Abstract
We present MassSieve, a Java-based platform for visualization and parsimony analysis of single and comparative LC-MS/MS database search engine results. The success of mass spectrometric peptide sequence assignment algorithms has led to the need for a tool to merge and evaluate the increasing data set sizes that result from LC-MS/MS-based shotgun proteomic experiments. MassSieve supports reports from multiple search engines with differing search characteristics, which can increase peptide sequence coverage and/or identify conflicting or ambiguous spectral assignments.
Keywords: Bioinformatics, MS, Parsimony, Protein identification
The data that result from the mass spectrometric analysis of enzymatically digested proteins are peptide sequences, not protein identifications. Connection back to proteins is inferred. This protein inference problem is succinctly summarized by Nesvizhskii and Aebersold [1]. The two main approaches to solve this problem use either probabilistic or discrete methods. The probabilistic approach is exemplified by ProteinProphet [2]. This program uses an expectation–maximization algorithm to determine correct and incorrect protein identifications. This method does not lend itself to an understanding of the structure of the relationships between the proteins and peptides. If two proteins have the same peptide evidence, then they will have the same resultant probabilistic score, barring some differences used by a heuristic rule to adjust the probability. This method could also lead to differentiating proteins based upon peptide evidence that would not be accepted as a valid identification by a reasonable observer.
Discrete methods for solving the protein inference problem depend upon a static list of identified peptides; either a peptide is identified or it is not. An attempt is then made to provide a minimal or maximal set of proteins that explain all of the peptides. A maximal list is trivially derived by non-redundantly listing all proteins that contain one or more peptides from the given set. This provides an upper bound on the set of proteins. The minimal set of proteins is the smallest number of proteins that explains all of the peptide evidence, and this provides a lower bound. There is often more than one minimal set since a given set of peptides may be explained by more than one protein from a set of proteins. One of the principle means of determining these minimal sets is parsimony, which is an interpretation of the data in accordance with Occam’s razor. The purposes of mass spectrometric based proteomic experiments are disparate; whether it is to verify the simple presence of a gene product, or to denote all possible isoforms present from a protein family. Simply providing a minimal list or the maximal list will not do. What is required is an interactive environment that may be used to explore the data at a high level, or drill down to view the source of any given identification and its other possible interpretations, depending upon the experimental requirements.
As applied to the protein inference problem, the principle of parsimony is simply a bijection of the set of proteins into those for which independent evidence exists, and those for which it does not. The idea of parsimony is not new; indeed, this protein inference effort is a continuation of the work that began in our lab with DBParser [3]. This was a web-based solution that did not scale well for larger experiments. Parsimony is also used by ID Picker [4], which is a similar web-based reporting solution that produces static parsimonious protein lists based upon peptide identifications verified by some means of estimating the false discovery rate.
The standards for accepting peptide identification may vary widely from investigator to investigator. It may be a standard cutoff score or one determined by an estimate of the false discovery rate. It may require more than one search engine to verify the identification, or require that the same peptide be identified more than once within the same sample. Permuting these parameters provides a deeper understanding of the data and of the identification standards themselves.
To facilitate communication, a standard nomenclature, as shown in Table 1, has been developed to describe the elements of a parsimonious set of proteins and peptides. Protein identifications that are discrete or differentiable by definition have independent evidence confirming their existence. The rest of the categories do not have independent validation, yet some of them must exist due to the pigeonhole principle (given n items placed in m boxes, if n > m then there exists a box containing more than one item). Strictly speaking, any of these remaining categories may refer to a group of one or more equivalent proteins. For example, if proteins A and B contain peptides x, y, and z, while protein C contains only peptide z, then A and B are supersets of protein C, yet they are still equivalent to each other. For determining minimal protein presence, subsets or subsumables are not considered since there exists more evidence for other proteins, while each group of indistinguishable proteins that are supersets or non-hierarchical equivalents are presumed to have at least one member present.
Table 1.
