for each species. The random distributions were then compared with the actual distributions of duplicate genes.
Gene Ontology Analysis
Lists of overrepresented Gene Ontology (GO) terms (Ashburner et al. 2000) were determined for all the duplicate genes and the LSDs in each strain. The hypergeometric distribution was used to calculate P values for the number of genes associated with each GO term. Duplicates were considered as a sample from all sequenced genes from each strain to account for incomplete genome sequences. The P values were corrected for multiple testing using a Monte Carlo approach (Hakes et al. 2007).
Inferring a Phenetic Tree
To determine whether strains from similar environments retain similar types of genes in duplicate, the over- and underrepresented “Biological Process” GO terms for each strain's duplicates and LSDs were compared using a semantic distance measure (Jiang and Conrath 1997), which has previously been applied to GO terms (Hakes et al. 2007). Briefly, the semantic distance d(t1, t2) between two terms t1 and t2 is given by
where p(t) is the fraction of all sequenced genes associated with that term and S(t1, t2) is the set of all parent terms shared by t1 and t2. We then define the semantic distance D(a, b) between two strains a and b with sets of over- and underrepresented terms A and B as
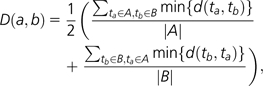 |
where |A| and |B| are the number of terms in the sets A and B, respectively. By incorporating the number of terms in each set A and B, the semantic distance measure provides a control for the differing number of over- and underrepresented terms for each strain.
The semantic distance between all pairs of strains was used to make a distance matrix, and neighbor joining was used to produce a phenetic tree. Only 21 S. cerevisiae and 18 S. paradoxus strains are included in the phenetic trees as these are the only strains showing over- and underrepresented “Biological Process” GO terms. To test whether any clusters in the tree were statistically significant, 1,000 Monte Carlo simulations were used to randomly assign strains to the phenetic tree. Each simulation was checked against the neighbor joining tree to see whether any of the original clusters were found. Therefore, the P values on the phenetic tree represent the probability of the clusters appearing by random chance.
Results
Identified Duplicates
We define LSDs as those genes that are found in duplicate in only one strain (lineage). We define “population duplicates” as those genes that are found in duplicate within only a subset of members of the population, which may be shared by two or more lineages. Were we to map these two sets to a phylogenetic tree, the LSDs would map to the terminal branches, whereas the population duplicates would map to internal nodes. These definitions are based only on the observation of duplicate sets and do not indicate where in the phylogenetic tree a duplication event has taken place. For example, duplicate genes may appear in only a single lineage due to an ancestral duplication, followed by lineage-specific loss.
Despite coverage of the genome sequences differing between strains, we are able to identify substantial evidence of gene duplication. Concerning coverage, the genome sequences have a coverage of 1- to 4-fold (Liti et al. 2009), and as a consequence, some genome sequences are incomplete and have a high number of contigs. These assembly problems limit the number of genes (additional data file 1, Supplementary Material online) and the number of duplicates identified (tables 1 and 2). Importantly, for each species there are several strains with >5,000 genes identified. Saccharomyces cerevisiae strains with >5,000 predicted genes have, on average, 21.79% of genes (1,220 ± 180) identified as duplicates. This figure is 9.17% (328 ± 98) and 6.79% (162 ± 59) for the <5,000 and >3,000, and <3,000 predicted gene categories, respectively.
Table 1.
