Abstract
Horizontal transfer (HT) of genes is known to be an important mechanism of genetic innovation, especially in prokaryotes. The impact of HT of transposable elements (TEs), however, has only recently begun to receive widespread attention and may be significant due to their mutagenic potential, inherent mobility, and abundance. Helitrons, also known as rolling-circle transposons, are a distinctive subclass of TE with a unique transposition mechanism. Here, we describe the first evidence for the repeated HT of four different families of Helitrons in an unprecedented array of organisms, including mammals, reptiles, fish, invertebrates, and insect viruses. The Helitrons present in these species have a patchy distribution and are closely related (80–98% sequence identity), despite the deep divergence times among hosts. Multiple lines of evidence indicate the extreme conservation of sequence identity is not due to selection, including the highly fragmented nature of the Helitrons identified and the lack of any signatures of selection at the nucleotide level. The presence of horizontally transferred Helitrons in insect viruses, in particular, suggests that this may represent a potential mechanism of transfer in some taxa. Unlike genes, Helitrons that have horizontally transferred into new host genomes can amplify, in some cases reaching up to several hundred copies and representing a substantial fraction of the genome. Because Helitrons are known to frequently capture and amplify gene fragments, HT of this unique group of DNA transposons could lead to horizontal gene transfer and incur dramatic shifts in the trajectory of genome evolution.
Keywords: Helitrons, insect viruses, transposable elements, lateral transfer
Introduction
The movement of genetic material between reproductively isolated species, known as horizontal transfer (HT), is known to be an important process in genome evolution. In eukaryotes, this has been shown in the case of genes (for review, see Anderson 2005; Keeling and Palmer 2008) and, more recently, with transposable elements (TEs) (e.g., Silva et al. 2004; Casse et al. 2006; Diao et al. 2006; de Boer et al. 2007; Loreto et al. 2008; Pace et al. 2008; Bartolome et al. 2009; Roulin et al. 2009). TEs are mobile, parasitic pieces of genetic material that can mobilize and replicate within the host genome. Their inherent ability to replicate and integrate into the genome is likely to make them prone to HT (Kidwell 1992). HT has been proposed as an essential part of the lifecycle of some types of TEs in order to avoid co-evolved host suppression mechanisms aimed at limiting their mobility within lineages (Hartl et al. 1997; Silva et al. 2004). It has also been proposed that the propensity for HT could be related to the mechanism of transposition used (see Schaack, Gilbert and Feschotte 2010 for review). TEs are classified based on whether they move via an RNA intermediate (Class 1) or a DNA intermediate (Class 2), with further divisions based on the mechanism of integration (Wicker et al. 2007).
A unique group of rolling-circle (RC) DNA transposons called Helitrons (with atypical structural characteristics including 5′ TC and 3′ CTRR termini and a 16 to 20-nt palindrome upstream of the 3′ end [Feschotte and Wessler 2001; Kapitonov and Jurka 2001]) have been described in a wide array of eukaryotes including fungi (Poulter et al. 2003; Cultrone et al. 2007), plants (Kapitonov and Jurka 2001; Lal et al. 2003; Rensing et al. 2008; Yang and Bennetzen 2009a), insects (Kapitonov and Jurka 2001; Poulter et al. 2003; Langdon et al. 2009; Yang and Bennetzen 2009a; The International Aphid Genomics Consortium 2010), nematodes (Kapitonov and Jurka 2001), and vertebrates (Poulter et al. 2003; Zhou et al. 2006; Pritham and Feschotte 2007). In some cases, Helitrons constitute a significant portion of the genomes (e.g.,Caenorhabiditis elegans, Arabidopsis thaliana, Myotis lucifugus). Helitrons, unlike most other DNA transposons that use transposase, putatively encode a protein with a rolling circle initiator motif and PIF1-like DNA helicase domains and are categorized in their own subclass (Kapitonov and Jurka 2001; Wicker et al. 2007). Homology of the Helitron-encoded protein to bacterial RC transposons (IS91, IS1294, IS801), which are well known for their propensity to shuttle antibiotic resistance genes between distinct bacterial species (Toleman et al. 2006), reveals a distant relationship (Kapitonov and Jurka 2001). Like their bacterial cousins, some Helitrons function as “exon shuffling machines” (Feschotte and Wessler 2001). This ability is particularly pronounced in maize where it is estimated that at least 20,000 gene fragments have been picked up and shuffled by Helitrons (Du et al. 2009; Feschotte and Pritham 2009; Yang and Bennetzen 2009b). The ability to seize and recombine exons from multiple genes to create novel genetic units (Brunner et al. 2005; Gupta et al. 2005; Lal and Hannah 2005; Morgante et al. 2005; Xu and Messing 2006; Pritham and Feschotte 2007; Jameson et al. 2008; Langdon et al. 2009) makes HT of Helitrons especially intriguing because they can shuttle gene fragments between genomes.
