Abstract
The glyoxalase pathway involving glyoxalase I (gly I) and glyoxalase II (gly II) enzymes is required for glutathione-based detoxification of methylglyoxal. We had earlier indicated the potential of gly I as a probable candidate gene in conferring salinity tolerance. We report here that overexpression of gly I+II together confers improved salinity tolerance, thus offering another effective strategy for manipulating stress tolerance in crop plants. We have overexpressed the gly II gene either alone in untransformed plants or with gly I transgenic background. Both types of these transgenic plants stably expressed the foreign protein, and the enzyme activity was also higher. Compared with nontransformants, several independent gly II transgenic lines showed improved capability for tolerating exposure to high methylglyoxal and NaCl concentration and were able to grow, flower, and set normal viable seeds under continuous salinity stress conditions. Importantly, the double transgenic lines always showed a better response than either of the single gene-transformed lines and WT plants under salinity stress. Ionic measurements revealed higher accumulation of Na+ and K+ in old leaves and negligible accumulation of Na+ in seeds of transgenic lines as compared with the WT plants. Comparison of various growth parameters and seed production demonstrated that there is hardly any yield penalty in the double transgenics under nonstress conditions and that these plants suffered only 5% loss in total productivity when grown in 200 mM NaCl. These findings establish the potential of manipulation of the glyoxalase pathway for increased salinity tolerance without affecting yield in crop plants.
Genetic manipulation of crop plants for enhanced abiotic stress tolerance holds great promise for sustainable agriculture (1). Abiotic stresses have been shown to have a quantitative character, and thus they are controlled by multiple genes (2, 3). However, there are number of instances where single-gene transfers have led to the development of tolerant plants (2, 4, 5). The increasing problem associated with the rise in soil and water salinity is a major threat to agricultural productivity worldwide. Recent reports have described production of transgenic plants with improved tolerance to salinity after transfer of a single gene such as Na+/H+ antiporter (6, 7). Recently, overexpression of trehalose biosynthetic genes has also contributed toward development of abiotic stress-tolerant genotypes in rice (8). But considering the complex metabolic reactions operating in the cell in response to abiotic stresses, there is a need to test the possible contribution of other candidate genes toward this multigenic trait.
The glyoxalase system has been long known in animal systems (9) and has been proposed to be involved in various functions that include regulation of cell division and proliferation, microtubule assembly, and protection against oxoaldehyde toxicity. Glyoxalase enzymes are important for the glutathione (GSH)-based detoxification of methylglyoxal (MG), which is formed primarily as a byproduct of carbohydrate and lipid metabolism. The reaction catalyzed by glyoxalase I (gly I) and glyoxalase II (gly II) is as follows:
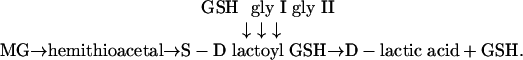 |
MG is a potent mutagenic and cytotoxic compound known to arrest growth, react with DNA and protein, and increase sister chromatid exchange (9). The genes encoding gly I and gly II have been isolated and characterized from microbial and animal systems and found to have significant protein sequence homology (ref. 10 and references therein). The physiological significance of the glyoxalase system has not been clearly defined in plants; however, this system has been often regarded as a “marker for cell growth and division” (11). Gly I has been shown to be up-regulated in tomato in response to salt and osmotic stress and to phytohormonal stimuli (12).
With a desire to investigate and analyze the effect of metabolic engineering of the glyoxalase pathway with respect to salinity tolerance, we transferred two enzymes, i.e., gly I and gly II, into the model plant, tobacco. We had earlier shown that overexpression of the gene for one of the enzymes of this pathway, gly I, resulted in not only improved tolerance against MG, but interestingly, the transgenic plants tolerated higher levels of salinity as compared with the nontransgenic plants (10). In a recent study, gly I has also been found to be one of several genes induced in response to drought and cold stresses in Arabidopsis (13). To perform metabolic engineering for the whole glyoxalase pathway, we have now isolated the gene for the second enzyme in the pathway, gly II, and overexpressed it in WT plants and the gly I transgenic background. In this article, we present the detailed investigation carried out for these different transgenic plants of tobacco pertaining to gene expression, protein synthesis, enzyme activity, performance under stress, and physiological investigations related to toxic ion content inside the plants. The sustained growth of the transgenic tobacco plants and their capability to yield seeds under salinity stress clearly demonstrate the potential of the glyoxalase pathway for improving salinity tolerance in crop plants.