Number of Predicted Genes, Duplicates, and LSDs for Saccharomyces cerevisiae
| Strain | Contigs | Predicted Genes | Duplicate Genes | LSDs |
|
| Inc. Trans.a | No Trans.b | ||||
| DBVPG6765 | 2,879 | 5,770 | 1,195 | 84 | 81 |
| RM11_1A | 384 | 5,501 | 1,353 | 86 | 77 |
| REF (S288c) | 18 | 5,464 | 1,356 | 149 | 100 |
| SK1 | 2,827 | 5,797 | 1,219 | 83 | 72 |
| W303 | 3,853 | 5,296 | 837 | 34 | 20 |
| Y55 | 2,751 | 5,875 | 1,282 | 94 | 92 |
| YJM789 | 207 | 5,437 | 1,292 | 29 | 15 |
| DBVPG1373 | 3,656 | 3,974 | 378 | 6 | 4 |
| DBVPG1788 | 3,617 | 3,621 | 340 | 2 | 2 |
| DBVPG6044 | 3,955 | 4,276 | 457 | 2 | 2 |
| L_1374 | 3,150 | 3,118 | 256 | 6 | 6 |
| L_1528 | 3,439 | 3,380 | 302 | 12 | 10 |
| S288c | 3,408 | 3,516 | 367 | 9 | 3 |
| UWOPS05_227_2 | 3,021 | 3,177 | 235 | 0 | 0 |
| YIIc17_E5 | 2,955 | 3,010 | 235 | 6 | 2 |
| YJM975 | 3,061 | 3,174 | 235 | 2 | 2 |
| YJM978 | 2,975 | 3,084 | 250 | 4 | 4 |
| YPS128 | 3,696 | 4,097 | 415 | 11 | 6 |
| YPS606 | 4,033 | 4,615 | 563 | 9 | 4 |
| YS4 | 3,021 | 3,119 | 280 | 26 | 24 |
| YS9 | 3,029 | 3,090 | 287 | 46 | 40 |
| 273614N | 2,591 | 2,485 | 159 | 2 | 2 |
| 322134S | 2,727 | 2,548 | 196 | 13 | 8 |
| 378604X | 3,014 | 2,998 | 255 | 14 | 10 |
| BC187 | 2,309 | 2,044 | 92 | 2 | 2 |
| DBVPG1106 | 2,013 | 1,802 | 86 | 4 | 2 |
| DBVPG1853 | 3,026 | 2,879 | 198 | 2 | 0 |
| DBVPG6040 | 2,487 | 2,384 | 210 | 16 | 14 |
| K11 | 2,657 | 2,629 | 163 | 2 | 2 |
| NCYC110 | 2,395 | 2,277 | 145 | 0 | 0 |
| NCYC361 | 1,873 | 1,301 | 83 | 16 | 14 |
| UWOPS03_461_4 | 2,927 | 2,969 | 223 | 4 | 4 |
| UWOPS05_217_3 | 2,454 | 2,591 | 241 | 8 | 8 |
| UWOPS83_787_3 | 2,713 | 2,721 | 206 | 19 | 16 |
| UWOPS87_2421 | 2,796 | 2,816 | 221 | 6 | 6 |
| Y12 | 2,584 | 2,472 | 147 | 6 | 6 |
| Y9 | 2,324 | 2,274 | 130 | 0 | 0 |
| YJM981 | 1,435 | 1,238 | 62 | 2 | 2 |
| YS2 | 2,294 | 1,639 | 112 | 8 | 6 |
Number of LSDs including transposable genes.
Number of LSDs excluding transposable genes.
Table 2.