This study expands our understanding of HT of TEs in several ways. First, we provide the first evidence for widespread, repeated HT of Helitrons, a distinctive group of transposons with a unique mechanism of replication. Second, in contrast to previous reports of widespread HT which have involved only hAT superfamily elements distributed largely among vertebrates (Pace et al. 2008; Gilbert et al. 2010), we show horizontally transferred Helitrons are frequently found in insect genomes. However, we have also identified cases of Helitron HT in vertebrates (bat, lizard, and jawless fish), a patchy distribution that indicates that certain host genomes are especially vulnerable to invasion. Third, this is the first report of Helitron HT in insect viruses, which could act as shuttle systems for the delivery of DNA between species (Loreto et al. 2008). Although HT has occasionally been invoked to explain discordant distributions in isolated cases (Kapitonov and Jurka 2003; Lal et al. 2009), our discovery of horizontally transferred Helitrons in viruses, insects, and vertebrates demonstrates the widest range of extensive HT among animals and possible vectors so far.
Materials and Methods
Helitrons identified in Myotis lucifugus (the little brown bat) were used as an initial query (BlastN using default parameters (BlastN… [Altschul et al.1990]) to find Helitrons in other genomes available at the National Center for Biotechnology Information, including the whole genome shotgun, nucleotide collection (nr/nt), genome survey sequences, high throughput genomic sequences, and expressed sequence tag databases. Hits that were ≥65% identical to the query over >300 bp were examined and, when possible, full-length Helitrons were manually extracted. These elements were used as queries to find additional related Helitrons; the resulting hits were examined, and full-length Helitrons were extracted to generate a library of Helitrons for each species (details on all methods are in supplementary Materials and Methods, Supplementary Material online). Helitrons were then classified into families based on the following criteria according to Yang and Bennetzen (2009a, 2009b). We established conservative criteria to identify cases of HT that could be fully analyzed, including >80% identity at the 3′ end, a >400 bp portion of the internal region that is >80% identical, and divergence estimates among species that exclude the possibility of vertical inheritance (supplementary materials and methods, Supplementary Material online). Helitrons that share high levels of identity (>80%) from the same family in multiple species were aligned using MUSCLE (Edgar 2004) and analyzed as a group (including calculations of pairwise divergence [MEGA 4.0.2; Tamura et al. 2007], abundance [RepeatMasker version 3.2.7; A. F. A. Smit, R. Hubley, and P. Green, www.repeatmasker.org], and, when possible, calculations of amplification date estimates [as in Pritham and Feschotte 2007; Pace et al. 2008]).
Results
Identification, Classification, and Characterization of Helitrons
In a previous study, Helitrons were reported only in the little brown bat, M. lucifugus, among the 44+ publicly available mammalian genome sequences (Pritham and Feschotte 2007) that suggested the acquisition of these elements via HT. Because M. lucifugus is a good candidate for investigating possible HT, a deeper survey of Helitrons was performed, a previously uncharacterized family (HeligloriaB_Ml) was identified, and was used as a starting point for a series of Blast searches. These searches led to the subsequent identification of Helitrons from animals and animal viruses which were then classified into families based on their identity at the 3′ end (for family designation) and 5′ end (for subfamily designation), as in Yang and Bennetzen (2009a, 2009b; see Materials and Methods): the families were named Heligloria, Helisimi,Heliminu, and Helianu. Cases of recent HT were identified and analyzed when Helitrons of the same family that exhibited >80% identity at the 3′ end and contained a >400 bp portion of the internal region with >80% identity (see Materials and Methods) were found in diverged species (>35 million years ago [Ma]). Helitrons demonstrating high levels of identity that were inconsistent with vertical descent were found in many taxa, including insect viruses, many invertebrates (e.g., insects, nematodes, annelids, molluscs, and planaria), and vertebrates (e.g., salamanders, lizards, snakes, jawless fish, and bat; see supplementary table S1, Supplementary Material online). Those cases for which there were sufficient data to fully analyze the evidence for HT include the little brown bat (M. lucifugus; Chiroptera, Mammalia), sea lamprey (Petromyzon marinus; Petromyzontiformes, Cephalaspidomorphi), green anole (Anolis carolinensis; Squamata, Reptilia), triatomine bug and aphid (Rhodnius prolixus, Acyrthosiphon pisum; Hemiptera, Insecta), fruit flies (Drosophila ananassae, D. willistoni, D. yakuba; Diptera, Insecta), silkworm moth (Bombyx mori; Lepidoptera, Insecta), and two polydnaviruses which are symbiotically associated with hymenopteran wasps (Hymenoptera, Insecta), Cotesia sesamiae Mombasa Bracovirus (CsMBV) and Cotesia plutella Bracovirus (CpBV; see table 1).