The ontology of parsimony
| Peptides | |
| Indeterminate | For a given scan, only the top scoring hit is used. If there is more than one match that ties for the top score, or it has been identified as more than one peptide by different sources, then the peptide is indeterminate. |
| Distinct | A peptide that is assigned to exactly one protein. |
| Shared | A peptide that is assigned to more than one protein. |
| Proteins | |
| Discrete | A protein identification that is identified by only distinct peptide(s). |
| Differentiable | A protein identification that can be distinguished from other proteins because it has at least one distinct peptide that is not present in other set of peptide(s) and at least one shared peptide that is present in other set of peptide(s). |
| Superset | A protein identification contains the shared peptides from at least one other subset protein. |
| Subsumable | A protein identification contains shared peptides that can be distributed as subsets two or more other proteins. Formally, subsumable proteins are simply another class of subsets. |
| Subset | A protein identification that contains peptides common to a larger set of peptides corresponding to another protein identification which is a superset. |
| Equivalent | Protein identifications that are based on the same set of shared peptide(s). Also known as indistinguishable. |
MassSieve is an open source rich client application developed in Java and has been tested on Windows, Mac OSX, and Linux. Several open source Java libraries are utilized, including BioJava [5] to read sequence databases, and provide graphical representation of protein sequence coverage, the MascotDatfile library [6] to parse MASCOT results, the Prefuse visualization toolkit to display protein–peptide relationship graphs, the ANTLR parser generator [7] to create a peptide selection set notation language, and the GlazedLists framework is used to display and manipulate tables.
The basic unit of analysis in MassSieve is the Experiment, which is a set of MS/MS peptide identification results all pertaining to one biological sample. Multiple fractions, or instrument runs, may be gathered into a single Experiment and the peptide evidence for each will be given equal weight for determining the protein list. In addition, multiple analyses of the same data (i.e. the same data searched on multiple search engines) may also be loaded into the same experiment.
The output from the various peptide identification algorithms is not standardized and does not agree in the amount and type of information to be provided. Therefore, adding support for a new source of data is the most labor-intensive part of the process. Currently, MassSieve supports Mascot DAT files, OMSSA XML, X!Tandem XML, and most other algorithm’s PepXML output (e.g. SEQUEST, SpectraST) including MASCOT, OMSSA, and X!Tandem PepXML.
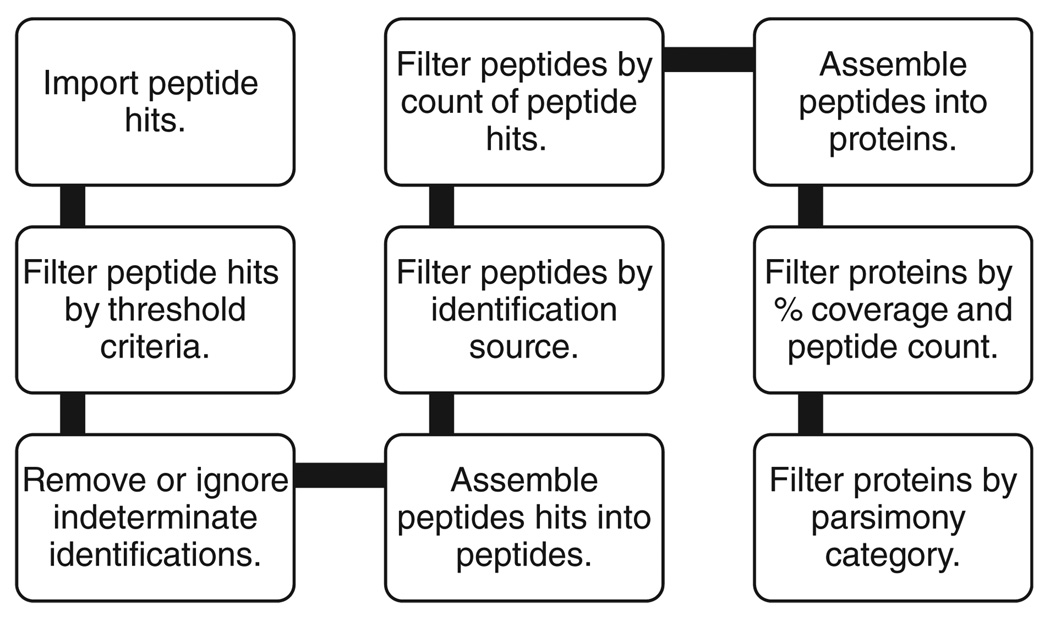
From each data source, peptide identifications for each MS/MS spectrum (scan) are loaded. These are known as peptide hits. Only the top scoring, or set of top scoring hits in the case where the best identification is indeterminate, is loaded. The peptide hits are then filtered as shown in the pipeline in Fig. 1. The ordering of the steps in the pipeline is important, as each step obviously affects the next step. First, each peptide hit is filtered based upon a user-configurable cut-off score depending upon the search algorithm used to determine the identification. For OMSSA and X!Tandem, this is the expectation score. For MASCOT, this is either the expectation score, or requiring the ion score to be greater or equal to the identity score, which by definition is equal to an expectation score of 0.05. SEQUEST results use the XCorrnorm, which is the XCorr normalized by the charge state. This is based upon the same criteria as DTASelect [8], where the normalized XCorr is (XCorr c)/c, where c = 1.8 for charge 1, c = 2.5 for charge 2, and c = 3.5 for charge 3+. If the data source is PepXML that has been processed by Peptide Prophet [9], then the Peptide Prophet probability may be used in lieu of the standard score. If the search algorithm format is not directly supported, then the Peptide Prophet score must be used.