Number of Predicted Genes, Duplicates, and LSDs for Saccharomyces paradoxus
| Strain | Contigs | Predicted Genes | Duplicate Genes | LSDs |
|
| Inc. Trans.a | No Trans.b | ||||
| CBS432 | 1,773 | 5,409 | 1,140 | 50 | 50 |
| REF (CBS432) | 17 | 5,348 | 1,269 | 31 | 31 |
| CBS5829 | 3,439 | 5,656 | 1,095 | 54 | 54 |
| N_17 | 3,606 | 5,797 | 1,163 | 116 | 114 |
| N_45 | 3,005 | 5,907 | 1,257 | 125 | 123 |
| UWOPS91_917_1 | 4,589 | 5,172 | 1,139 | 543 | 541 |
| A12 | 3,709 | 3,767 | 388 | 14 | 12 |
| A4 | 3,745 | 3,935 | 346 | 5 | 5 |
| DBVPG4650 | 4,082 | 4,381 | 508 | 0 | 0 |
| DBVPG6304 | 4,094 | 4,617 | 536 | 19 | 19 |
| N_43 | 3,801 | 4,663 | 583 | 8 | 8 |
| N_44 | 3,704 | 4,091 | 421 | 2 | 2 |
| Q32_3 | 3,919 | 3,924 | 399 | 0 | 0 |
| Q59_1 | 3,856 | 3,871 | 393 | 4 | 4 |
| Q62_5 | 4,064 | 3,981 | 408 | 6 | 6 |
| Q95_3 | 4,029 | 4,411 | 484 | 6 | 6 |
| T21_4 | 3,953 | 4,106 | 474 | 6 | 6 |
| UFRJ50816 | 3,649 | 3,602 | 345 | 12 | 10 |
| Y6_5 | 3,305 | 3,103 | 277 | 2 | 2 |
| Y7 | 3,805 | 3,673 | 363 | 0 | 0 |
| YPS138 | 3,847 | 4,093 | 432 | 5 | 5 |
| IFO1804 | 2,668 | 2,564 | 160 | 0 | 0 |
| KPN3828 | 2,700 | 2,545 | 137 | 0 | 0 |
| KPN3829 | 2,666 | 2,502 | 162 | 4 | 2 |
| Q89_8 | 2,816 | 2,526 | 153 | 2 | 0 |
| S36_7 | 1,642 | 1,239 | 43 | 0 | 0 |
| UFRJ50791 | 2,315 | 2,130 | 103 | 0 | 0 |
| Z1_1 | 3,073 | 2,847 | 186 | 2 | 2 |
Number of LSDs including transposable genes.
Number of LSDs excluding transposable genes.
The strain with the largest number of duplicate genes is the S. cerevisiae reference strain (S288c) with 1,356 duplicate genes (table 1). These genes may have arisen from either whole-genome duplication or small-scale duplication. Previous studies identified 1,102 duplicate genes arising from whole-genome duplication alone in S. cerevisiae (Byrne and Wolfe 2005). We conclude, therefore, that the number of duplicates found in the S. cerevisiae reference strain is broadly in line with previous studies, whereas the number found in the more poorly sequenced strains will tend to be underestimates.
The effects of varying sequence coverage are also limited when identifying LSDs as a result of the availability of reference strains and several other well-covered genomes. For example, in the extreme case where only a single well-sequenced reference strain and a single low-coverage strain were available, the total number of duplicates in the high-coverage strain will be correct, whereas the total number in the low-coverage strain will be underestimated. To determine whether these duplicates are lineage specific, we must compare these two strains. The number of duplicates counted as lineage specific in the reference strain may be overestimated because duplicate pairs that are actually present in the low-coverage strain would be missed due to data incompleteness. By contrast, the number of LSDs identified in the low-coverage strain would probably be underestimated because the total number of genes is underestimated. Continued addition of further data from other strains will not change the total number of genes in well-sequenced strains but will reduce the number of those counted as lineage specific. The number of LSDs in well-sequenced strains will converge to the correct number, whereas those on low-coverage strains will always be underestimated. In addition to the reference strains, there are six well-sequenced strains (>5,000 genes annotated) for S. cerevisiae and five for S. paradoxus, and so it is likely that for these strains, the number of LSDs estimated is reasonably accurate.
Despite the possibility of underestimating LSDs in low-coverage strains, we are able to detect a large number of LSDs in the majority of strains for both species: the mean for the best sequenced S. cerevisiae and S. paradoxus strains is 80 and 75, respectively. The strains sequenced to lower coverage have fewer LSDs, although these are still substantial numbers. We find as many as 40 LSDs in the lower coverage S. cerevisiae strains and up to 19 in the lower coverage S. paradoxus strains. These figures exclude the S. paradoxus strain UWOPS91_917_1, which has an anomalously large number of LSDs; this strain contains S. cerevisiae-like reads, indicative of a hybrid origin (Liti et al. 2009). The number of LSDs for UWOPS91_917_1 is, therefore, probably an overestimate.