Table 1.
Distribution and Characterization of Four Helitron Families Including Evidence for HT Across Taxa
| Average % Identity |
||||
| Group | 5′ (30 bp) | 3′ (30 bp) | Internal (Length bp) | Copy Number (% Genome) |
| Species | ||||
| HeligloriaAi | ||||
| Drosophila yakuba | 91a | 93b | 96 (562) | 6 (0.02) |
| D. ananassae | 117 (0.14) | |||
| D. willistoni | 4 (0.02) | |||
| Rhodnius prolixus | 184 (0.01) | |||
| Acyrthosiphon pisum | 8 (0.00) | |||
| Bombyx mori | 89 (0.04) | |||
| HeligloriaAii | ||||
| B. mori | 89 (307) | * | ||
| Cotesia plutella BV | 1 (0.14) | |||
| HeligloriaB | ||||
| Myotis lucifugus | 95 | 88 (579) | 51 (0.0) | |
| R. prolixus | * | |||
| Anolis carolinensis | 667 (0.02) | |||
| Petromyzon marinus | 32(0.00) | |||
| Helisimi | ||||
| D. willistoni | 89 | 95 | 88 (463) | 2 (0.09) |
| D. ananassae | 2 (0.08) | |||
| A. pisum | 31 (0.02) | |||
| B. mori | 65 (0.20) | |||
| C. sesamiae MBV | 2 (n/a) | |||
| R. prolixus | 15 (0.00) | |||
| Heliminu | ||||
| D. willistoni | 90 | 87 | 93 (1,378) | 3 (0.01) |
| D. ananassae | 12 (0.03) | |||
| A. pisum | 1 (n/a) | |||
| B. mori | 9 (0.01) | |||
| Helianu | ||||
| D. willistoni | 93 | 93 | 97 (1,894) | 57 (0.10) |
| D. ananasse | 1 (0.10) | |||
| B. mori | 67 (0.03) | |||
NOTE.—n/a, not applicable, as the data were obtained from BAC clones deposited in the nucleotide collection (nr) database; Family names include, where applicable, subfamily designation (capital letter) and exemplar identification (roman numeral). Percent identity across species provided for 30 bp of 5′ and 3′ ends, respectively, and aligned internal regions are reported as well as copy number and percent of genome occupied (see supplementary Materials and Methods, Supplementary Material online). Asterisk (*) indicates copies that were analyzed as part of the other subfamily.
This value is applicable to both Heligloria Ai and Heligloria Ai as the pariwise identity of the 30 bps at the 5′ end was analyzed together as they belong to the same subfamily.
This value is applicable to the HeligloriaAi, HeligloriaAii, and HeligloriaB as the pairwise identity of the 30 bps at the 3′ end was analyzed together as they belong to the same family.
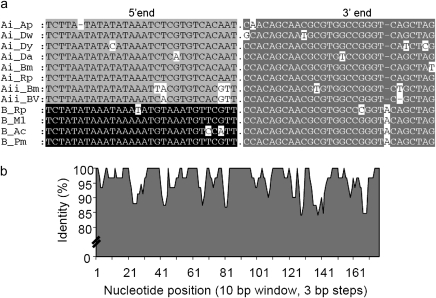
Among the four families of Helitrons, Heligloria (which contains two subfamilies based on divergence at the 5′ end, HeligloriaA and HeligloriaB [fig. 1a]) is the most widely distributed across taxa. Furthermore, the subfamily HeligloriaA includes two exemplars (HeligloriaAi and HeligloriaAii) based on the presence of two unique internal regions. A putative autonomous representative (HeligloriaAi) was found in six different insect species (D. yakuba, D. ananassae, D. willistoni, A. pisum, R. prolixus, and B. mori) with levels of sequence identity ≥90% (over 768–3,927 bp) based on pairwise comparisons of the internal region (see supplementary Dataset S1, table S3.1, fig 1b, Supplementary Material online). HeligloriaAii is present in B. mori and two polydnaviruses (CpBV and CsMBV), which have a segmented genome in viral particles and an integrated form (provirus) in the genome of parasitic hymenopteran wasps in which they reside, C. plutella and C. sesamiae (Dupuy et al. 2006; see Discussion). These two polydnaviruses are associated with the braconid wasps, C. plutella and C. sesamiae, and both contain short, nonautonomous copies of HeligloriaAii. Because the Cotesia genus is 10 My old (Dupuy et al. 2006), we included only one of the two species in our analysis of HT (table 1). The subfamily HeligloriaB is present in M. lucifugus, A. carolinensis, R. prolixus, and P. marinus. It has a unique 5′ end and internal region compared with HeligloriaAi and HeligloriaAii. HeligloriaB is 88% identical over 579 bp between these four species (see alignment, supplementary fig. S1, Supplementary Material online). Fragments of HeligloriaB with high sequence identity (84–96%) are also present in mole salamanders, snakes, and in nematodes (see supplementary table S1, Supplementary Material online).