Figure 1.
The MassSieve pipeline filters for processing peptide identifications into protein identifications.
At this point, a peptide hit may be indeterminate because the spectrum that it identifies is also identified as a different peptide by either the same algorithm with the same score (for example, I/L), or by a different algorithm. The choice is to either retain all identifications or remove those spectra and their indeterminate identifications. If the choice is to retain indeterminate identifications, then they are subsequently used as if all of them are independently present and valid, but their presence is noted and displayed for any dependent peptide or protein identification.
All peptide hits are grouped by their identified unique peptide. If a user has loaded data from multiple search algorithms, these peptides may then be filtered by the search algorithms which identified it. This filtering is based upon a modified set notation. This notation may be used to specify any desired combination of results from the input. For example, given the filter string, (M&X)+O, the resultant list will include any peptide that has a peptide hit from OMSSA, or has been identified at least once by both MASCOT and X!Tandem. Note the use of parenthesis to explicitly differentiate the string from M&(X+O), which would result in a very different list of peptides.
The next step is to filter peptides by the number of peptide hits, independent spectra (scans) that were identified as that peptide. For a multiple search engine experiment, this is not the same as the number of peptide identifications. If both OMSSA and MASCOT identified spectrum 506 as ABLLAYLK, then this would still be only one peptide hit. In a similar fashion, proteins may be filtered by the number of member peptides, by the percent sequence coverage, or by both criteria.
Once the filtered list of peptide hits, peptides, and associated proteins is available, then each protein can be assigned to the correct parsimony category, as summarized in Table 1. Parsimony analysis is equivalent to the vertex cover problem, which is classic example of an NP-complete problem. Simply stated, a vertex cover is a subset S of a set of vertices V, where the edges E are contained in the graph G = (V,E), then each edge in E has at least one endpoint in S. To find a minimal S for any given G is generally intractable; however, the problem as applied to proteins and peptides may be mapped to a bipartite graph, for which efficient algorithms do exist. The algorithm described here is slightly modified to account for the parsimony categories.
First, a breadth-first search of every protein to compose a list of proteins related to it by shared peptides, and to assign each protein to a cluster. The cluster has no effect on the algorithm; it is for convenience when displaying graphs of related proteins and peptides. Second, each protein on this list is compared by its set of peptides to separate the list into differentiable, superset, subset, or equivalent proteins. Then, each protein is examined in the following order to determine its parsimony category. If it contains only distinct peptides, then it is discrete. If it contains at least one distinct peptide, and one shared peptide, then it is differentiable. If it has at least one superset protein, then it is a subset. If the protein is linked by shared peptides to two or more differentiable proteins that are themselves differentiable from each other, and all of the protein’s peptides form a subset of the set of peptides of all of its differentiable proteins, then it is subsumable. If none of the preceding applies and it has at least one subset protein, then it is a superset. Finally, if nothing else, it must be an equivalent protein. The time complexity for this algorithm is O(n2m), where n is the number of proteins and m the number of peptides. The space requirement of this algorithm is O(n+m+p), where p is the number of peptide hits.