Full details of numbers of duplicates and LSDs are given in tables 1 and 2. LSDs are shown with and without the inclusion of duplicate pairs composed of retrotransposons. The removal of retrotransposons from the reference strain (S288c) LSDs reduces the number of LSDs by 2-fold, although the effect of this exclusion is smaller for the other strains. A higher number of retrotransposons might be expected in the reference strain which is completely sequenced and assembled. Assembly of incomplete genome sequences might miss some repetitive sequences such as retrotransposons. Table 2 shows that the number of contigs, the total number of duplicates, and the number of LSDs in S. paradoxus are similar to S. cerevisiae.
Chromosomal Position of Duplicate Genes
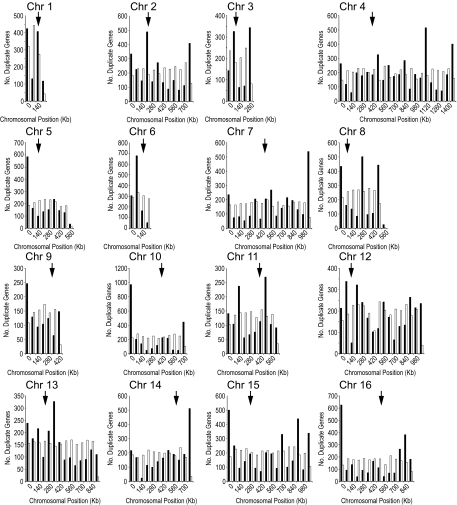
Duplicate genes are not randomly distributed along chromosomes in either S. cerevisiae (fig. 1) or S. paradoxus (additional data file 2, Supplementary Material online). For all chromosomes in both species, the differences between the observed position of duplicates and randomly placed duplicates are statistically significant (P < 2.2 × 10−16, chi-squared test). This remains the case after correction for the nonrandom position of all genes in the genome.
FIG. 1.—
Chromosomal distribution of duplicate genes. The graphs show the distribution of duplicate genes (black) and randomly generated duplicate genes (white) for 16 Saccharomyces cerevisiae chromosomes. Arrows indicate positions of centromeres.
Several chromosomes have an abundance of duplicates located in the subtelomeric regions: V, VII, X, XIV, XV, XVI in S. cerevisiae and IV, VII, IX, X, XIV, XV, and XVI in S. paradoxus. Interestingly, chromosomes VII, X, XIV, XV, and XVI all have duplicated genes in similar location between the two species. This indicates that these regions of chromosomes are more prone to duplication events and/or the biased retention of duplicated genes. Genomic rearrangement can explain the location of duplicated chromosomal regions in S. cerevisiae (Seoighe and Wolfe 1998). In S. cerevisiae, the regions near telomeres frequently undergo rearrangement (Horowitz et al. 1984), and so the frequent rearrangements near telomeric regions might account for the accumulation of duplicate genes in these regions.
The yeast telomeric position effect (TPE) is the repression of genes in close proximity to the telomeres (Gottschling et al. 1990). The repression effect has been demonstrated up to ∼20 kb from the telomere with URA3 (YEL021W). We find that on average 12.29 ± 5.2% (S. cerevisiae) and 11.40 ± 3.1% (S. paradoxus) of duplicate genes are located in TPE regions. In addition, 41.1% (S. cerevisiae) and 35.5% (S. paradoxus) of all genes in the TPE regions are identified as duplicates. The average proportion of genes that are duplicates in TPE regions is, therefore, higher than the proportion of genes that are duplicates over the entire genome, demonstrating that duplicates tend to aggregate in these regions.
The TPE may act to silence some duplicates, and this could be a mechanism of nonfunctionalization. Interestingly, S. paradoxus strain UFRJ50816 contains two copies of IFH1 (YLR223C), an essential protein that interferes with silencing at telomeres when overexpressed (Singer et al. 1998). This duplicate is lineage specific and may confer a resistance to the TPE in this strain.