Materials and Methods
Generation of Transgenic Tobacco Plants. The gly I and II ORFs were cloned independently in separate plant transformation vectors. Gly I from Brassica juncea (GenBank accession no. Y13239) was cloned in pBI121 binary vector (Clontech) to give rise to pBI-S1, where both the gly I and the reporter uidA genes are kept under the control of separate CaMV 35S promoters with npt II as the selectable marker as described (10). The gly II gene isolated from Oryza sativa L cv IRBB10 (GenBank accession no. AY054407) was cloned in pCAMBIA1304 as NcoI fragment with hptII as the selectable marker to get pCAM-glyII. In this case, both gly II and the reporter gene gfp:gusA are driven by a single CaMV 35S promoter; however, a stop codon has been inserted in between gly II and the reporter gene to avoid translational fusions. Importantly, different selectable markers for both gly I and gly II gene constructs were used to enable screening for independent single-gene transformants and double transgenics.
For tobacco transformation, the recombinant plasmids were transferred into Agrobacterium tumefaciens (LBA4404) by the liquid nitrogen freeze-thaw method. Tobacco leaf discs (Nicotiana tabaccum cv Petit Havana) were transformed (14) with either gly I or gly II genes independently, or by transferring gly II gene in gly I transgenic plants so as to get the double transformants. The single-gene transformants were selected on appropriate antibiotics, whereas the double transformants were selected on the mixture of kanamycin (50 mg/liter) and hygromycin (25 mg/liter).
PCR and Southern Hybridization. Putative transformants were screened by PCR analysis using tobacco genomic DNA from WT and various transgenic lines as template and 5′ (5′-ATGCGGATGCTGTCCAAGGCG-3′) and 3′ (5′-TTAAAAGTTATCCTTCGCTCG-3′) gly II end primers. For Southern hybridization, 20 mg of genomic DNA from the tobacco lines testing positive for PCR above was digested with NcoI enzyme (cloning site for gly II gene in pCAM-glyII vector), blotted, and probed by using the rice gly II gene according to the standard protocol.
Antibodies, Western Analysis, and Enzyme Assay. The gly II gene was overexpressed in Escherichia coli BL21 (DE3) strain by cloning it in pET28a vector (Novagen). The gly II protein was purified as described (15) and used to raise polyclonal antibodies in rabbits (New Zealand White) as per standard procedures (16). For Western blotting, extraction of soluble proteins from tobacco was essentially carried out as reported (17). The amount of protein was estimated by the Bradford method (18). Twenty micrograms of soluble proteins was resolved on 1D SDS/PAGE and transferred onto nitrocellulose membrane. The specific position of antigen–antibody complex on the membrane was visualized by using alkaline phosphatase linked to secondary antibodies.
For enzyme assays, protein extract was prepared by homogenizing leaf tissue in liquid nitrogen and then resuspending the powder in 2 vol (wt/vol) of extraction buffer (0.1 M sodium phosphate buffer, pH 7.0/50% glycerol/16 mM MgSO4/0.2 mM PMSF/0.2% polyvinyl polypyrrolidone). The enzyme activity for gly I and gly II was determined as described (19, 20). Three different enzyme extractions were done per sample for three independent transgenic lines of each of the single (gly II) and double transformants (gly I+II). The specific activity for both enzymes is expressed in units per mg–1 of protein.