Fig. 1.—
(a) Alignment of 5' and 3' ends (30 bp) of Heligloria (Hg) elements from species into which the element has horizontally transferred (for criteria, see Materials and Methods and supplementary table S1, Supplementary Material online for additional species not included in the analysis). Similarity at the 3' end (>80%) is used to determine family designation (shown in gray with white letters). Similarity at the 5' end (>80%) is used to designate subfamilies (two shown here: A, light gray with black letters and B, black with white letters). Species names denoted with the first letter of the genus and species (Acyrthosiphon pisum [Ap], Anolis carolinensis [Ac], Bombyx mori [Bm], Drosophila willistoni [Dw], D. ananassae [Da], D. yakuba [Dy], Myotis lucifugus [Ml], Petromyzon marinus [Pm], Rhodnius prolixus [Rp], Cotesia plutella Braco Virus [BV]). (b) Sequence identity (%) among copies of HeligloriaAi elements from the six species (table 1) in which they were identified (see supplementary Materials and Methods, Supplementary Material online); sequence identity was calculated using 10 bp windows with 3-bp stepwise increments over the internal region.
The second family of Helitrons, called Helisimi, was identified in D. ananassae, D. willistoni, A. pisum, R. prolixus, B. mori, and CsMBV. On average, these elements are 87% identical over 463 bp across species that diverged >300 Ma (fig. 2). Pairwise comparisons of individual elements reveal high sequence identity (81–96%) over 469–4,548 bp (supplementary Dataset S1, Supplementary Material online). Although there are no subfamilies within this family, there are two clusters, each of which are 98% identical (supplementary table S3.2, Supplementary Material online), with 81–83% identity between groups. Two copies of Helitrons were found in the C. sesamiae Mombasa bracovirus genome, one copy is intact with a 5′ and 3′ end but has captured host genomic sequence and the other copy is truncated at the 5′ end. The Helitron copy with the intact ends was also found at the orthologous position in the Kitale strain of the virus (supplementary fig. S5 and table S2, Supplementary Material online). Fragments of Helisimi were identified in several other Drosophilids as well; however, the short divergence times between these species prevent us from ruling out the possibility of vertical inheritance, and thus they were excluded from this analysis (see supplementary table S1, Supplementary Material online).
FIG. 2.—
Schematic representation of phylogenetic relationships among animal lineages and estimated divergence times (Ma). Presence of horizontally transferred Helitrons from four different families in each lineage are denoted by rectangles (not placed relative to the timescale). Numbers in parentheses on the right indicate the number of species (when >1) for which whole genome sequence data are publicly available in the whole genome shotgun (National Center for Biotechnology Information).
The families Heliminu and Helianu were identified in insects only. Heliminu is 93% identical over a region of 1,378 bp across species (table 1 and supplementary table S3.3, Supplementary Material online). A pairwise comparison of copies from A. pisum and B. mori reveals that elements are 95% identical over 4,000 bp (supplementary Dataset S1, Supplementary Material online). The fourth family, Helianu, was identified in D. willistoni, D. ananassae, and B. mori. Across these three species, Helianu is 97% identical over 1,894 bp (table 1 and supplementary table S3.4, Supplementary Material online) and pairwise comparisons between species extend the region of identity to 2,600 bp (supplementary Dataset S1, Supplementary Material online). Fragments of copies of Heliminu and Helianu (>90% identical) are also present in a variety of other insects, including butterflies, moths, flies, and fleas (see supplementary table S1, Supplementary Material online). Paralogous or orthologous empty sites were identified for at least one member from each family to confirm the mobility of these elements (supplementary fig. S2, Supplementary Material online). The putative autonomous elements encode all the expected motifs and domains consistent with other described animal protein-coding Helitrons (Rep and helicase; supplementary fig. S3a, b, Supplementary Material online).