Evolutionary Divergence of Duplicates
LSDs and population duplicates may arise through two mechanisms. First, there may be an ancestral duplication event, and both genes have become fixed. Subsequent gene loss in one of the lineages will give rise to the observation of LSD. Alternatively, duplication events occur within a specific lineage. Ancient duplications will be accompanied by greater sequence divergence between the two members of the duplicate pair and so will be distinguishable from more recent LSDs.
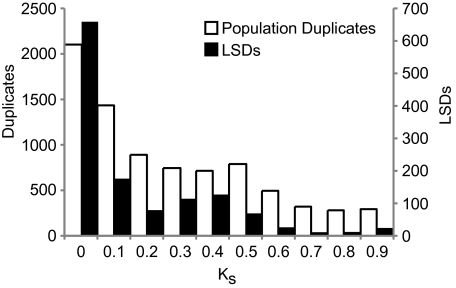
Using the number of synonymous substitutions per site (Ks) as a proxy for time since the duplication event (fig. 2), it is evident that both LSDs and population duplicates display a range of divergences and, therefore, probably have a range of times since duplication. Saccharomyces cerevisiae LSDs have a lower average Ks when compared with the remaining duplicates (mean Ks: 0.34 for LSDs, 2.35 for other duplicates; P < 2.2 × 10−16, Wilcoxon rank sum). The same pattern is shown when comparing S. paradoxus LSDs with the remaining duplicates (mean Ks: 0.38 for LSDs, 3.06 for other duplicates; P < 2.2 × 10−16, Wilcoxon rank sum).
FIG. 2.—
The age of population duplicates (white) and LSDs (black) is measured by the number of synonymous mutations (Ks). A higher value of Ks indicates that the duplicate genes have diverged and are therefore older. Genes from both Saccharomyces cerevisiae and S. paradoxus are shown.
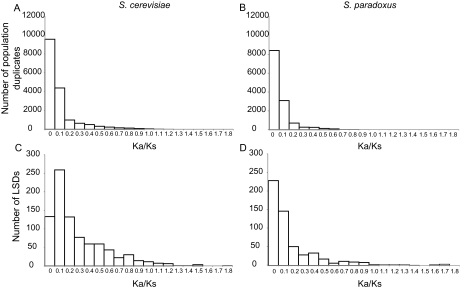
The Ka and Ks ratio can be used as a direct measure of selection on duplicate genes and LSDs. Because we would expect on average that synonymous substitutions are more likely to be neutral, a relative increase in the proportion of nonsynonymous substitutions is likely to be related to functional change. For both the population duplicate set (fig. 3A and B) and the LSDs (fig. 3C and D), the majority of sites display relatively few nonsynonymous substitutions. However, the set of LSDs display considerably more nonsynonymous substitutions, as indicated by the higher overall Ka/Ks values (fig. 3). This indicates that LSDs are undergoing elevated evolutionary change, presumably because they are less prone to purifying selection, which is consistent with greater levels of functional redundancy. This is common for newly created genes (Wagner 2002; Scannell and Wolfe 2008).
FIG. 3.—
Signs of selection acting on duplicate genes. (A) The number of Saccharomyces cerevisiae duplicate genes; (B) the number of S. paradoxus duplicates; (C) the number of S. cerevisiae LSDs; and (D) the number of S. paradoxus LSDs. Here selection is measured by the ratio of nonsynonymous (Ka) to synonymous mutations (Ks). A higher Ka/Ks ratio indicates that one member of a duplicate pair has more nonsynonymous substitutions.
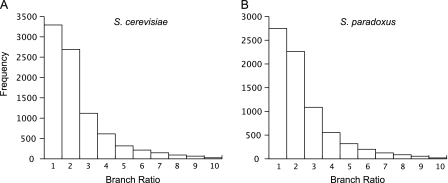
Functional evolution in one member of a duplicate gene may lead to an increase in evolutionary rate when compared with its duplicate partner. Alternatively relaxed selection in one member may also lead to a difference in evolutionary rates between duplicate pairs. Such an increase in rate for one gene can be detected by calculating phylogenetic branch lengths, with asymmetric branch lengths indicating a difference in the evolutionary rate. The distribution of branch rations (fig. 4) shows that the majority of genes are evolving at a similar rate. There is, nevertheless, a significant tail for both distributions, indicating accelerated evolution in one member of a duplicate pair for a significant minority of genes. Some examples are shown in additional data file 3, Supplementary Material online.