Leaf Disc Assay for Tolerance Against MG and Salinity Stress. Healthy and fully expanded leaves (of similar age) from WT and transgenic plants (60 days old) were briefly washed in deionized water, and leaf discs of 1 cm diameter were punched out and floated in a 6-ml solution of MG (5 and 10 mM, 48 h) or NaCl (400–800 mM, 3 and 5 days) or sterile distilled water (which served as experimental control). The chlorophyll content was measured as described (21). The experiment was repeated thrice with three different transgenic lines.
Transgenic Plants and Salinity Stress Tolerance. To assess the relative salinity tolerance of various plants, WT and T1 generation transgenic seeds overexpressing gly I, gly II, or both were germinated in the presence of respective antibiotics. The surviving seedlings (7 days old) were transferred to either Murashige and Skoog (MS) medium (Sigma) supplemented with 100, 200, or 400 mM NaCl for imposing salinity stress or onto plain MS medium that served as the experimental control. The seedlings were maintained under culture room conditions, and their growth was monitored for 25 days under stress.
In addition to the experiments with seedlings, we carried out the assessment of the transgenic plants for their tolerance toward salinity stress throughout their life cycle. For this purpose, WT and T1 transgenic seeds overexpressing gly genes were germinated on Murashige and Skoog medium containing appropriate antibiotics. The surviving seedlings were transferred to earthen pots and grown in a greenhouse (16 h light/8 h dark, 25°C ± 2°C). Starting 2 weeks after transfer, the plants were watered biweekly with a 200 mM NaCl solution. Three WT and three independent transgenic lines of each type (i.e., gly I, gly II, and double transformants) with three plants each were distributed in two groups, and each group was watered either with 200 mM NaCl solution or water.
Endogenous Ion Content Determination. Mature WT and transgenic plants grown under water or in 200 mM NaCl in a greenhouse for 150 days were used. Roots, old leaves, young leaves, and seeds were collected from three different plants of each type and thoroughly rinsed in deionized water, and the fresh weight of each sample was determined. The samples were dried at 70°C for 48 h, and the dry weight of each sample was measured. The material was ground to a fine powder and digested in concentrated HNO3 overnight at 120°C. Samples were then dissolved in HNO3/HCLO4 (1:1, vol/vol) at 220°C, resuspended in 5% (vol/vol) HNO3, and analyzed for sodium (Na+), potassium (K+), and calcium (Ca2+) content by using simultaneous inductively coupled argon-plasma emission spectrometry (ICP trace analyzer, Labtam, Australia).
Results and Discussion
Metabolic activities in plant cells are very complex, and various biochemical pathways are interconnected with each working in coherence toward cellular homeostasis. Thus, understanding and ultimately modifying these processes may prove to be useful for developing stress-tolerant plants. In this context, we have previously shown that overexpression of one of the enzymes (gly I) of the glyoxalase pathway in tobacco results in not only improved survival under MG but transgenic plants were found to tolerate high levels of salinity (10). The T1 generation of these transgenic plants was analyzed, and it was found that the gly I-overexpressing plants withstand and complete their life cycle under 200 mM NaCl stress (data not shown). This finding indicates that the introduced trait is functionally and genetically stable. This observation prompted us to genetically manipulate the entire glyoxalase pathway.
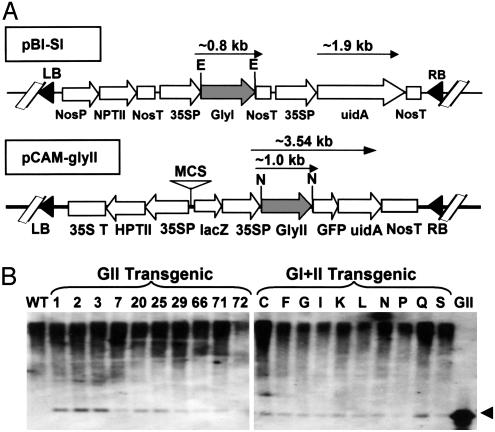
For this purpose, we isolated the full-length gene for the second enzyme of this pathway, gly II, from O. sativa (GenBank accession no. AY054407). This gene showed significant sequence homology to other gly II genes in the data bank (data not shown). To analyze the contribution of gly I and gly II transgenes individually and in concert with each other toward stress tolerance, rice gly II cDNA was introduced into tobacco plants in the WT background or the gly I transgenic plant line (to produce double transgenics). The gene constructs developed for plant transformation are shown in Fig. 1A.