Species-Specific Proliferation of Helitrons and Timing of Amplification
In the case of all four families, Helitrons have proliferated via amplification of nonautonomous copies. In the case of HeligloriaB, the autonomous partner responsible for the amplification of the non-autonomous elements was not identified in the genome sequences of bat, lizard, and insect. However, we were able to detect autonomous copies of HeligoriaB in the jawless fish genome sequence (supplementary table S2, Supplementary Material online). However, we were able to detect autonomous copies of HeligloriaB in jawless fish in the UCSC genome browser (supplementary table S2, Supplementary Material online). The discovery of autonomous partners for this family was likely hindered by low genome coverage and the older age of the family. It may be that with higher sequencing coverage or examination of additional genomes that the autonomous copies might be discovered.
Copy number varies across species but in some cases is high (up to 677 copies; table 1). Because we used the last 30 bp of the 3′ end, copy number estimates include all subfamilies. To estimate how much of the genome is occupied by each Helitron family, individual genomes were masked by the four families of Helitrons (not only the last 30 bp but with the entire element [table 1]). The apparent discrepancy in the copy number estimation and percent genome occupied is due to the difference in the methods employed. Some Helitrons tend to capture new 3′ ends, retaining the 5′ end and internal region. In those cases, copy number estimate (based on Blast with 3′ end) will be lower than the RepeatMasker estimate (based on the entire element). Helitron families appear to have differentially amplified or been retained in each host species (fig. 3), Helisimi is the most “successful,” having amplified in B. mori to such an extent that it constitutes 0.2% of the genome and contributes almost 0.8 Mb of DNA (table 1). The timing of amplification of HeligloriaB_Ml in bat was estimated based on the average divergence of copies from the consensus sequence (3.8%) to be 14.1 Ma based on the neutral substitution rate as in Pace et al. (2008). In most of the cases, it was not possible to use this method because of difficulty reconstructing a consensus to estimate the ancestral copy and the lack of data on mutation rates. In these cases, the percent divergence between a given Helitron copy (representative of a particular family) and its second-best hit (not with itself) were used as a proxy to estimate the relative timing of amplification (see supplementary table S4, Supplementary Material online). Even though Helitrons appear to be recently active in many genomes (≥99% identity between copies of some families in R. prolixus, A. pisum, and B. mori), there were other cases with no signs of recent activity (as low as 75% identity between copies).
Fig. 3.—
Distribution of Helitron families (Heligloria, Helisimi, Heliminu, and Helianu) across species and their contribution (shown in Kbp) toward the host genome.
Evidence for HT
The high sequence identity (80–97%) of the Helitrons is not limited to the 5′ and 3′ ends but is also observed in the internal regions of all families (fig. 1a and b and supplementary table S3, Supplementary Material online). In many cases, the sequence identity of the Helitrons is exceptionally high compared with the divergence of the hosts (fig. 2). For example, there is 88% sequence identity between Helitrons in the mammal, M. lucifugus, and the lizard, which diverged 360 Ma and these diverged from the common ancestor of the jawless fish and the insect R. prolixus >600 and >750 Ma, respectively (fig. 2; Hedges et al. 2006). Similar patterns of sequence identity of Helitrons (86–97%) can be observed among insects of different orders (Lepidoptera, Diptera, Hemiptera) and the polydnaviruses inhabiting the hymenopteran parasitic wasps. The insects belonging to these orders diverged from their common ancestor >200 Ma (in the case of Diptera and Lepidoptera) and up to 350 Ma (in the case of Hemiptera). Previous work on TEs suggests that that these elements are not under host selective constraints (Silva and Kidwell 2000; Pace et al. 2008), and instead, TEs evolve neutrally upon inactivation of their transposition in the host genomes. The highly fragmented nature and lack of intact open reading frames of the Helitrons identified further supports the idea of lack of active transposition. The levels of divergence observed among Helitrons in these species are much lower than what would be expected based on direct estimates of neutral substitutions rates (e.g., 5.8 × 10−8 mutations per site per year in Drosophila [Haag-Liautard et al. 2007]) given the current estimates of their divergence times (Hedges et al. 2006). Thus, HT is the best explanation for the exceedingly high sequence identity displayed by these TEs across widely diverged species. Another line of evidence that can be used to exclude the possibility of vertical transfer is the discontinuous presence of these elements across different species represented in the database. All four families of Helitrons have a patchy distribution with high sequence identity among vertebrates and insects (figs. 2 and 3). Although, it should be noted that false negative results might occur in genomes with low sequencing coverage and few copies. However, to attribute the patchy distribution observed here to vertical inheritance would require a nonparsimonious scenario of many cases of independent loss and intense activity in a small subset of lineages.