FIG. 4.—
The distribution branch length ratios for (A) Saccharomyces cerevisiae and (B) S. paradoxus. The branch ratio is defined as the ratio between the branch lengths on a phylogenetic tree of each duplicate pair rooted by a Kluyveromyces waltii outgroup.
Specific Retention of Duplicate Genes
If selection is operating to differentially retain duplicate genes, we would expect genes with functions that increase fitness to be retained more frequently than a randomly selected set. This would manifest itself as overrepresentation of specific functions in different lineages. In order to determine whether there are overrepresented functions, we used the GO (Ashburner et al. 2000).
We find that a highly nonrandom set of functions is associated with the population duplicates (additional data file 4, Supplementary Material online). From the “Molecular Function” ontology, 26 S. cerevisiae strains show an enrichment of terms relating to sugar transport. Genes relating to catalytic activity such as hexase and helicase activities are also found across many strains.
A large number of S. cerevisiae strains show an overrepresentation for transposon-related GO terms. This result might be expected due to the large number of transposable genes identified in this study and their high similarity making their identification as duplicates more reliable.
Overrepresentation of some GO terms is also seen in the LSD sets (additional data file 4, Supplementary Material online), indicating selection acting on the youngest duplicates. The functions of the LSDs may also give an insight into the recent adaptation of a particular strain. Indeed, we see the overrepresentation of sugar transporters in the S. cerevisiae strain 378604X and genes involved in response to toxins in the reference strain.
Overrepresented genes were also determined for S. paradoxus population duplicates and LSDs (additional data file 5, Supplementary Material online). As with the S. cerevisiae data, sugar transporters are the most commonly overrepresented genes, occurring in 24 strains.
Clustering of Duplicate Genes Leads to Recapitulation of Environment Grouping
The various strains of S. cerevisiae and S. paradoxus have been isolated from a range of environments (Liti et al. 2009). The S. cerevisiae strains are derived from natural environments such as tree bark and soil and artificial environments such as the brewing and baking industries and various laboratory and medical isolates. Saccharomyces paradoxus isolates are mostly derived from natural environments, including 18 strains the bark of Quercus spp from two UK parks. The range of environmental conditions from which S. cerevisiae strains have been isolated is therefore larger than for the S. paradoxus strains.
According to our hypothesis, selection from the environment will alter the gene content of the genomes and the population. From this hypothesis, we predict that retention of duplicates will show correlation with adaptation to the environment from which they were isolated. Such a correlation is found between single nucleotide polymorphisms (Schacherer et al. 2009). In order to test the hypothesis, we inferred a phenetic tree, that is, one that represents observed characteristics rather than evolutionary relationships.
The tree is based on over- and underrepresented “Biological Process” GO terms of both population duplicates and LSDs, where semantic similarity was used to generate a distance between each strain. This distance matrix is then used to create a neighbor joining tree. Thus, the tree topology reflects differences in functional annotations of sets of duplicate genes between strains. Only 21 S. cerevisiae and 18 S. paradoxus strains are included in the phenetic trees as these are the only strains showing over- and underrepresented “Biological Process” GO terms.
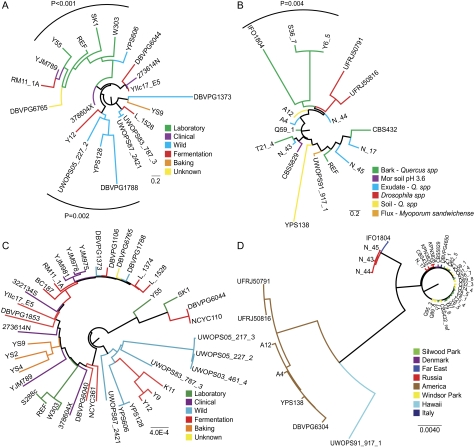
We find that the S. cerevisiae phenetic tree recapitulates the known environments from which the strains were isolated (fig. 5A). In particular, we see a group of laboratory strains showing over- and underrepresentation for more similar GO terms than strains from the other environments. Monte Carlo simulation shows that this clustering is significant (P < 0.001) and would not be expected by chance. Several strains isolated from the wild also form a significant cluster (P = 0.002). This is direct evidence of correlation between the functional classifications of duplicate genes in a population and the environment from which the members of that population were isolated, indicative of adaptation to the environment.