Fig. 1.
Transformation of tobacco by using glyoxalase pathway genes. (A) Schematic representation of various glyoxalase constructs used to overexpress gly I enzyme (pBI-SI) and gly II enzyme (pCAM-glyII) in tobacco plants. (B) Testing of various gly II (GII) and double (GI+II) transgenic lines for the presence of gly II transgene by Southern hybridization. The line number of each type of transgenic is given at the top. The presence of a 1.0-kb band on the Southern blot is shown by an arrowhead.
The Single and Double Glyoxalase Transgenic Plants Are Phenotypically Similar and Maintain the Transgenes. The transgenes did not exhibit any effect on the regeneration, growth, or morphology of the resultant single or double transgenic plants. Thirty putative independent transgenic lines of gly II and double transformants, growing on the antibiotic selection medium, were initially screened by using the histochemical β-glucuronidase (GUS) assay. It was observed that in some of the hygromycin-resistant plants GUS expression was not seen. The transgenic nature of the plants was also checked by PCR using tobacco genomic DNA as the template and rice gly II-specific primers (data not shown).
A total of 10 independent PCR-positive plants were further confirmed for the stable integration of the transgenes by Southern hybridization using equal amounts of transgenic tobacco genomic DNA digested with NcoI and probed with rice gly II cDNA. The presence of 1.0-kb gly II DNA in gly II and double transgenic plants was observed (Fig. 1B), because digestion with NcoI released the rice gly II DNA fragment from the pCAM-gly II construct used in the transformation experiment. The intensity of the hybridized signal in each lane was variable, indicating the multiple insertion of the gly II gene in the tobacco genome.
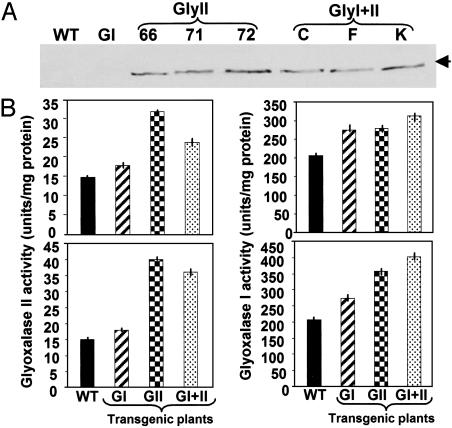
Glyoxalase Transgenic Plants Show Increased Levels of Gly I and II Protein and Enzyme Activity. Western blot analysis of selected single or double transgenic plants using the antibodies raised against rice gly II protein indicated the functional activity of the foreign gene in transgenic plants, leading to accumulation of foreign protein gly II (Fig. 2A). The size of the gly II protein in the two types of plants is also comparable (36 kDa), indicating that functional aspects of the gly II gene are not affected by the constitutive overexpression of the gly I gene. Moreover, different amounts of the gly II protein were detected in different transgenic lines when an equal amount of total protein was loaded for Western blotting, whereas no crossreaction was detected in the WT and gly I transgenic plants. Three independent lines of gly II (66, 71, and 72) and double transgenics (C, F, and K) that showed significant accumulation of gly II protein (Fig. 2 A) were used for all further analysis. Further, both gly I and gly II enzyme activity was measured in the same three transgenic lines for each of the transformant types. The data for two such lines show that on the basis of total protein the gly II enzyme activity was higher in line 72 than in line 66 (60% and 55% more, Fig. 2B Lower Left and Upper Left, respectively) as compared with WT levels. In the case of double transgenics, the gly II activity in line K>C was 58% and 38% (Fig. 2B Lower Left and Upper Left, respectively). Interestingly, the gly II activity in gly I plants was found to be at least 12% more than in WT.
Fig. 2.