Discussion
This is the first report of the HT of Helitrons among a diverse array of animal species. We identified 25 definitive cases of HT involving four families of Helitrons and nine animal species, including vertebrates and invertebrates that diverged, in some cases, more than 700 Ma (fig. 2 and table 1; for additional cases, see supplementary table S1, Supplementary Material online). Very high sequence identity among species (80–97%), in conjunction with the extremely fragmented nature of the Helitrons identified, preclude the possibility of vertical inheritance and selective constraint as an explanation for the similarity observed between elements across species. Our data reveal interesting patterns within the patchy distribution among animals, including the repeated invasion of some genomes by multiple Helitron families (figs. 2 and 3). Although some families (Heliminu and Helianu) are restricted to insects, HeligloriaB has invaded mammals, reptiles, and jawless fish, in addition to several insect species (table 1 and supplementary table S1, Supplementary Material online). Remarkably, two of the four Helitron families were also found in polydnaviruses that are involved in facilitating the parasitism of lepidopterans by hymenopteran wasps. We propose that the presence of Helitrons in viruses may reflect their role as vectors for HT between parasitic wasps and their hosts, although other routes of HT also likely exist.
Mechanisms of Transfer
The remarkable breadth of species involved in these cases of HT (including not only bat, lizard, jawless fish but also triatomine bug, silkworm, aphid, drosophilids, and bracoviruses) suggests multiple mechanisms may underlie the horizontal spread of TEs. The identification of Helitrons in bracoviruses (double-stranded DNA viruses; Polydnaviridae family) is of particular interest as a potential vector for the delivery of TEs among species. These viruses have an obligatory relationship with parasitic wasps belonging to the Braconidae family, replicating only in wasp ovary cells and releasing fully formed viral particles during oviposition by the wasp into the lepidopteran larvae. The viral particles encode virulence factors that suppress the immunity of the lepidopteran (e.g., for review, see Webb et al. 2009), facilitating the growth of the wasp larvae. Yoshiyama et al. (2001) suggested that the close association between the parasitoid wasp and moth facilitates the HT of TEs, as in the case of the “mariner” element transferred between the braconid parasitoid wasp, Ascogaster reticulatus, and its moth host, the smaller tea tortrix, Adoxophyes honmai. There have been several reports of TE-like sequences in the genomes of DNA viruses (Miller DW and Miller LK 1982; Fraser et al. 1983; Fraser 1986; Friesen and Nissen 1990; Jehle et al. 1998; Drezen et al. 2006; Piskurek and Okada 2007; Desjardins et al. 2008; Marquez and Pritham 2010). If viruses shuttle TEs from one species to another, we might expect to see biased distributions of horizontally transferred TEs based on host susceptibility to a particular virus group. In fact, our data reveal biased distributions (e.g., Helisimi and Heliminu are only found in insects, whereas HeligloriaB is frequently found in vertebrates); however, the sampling bias of the available databases also influences our ability to detect patterns or identify mechanisms based on distribution.
In addition to viruses, some parasitic insects have also been implicated as agents of HT because of their intimate association with their hosts (e.g., Houck et al. 1991). Gilbert et al. (2010) recently found evidence for the HT of four DNA transposon families in R. prolixus and a wide array of tetrapods. Because R. prolixus is a sanguivorous parasite of mammals and vertebrates, transfer of DNA could occur through salivary deposition or blood intake by this species. The presence of closely related Helitrons in R. prolixus and M. lucifugus, a host of R. prolixus, further indicates this bug may be a candidate vector for transferring TEs. Other proposed mechanisms of transfer include endosymbiotic bacteria such as Wolbachia (Hotopp et al. 2007). It is known that Wolbachia infect C. sesamiae wasps (Mochiah et al. 2002), drosophilids, aphids (Jeyaprakash and Hoy 2000; The International Aphid Genomics Consortium 2010), Rhodnius sp. (Espino et al. 2009), and even nematodes (Fenn et al. 2006). In addition to the possibility of HT through Wolbachia, the bacteriophage of Wolbachia is also a potential vector for HT (Gavotte et al. 2007; Loreto et al. 2008). Additional experiments and taxon sampling are necessary to further delineate the role of host–parasite interactions and other intermediates such as bacteria and viruses, in the direction and frequency of HT of TEs and the as of yet unknown mechanisms underlying this process.