FIG. 5.—
Phenetic and phylogenetic trees for Saccharomyces cerevisiae and S. paradoxus. (A) The 21 S. cerevisiae strains with over- or underrepresented “Biological Process” GO terms. (B) The 18 S. paradoxus strains with over- or underrepresented “Biological Process” GO terms. Distances between strains were determined using the semantic distance between the over- and underrepresented “Biological Process” GO terms of each strain. Branches leading to each strain were then colored according to environmental background. Strains from similar backgrounds have similar overrepresented GO terms, indicating selection for similar types of duplicate genes. (C) Phylogenetic for all S. cerevisiae strains. (D) Phylogenetic tree for all S. paradoxus strains. Phylogenetic trees are taken from Liti et al. (2009) and are based on single nucleotide polymorphism data.
The S. paradoxus phenetic tree shows very little correlation between clusters and environment (fig. 5B). There is a significant cluster of strains isolated from the bark of Quercus spp (P = 0.004). However, there are several other strains isolated from the bark of Quercus spp, which group together with strains isolated from exudate and soil of Quercus spp. A lack of significant clustering for the remainder of the tree might be expected as the majority of strains are isolated from similar environments (oak bark) in different geographical regions.
It should be noted that the environments from which the strains were isolated are somewhat limited. The 39 S. cerevisiae strains are six basic environment types (laboratory, wild, fermentation, clinical, baking, and unknown). The S. paradoxus strains are predominantly isolated from oak bark. There may well be whole groups of yeasts from very different environments that are yet to be isolated. We predict that these as-yet undiscovered strains are likely to differ markedly at the genomic level.
Discussion
Here we have quantified the extent of lineage-specific duplication in 39 strains of S. cerevisiae and 28 strains of S. paradoxus from the Saccharomyces Genome Resequencing Project (Liti et al. 2009). We demonstrate that LSDs and population duplicates are abundant and that the overrepresented functions in each strain's duplicate genes correlates with environment.
The types of genes that have a tendency to be retained as duplicates for each strain are not random (additional data files 4 and 5, Supplementary Material online). Previous studies (Guan et al. 2007; Hakes et al. 2007) identified specific types of genes that are retained in duplicate. Similarly, we find sugar transporters and genes with catalytic activity to be overrepresented in duplicate sets. Such a nonrandom distribution may arise either from nonuniform duplication generation or from nonrandom retention of duplicates. Nonrandom duplication is certainly possible: subtelomeric regions are known to have enriched numbers of duplicates (Horowitz et al. 1984). Indeed, the duplicated genes identified in this study have nonrandom positions; they are found throughout the genome but are overrepresented in the subtelomeric regions. In addition, the rate of transposable element transposition may differ between strains or the rate of duplication may be higher for genes that are already tandemly duplicated because this could increase the chance of unequal crossing over. However, the known mechanisms that give rise to nonrandom duplication would not result in enrichment of specific functions that correlate with environment. A more likely explanation for the nonrandom functions of observed duplicates is that duplicate genes are preferentially retained if they confer a fitness advantage.
Gene duplication has long been known to be an important factor in genome evolution (Ohno 1970). In S. cerevisiae, the rate of duplication is thought to be as high as ≈0.01 duplications per gene per million years (Lynch and Conery 2000). The rate of duplicate gene loss has also been shown to be high with 88% of duplicated genes being lost after a whole-genome duplication event in an ancestor of S. cerevisiae (Kellis et al. 2004; Scannell et al. 2006). Genes both gained and lost since the whole-genome duplication can be inferred from the reconstruction of the ancestor genome (Gordon et al. 2009).