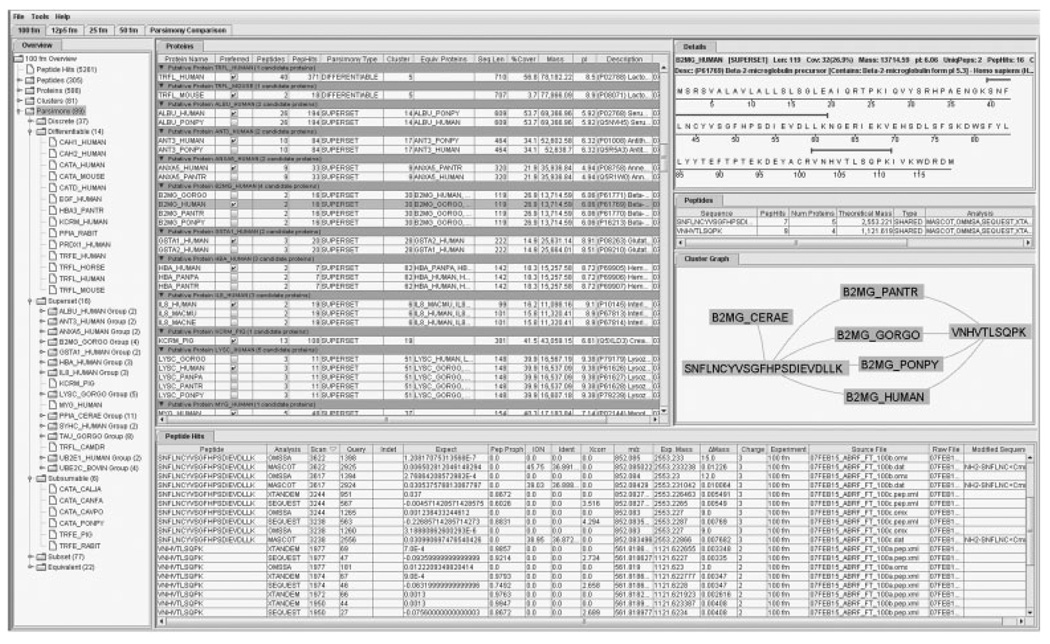
Each experiment contains six sub-windows that can be rearranged at will, as shown in Fig. 2: an overview, three lists windows for proteins, peptides, and peptide hits, respectively, a graphical display of the protein and peptide clusters, and a detailed view. Each view is contextual; if an element is chosen in one window, then the other windows display related information. For example, if a protein is selected in the protein list, then the peptide list displays its list of peptides, the peptide hit list displays all of that protein’s peptide hits, the appropriate cluster graph is displayed, and the detailed view shows the protein sequence coverage. The same is true if a set of proteins or peptides is selected, the remaining views are updated as appropriate.
Figure 2.
Main window of MassSieve. All display areas are moveable and all columns are sortable and moveable. The tree on the left expands to show peptide and protein lists and parsimony hierarchy. Selection of a specific peptide or protein triggers an update of related information in corresponding windows. Information can be shown for total experiment and individual protein. Upper most tabs denote individual experiments or groups, with each experiment containing culled results for each group and corresponding parsimonious protein lists.
The protein list has three different states: it may show the maximal list of proteins, it may show the maximal list grouped by cluster, or it may show the proteins divided into their parsimony categories. Any list view may be exported as a comma separated value (CSV) formatted file, suitable for further processing by other programs. In addition, the protein parsimony list has an additional selection method to choose a representative protein for each equivalent group to facilitate manual designation of a minimal list to be exported as a CSV file. Export options are accessed from a contextual menu displayed by clicking the right mouse button on the selected table.
Once the experimental data have been loaded, the filter criteria may be changed on the fly. The data will be re-evaluated and all the displays updated. Multiple experiments can be loaded and their results compared. There are two types of comparisons: a straightforward listing of proteins, and their status in each experiment, or a more subtle parsimony comparison, which loads all of the peptide hits, peptides, and proteins from all of the experiments to be compared into a new experiment that does a parsimony analysis as it would for a standard experiment, yet adds a new column to each list to keep track of the origin of the data.
MassSieve also facilitates label-free relative quantification based on peptide hits. Peptide hits are output for each peptide and/or as the total peptide hits for each protein. The most visually useful format for peptide hit quantification is the parsimony comparison. Each sample type to be compared is loaded into separate experiments and compared by parsimony comparison. The result is one report that contains a comparative parsimonious minimal protein list with all relevant peptide hit information listed individually for each experiment. The user can output CSV reports with all associated information and choose either protein level information only or include experiment-specific peptide hit counts for each peptide at each time point. Output files are suitable for submission to data repositories and journal supplements.
The source and binary for MassSieve may be downloaded from http://www.ncbi.nlm.nih.gov/staff/slottad/MassSieve/. This site also contains additional information and documentation about the program.
Acknowledgments
The authors thank Dr. J. Kowalak and A. Makusky for helpful discussions throughout this project. This work was supported in part by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health.
Abbreviation
- CSV
comma separated value
Footnotes
The authors have declared no conflict of interest.
References
- 1.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Dondeti V, Dezube R, Maynard DM, et al. DBParser: web-based software for shotgun proteomic data analyses. J. Proteome Res. 2004;3:1002–1008. doi: 10.1021/pr049920x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holland RCG, Down T, Pocock M, Prliæ A, et al. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24:2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helsens K, Martens L, Gevaert K, Vandekerckhove J. MascotDatfile: an open-source library to fully parse and analyze Mascot MS/MS search results. Proteomics. 2007;7:364–366. doi: 10.1002/pmic.200600682. [DOI] [PubMed] [Google Scholar]
- 7.Parr TJ, Quong RW. ANTLR: a predicated-LL(k) parser generator. Softw. Pract. Exp. 1995;25:789–810. [Google Scholar]
- 8.Tabb DL, McDonald WH, Yates JR. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]