Impact on Genome
Diverse mechanisms of HT can lead to recurrent invasions of genomes by Helitrons, thereby increasing the dynamic portion of the genome. The proposed rolling circle-like transposition mechanism could explain the tandem duplicates and arrays generated by Helitrons (supplementary fig. S4, Supplementary Material online, Pritham and Feschotte 2007; Schaack et al. 2010, Choi et al. 2010). The frequent capture of new 3′ and 5′ ends without disrupting their ability to transpose could extend the lifespan of Helitrons in the host genome and generate genetic diversity among elements. Their proposed replication mechanism also likely explains their unique propensity to capture host gene fragments, which could have a tremendous impact on the genome (e.g., Brunner et al. 2005; Gupta et al. 2005; Morgante et al. 2005; Xu and Messing 2006; Jameson et al. 2008; Du et al. 2009; Langdon et al. 2009; Yang and Bennetzen 2009b). Indeed, in M. lucifugus, HelibatN3 has captured the promoter and first exon of the NUBPL (a single copy gene which is highly conserved in mammals) and amplified it to high copy number (>1,000; Pritham and Feschotte 2007). Amplification is thought to closely follow invasion of a naive genome (Pace et al. 2008) and results in opportunities for genetic innovation. Genetic innovation, in turn, leads to diversification within the lineage, a possibility supported by the occurrence of multiple waves of TE invasion in the bat lineage around the time of their rapid diversification, 16–25 Ma (Teeling et al. 2005; Pritham and Feschotte 2007; Ray et al. 2008; Oliver and Greene 2009; Zeh et al. 2009; Gilbert et al. 2010). We conclude that the HT, colonization, and amplification of Helitrons are rampant and widespread across animals and can play a major role in genome evolution.
Supplementary Material
Supplementary dataset, materials and methods, figures S1–S5, and tables are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by start-up funds from the University of Texas-Arlington to E.J.P. and National Science Foundation award 0805546 to S.S. We would like to acknowledge the genome sequencing consortiums for sequencing the M. lucifugus, A. carolinensis, P. marinus, and R. prolixus and C.sesamiae bracovirus genomes. We would also like to thank Brian Fontenot, Matt Carrigan, and two anonymous reviewers for helpful comments on the manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:2. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Pea G, Rafalski A. Origins, genetic organization and transcription of a family of non-autonomous Helitron elements in maize. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- Casse N, Bui QT, Nicolas V, Renault S, Bigot Y, Laulier M. Species sympatry and horizontal transfers of Mariner transposons in marine crustacean genomes. Mol Phylogenet Evol. 2006;40:609–619. doi: 10.1016/j.ympev.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Cultrone A, Dominguez YR, Drevet C, Scazzocchio C, Fernandez-Martin R. The tightly regulated promoter of the xanA gene of Aspergillus nidulans is included in a Helitron. Mol Microbiol. 2007;63:1577–1587. doi: 10.1111/j.1365-2958.2007.05609.x. [DOI] [PubMed] [Google Scholar]
- de Boer JG, Yazawa R, Davidson WS, Koop BF. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics. 2007;8:442. doi: 10.1186/1471-2164-8-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, et al. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 2008;9:12. doi: 10.1186/gb-2008-9-12-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drezen JM, et al. The few virus-like genes of Cotesia congregata bracovirus. Arch Insect Biochem Physiol. 2006;61:110–122. doi: 10.1002/arch.20108. [DOI] [PubMed] [Google Scholar]
- Du C, Fefelova N, Caronna J, He LM, Dooner HK. The polychromatic Helitron landscape of the maize genome. Proc Natl Acad Sci U S A. 2009;106:19916–19921. doi: 10.1073/pnas.0904742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy C, Huguet E, Drezen JM. Unfolding the evolutionary story of polydnaviruses. Virus Res. 2006;117:81–89. doi: 10.1016/j.virusres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino CI, et al. Detection of Wolbachia bacteria in multiple organs and feces of the triatomine insect Rhodnius pallescens Hemiptera, Reduviidae. App Environ Microbiol. 2009;75:547–550. doi: 10.1128/AEM.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn K, et al. Phylogenetic relationships of the Wolbachia of Nematodes and Arthropods. PLoS Pathog. 2006;2(10):e94. doi: 10.1371/journal.ppat.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. A cornucopia of Helitrons shapes the maize genome. Proc Natl Acad Sci U S A. 2009;106:19747–19748. doi: 10.1073/pnas.0910273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Wessler SR. Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes. Proc Natl Acad Sci U S A. 2001;98:8923–8924. doi: 10.1073/pnas.171326198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ. Transposon-mediated mutagenesis of baculoviruses: transposon shuttling and implications for speciation. Ann Entomol Soc Am. 1986;79:773–783. [Google Scholar]