The evolutionary divergence between the various duplicate pairs, both LSDs and population duplicates, suggests that at least some of the duplication events are relatively old, allowing time for accumulation of substantial substitution. Even in these older duplicate pairs, we find that the number of copies of genes varies between strains, indicating lineage-specific gene loss. Duplicate pairs in both S. cerevisiae and S. paradoxus display signs of asymmetric divergence when their branch lengths are compared on a phylogenetic tree (fig. 4). This finding is in agreement with previous work that shows asymmetric divergence of genes derived from the whole-genome duplication (Scannell and Wolfe 2008).
The LSDs offer an important insight into gene content variation at a population level. LSDs often have the same functional annotation and show little or no sequence divergence (measured with Ks). They therefore frequently represent relatively recent duplications that are not found in the other strains. At the extreme, these duplications may have occurred only in single individuals and so would represent copy-number variation. Alternatively, they could be somewhat older, becoming established in individual lineages within the population. Because we have no direct evidence as to them being actual copy-number variants, we are conservative and term them lineage specific.
In addition to low divergence measured by Ks, LSDs also have higher Ka/Ks ratios than the population duplicate set. These two observations taken together suggest that, on average, LSDs are relatively young compared with population duplicates, and subsequent to their generation, they display a burst of evolutionary change. Other work (Wagner 2002; Scannell and Wolfe 2008) has shown that newly created duplicates often display rapid evolutionary change and contribute to functional divergence. There is an ongoing process of duplication, loss, selective retention, and evolutionary divergence. In LSDs, we detect those duplicate genes still in this state of flux as they are not yet fixed in the population. For these reasons, LSDs provide a view into the early stages of functional evolution and adaptation at the population level.
The LSDs permit us to distinguish evolutionary contingency from functional necessity. By definition, each particular LSD gene is found in duplicate in only a single lineage. Any similarities, therefore, cannot be due to shared evolutionary history and must have arisen through either independent gains or independent losses. Nevertheless, there are whole classes of functions that are overrepresented. These functions must have arisen and/or been retained multiple times independently. This represents strong evidence that the bias toward specific functions is adaptive rather than being contingent on the specific details of duplication or loss/retention events. Indeed, if we view each of these lineages as an independent “running of the tape of evolution,” we find that the same functions arise repeatedly and independently.
Supplementary Material
Additional data files 1–5 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
The authors would like to thank Casey Bergman for supplying the PCAP assemblies, John Pinney for help calculating the hypergeometric distribution, Jamie MacPherson for help with calculating semantic distances and Daniel Money and Simon Whelan for scientific discussion and helpful suggestions. This work was funded by BBSRC grant BB/F007620/1.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AB, Pekosz MR, Kurvathi RS, Kaback DB. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics. 2008;179:1221–1235. doi: 10.1534/genetics.107.083493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner A. GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res. 2002;30:3378–3386. doi: 10.1093/nar/gkf449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:e55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D, Aparicio O, Billington B, Zakian V. Position effect at S. cerevisiae telomeres: reversible repression of POL II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney J, Lovell S, Oliver S, Robertson D. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H, Thorburn P, Haber J. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang J, Aluru S, Yang SP, Hillier LD. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JJ, Conrath DW. Proceedings of International Conference on Research in Computational Linguistics. Taiwan: 1997. Semantic similarity based on corpus statistics and lexical taxonomy; pp. 19–33. [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:0008.1–0008.9. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer; 1970. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Polakova S, et al. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Wolfe KH. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Extent of genomic rearrangement after genome duplication in yeast. Proc Natl Acad Sci U S A. 1998;95:4447–4452. doi: 10.1073/pnas.95.8.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Curr Opin Microbiol. 1999;2:548–554. doi: 10.1016/s1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- Singer MS, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford JB. Heterosis and the evolution of duplications. Am Nat. 1969;103:407. [Google Scholar]
- Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]