(A) Western blot analysis of WT and various glyoxalase transgenic plants (GI, gly I; GlyII, gly II; and GlyI+II, double transgenics) carried out with antibodies raised against rice gly II protein. The line number of each type of transgenic is given at the top. The presence of 36-kDa gly II protein is marked with an arrow. (B) Histograms showing the activity of the gly II (Left) and gly I (Right) enzymes in the selected transgenic lines: NtBIS-11 for gly I; line 66 (Upper Left) and 72 (Lower Left) for gly II; and line C (Upper Right) and K (Lower Right) for glyI+II. The standard deviation is indicated by each bar in the graph (n = 3).
Further, in the same set of transgenic lines, gly I activity was measured (Fig. 2B Right). An average of 25% enhancement in gly I activity over the WT was noted in gly I transgenic lines on the basis of total protein content. In gly II transgenic lines, gly I activity was significantly high (43% in line 72 and 27% in line 66, Fig. 2B Lower Right and Upper Right, respectively), indicating that overexpression of gly II enzyme probably leads to an increase in the endogenous gly I enzyme. A similar enhancement of gly I activity was noted in double transgenic plants with line K showing 59% and line C showing 35% (Fig. 2B Lower Right and Upper Right, respectively) as compared with their untransformed counterpart.
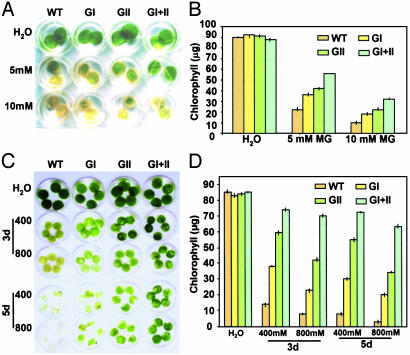
Glyoxalase Transgenic Plants Can Tolerate High MG Levels. We conducted studies to determine whether overexpression of gly I and gly II enables the transgenic plants to survive better under high MG. These experiments were carried out on the three lines tested for enzyme activity; however, data for only one of the lines of each type are presented (line 72 for gly II and line K for double transgenic). To make functional correlations in vivo, we checked the capability of double transgenic plants to tolerate MG. For this purpose, leaf discs from WT, gly I, gly II, and double transgenic plants were floated in 5 and 10 mM solution of MG for 48 h; leaf discs floated in water served as experimental control. Fig. 3A shows the phenotypic differences among WT, gly I, gly II, and double transgenic plants after 48 h of MG treatment. Based on visual observations, it was found that double transgenics could tolerate MG at higher concentrations than the gly I and gly II individual transformants (Fig. 3A). Measurement of chlorophyll content indicated that the leaf discs of the double transgenics retained 64% of chlorophyll, whereas the WT plants retained only 33% of their chlorophyll in 5 mM MG (Fig. 3B). There was a minimum loss of chlorophyll in the WT, gly I, gly II, and double transgenics incubated in water for 48 h, suggesting that overexpression of glyoxalase pathway enzymes does not impose a metabolic stress on the transgenic plants. The ability of the transgenic plants to maintain physiological levels of chlorophyll under this stress condition was taken as an index for the measurement of the injury caused by the stress. At 5 or 10 mM MG, the double transgenics performed better (i.e., they were able to maintain higher levels of chlorophyll contents in the tissue subjected to stress) than the single transformed lines.
Fig. 3.
Retardation of MG- and salt stress-promoted senescence in transgenic tobacco plants overexpressing either gly I (GI), gly II (GII), or both gly I and II in double transgenics (GI+II), indicating the tolerance at cellular levels toward toxic levels of MG and salt. Phenotypic differences (A) and chlorophyll content (B) (μg/g of fresh weight) from MG-treated leaf discs of WT and various transgenic plants (GI, GII, and GI+II) after incubation in 5 and 10 mM solutions of MG for 48 h are shown. Discs floated in water served as the experimental control. Phenotypic differences (C) and chlorophyll content (D)(μg/g of fresh weight) from sodium chloride-treated leaf discs of WT and various transgenic plants (GI, GII, and GI+II) after incubation in 400 and 800 mM solutions of NaCl for 3 and 5 days are shown. Discs floated in water served as the experimental control. The standard deviation in each case is represented by the vertical bar in each graph (n = 3). Note the difference in retention of chlorophyll in WT and transgenic plants.