- Fraser MJ, Smith GE, Summers MD. Acquisition of host-cell DNA-sequences by baculoviruses—relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen PD, Nissen MS. Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L, et al. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, II, Brindley PJ, Feschotte C. A role for host–parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol Biol. 2005;57:115–127. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Hartl DL, et al. What restricts the activity of mariner-like transposable elements? Trends Genet. 1997;13:197–201. doi: 10.1016/s0168-9525(97)01087-1. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hotopp JCD, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Houck MA, et al. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991;253:1125–1129. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- The International Aphid Genomics Consortium. Genome sequence of the pea aphid acyrthosiphon pisum. PLoS Biol. 2010;82:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson N, et al. Helitron mediated amplification of cytochrome P450 monooxygenase gene in maize. Plant Mol Biol. 2008;67:295–304. doi: 10.1007/s11103-008-9318-4. [DOI] [PubMed] [Google Scholar]
- Jehle JA, Nickel A, Vlak JM, Backhaus H. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J Mol Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: WSP sequence found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Horizontal transfer of P-elements and other short inverted repeat transposons. Genetica. 1992;86:275–286. doi: 10.1007/BF00133726. [DOI] [PubMed] [Google Scholar]
- Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. The maize genome contains a Helitron insertion. Plant Cell. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal SK, Hannah LC. Helitrons contribute to the lack of gene colinearity observed in modern maize inbreds. Proc Natl Acad Sci U S A. 2005;102:9993–9994. doi: 10.1073/pnas.0504713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal SK, Oetjens M, Hannah LC. Helitrons: enigmatic abductors and mobilizers of host genome sequences. Plant Sci. 2009;176:181–186. [Google Scholar]
- Langdon T, et al. Fragments of the key flowering gene GIGANTEA are associated with Helitron-type sequences in the Pooideae grass Lolium perenne. BMC Plant Biol. 2009;9:70. doi: 10.1186/1471-2229-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto ELS, Carareto CMA, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- Marquez CP, Pritham EJ. Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics. 2010;185:1507–1517. doi: 10.1534/genetics.110.116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DW, Miller LK. A virus mutant with an insertion of a Copia-like transposable element. Nature. 1982;299:562–564. doi: 10.1038/299562a0. [DOI] [PubMed] [Google Scholar]
- Mochiah MB, Ngi-Song AJ, Overholt WA, Stouthamer R. Wolbachia infection in Cotesia sesamiae (Hymenoptera: Braconidae) causes cytoplasmic incompatibility: implications for biological control. Biological Control. 2002;25:74–80. [Google Scholar]
- Morgante M, et al. Gene duplication and exon shuffling by Helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U S A. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci U S A. 2007;104(29):12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter RTM, Goodwin TJD, Butler MI. Vertebrate helentrons and other novel Helitrons. Gene. 2003;313:201–212. doi: 10.1016/s0378-1119(03)00679-6. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, et al. Multiple waves of recent DNA transposon activity in the bat Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Roulin A, et al. Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol Biol. 2009;9:58. doi: 10.1186/1471-2148-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Choi E, Lynch M, Pritham EJ. DNA transposons and the role of recombination in mutation accumulation in Daphnia pulex. Genome Biol. 2010;11:R46. doi: 10.1186/gb-2010-11-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feshotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements. Mol Biol Evol. 2000;17:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teeling EC, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Fisher T, Nusawardani T. The natural genetic engineering of polydnaviruses. Ann N Y Acad Sci. 2009;1178:146–156. doi: 10.1111/j.1749-6632.2009.05023.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Xu JH, Messing J. Maize haplotype with a helitron-amplified cytidine deaminase gene copy. BMC Genet. 2006;7:52. doi: 10.1186/1471-2156-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LX, Bennetzen JL. Structure-based discovery and description of plant and animal Helitrons. Proc Natl Acad Sci U S A. 2009a;106:12832–12837. doi: 10.1073/pnas.0905563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LX, Bennetzen JL. Distribution, diversity, evolution, and survival of Helitrons in the maize genome. Proc Natl Acad Sci U S A. 2009b;106:19922–19927. doi: 10.1073/pnas.0908008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama M, et al. Possible horizontal transfer of a transposable element from host to parasitoid. Mol Biol Evol. 2001;18:1952–1958. doi: 10.1093/oxfordjournals.molbev.a003735. [DOI] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA, Ishida Y. Transposable elements and an epigenetic basis for punctuated equilibria. Bioessays. 2009;31:715–726. doi: 10.1002/bies.200900026. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. Helitron transposons on the sex chromosomes of the platyfish Xiphophorous maculates and their evolution in animal genomes. Zebrafish. 2006;31:39–52. doi: 10.1089/zeb.2006.3.39. [DOI] [PubMed] [Google Scholar]