Glyoxalase Transgenic Plants Can Tolerate High Levels of Salinity. For assessing the potential of transgenic plants for relative tolerance toward salinity stress, tests were performed on isolated leaf discs, seedlings of certain age, and complete mature plants. In all of these analyses, comparison of the single-gene transformants with the WT genotype and the double transgenic plant was conducted.
Incubation of the leaf discs obtained from WT, gly I, gly II, and double transgenic plants in 400 and 800 mM sodium chloride showed an early bleaching of WT leaf discs compared with transgenic plants, as shown in Fig. 3C. At both salt concentrations (400 and 800 mM), the decrease in chlorophyll content in all three types of transgenic plants was less than in the WT plants (Fig. 3D). The double transgenic plants exhibited more improved salinity tolerance than any of the single-gene transformants (Fig. 3 C and D). There was a total loss in chlorophyll content in the WT plants (≈85% loss), whereas the double transformants experienced a 14% decline in chlorophyll at 800 mM NaCl concentration (Fig. 3D). These observations establish a positive relationship between the overexpression of glyoxalase pathway enzymes and salinity stress tolerance in leaf tissues.
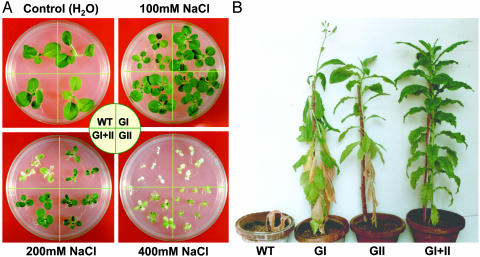
Salt tolerance of T1 generation transgenic seedlings was checked by transferring them to various concentrations of NaCl and monitoring their growth for 25 days. All four types of plants, i.e., WT, gly I, gly II, and double transgenics, showed comparable growth in the absence of NaCl (Fig. 4A Upper Left). However, under 100 mM NaCl, there was a reduction in growth of the WT plants, but the other three types of transgenic plants grew very well (Fig. 4A Upper Right). When exposed to 200 mM NaCl, the WT plants showed drastic reduction in growth, whereas the transgenic lines tolerated this degree of salinity (Fig. 4A Lower Left). Although there was a slight reduction in the overall growth of all of the transgenic plants, growth of double transgenics under salinity seemed better as compared with either of the individual transformants. Even when subjected to 400 mM NaCl, the bleaching of leaves in double transgenics was the lowest, whereas the WT seedlings became totally bleached out and showed no growth (Fig. 4A Lower Right). This observation indicates that overexpression of the entire glyoxalase pathway confers a high degree of salinity tolerance in transgenic plants. At this stage, gly I and II enzyme activities were measured in the T1 generation transgenic plants growing under water and in saline conditions. For both enzymes, a further enhancement in their activity was noted in response to NaCl, thereby reflecting the regulation of endogenous glyoxalase enzymes by salt (data not shown).
Fig. 4.
Relative salt tolerance of WT and glyoxalase-overexpressing transgenic T1 generation tobacco plants (GI, gly I; GII, gly II; and GI+II, double transgenics) at seedling and whole mature plant level. (A) Seedlings were grown on medium supplemented with 0, 100, 200, and 400 mM NaCl for 25 days. (B) WT and transgenic plants were grown in the continued presence of 200 mM NaCl for 98 days. Note that WT plants could not sustain growth under this condition.
To assess whether the enhanced expression of the glyoxalase enzymes would allow plants to grow, mature, and set seeds under high-salt conditions, all four types (WT, gly I, gly II, gly I+II) of plants were grown in the continued presence of 200 mM NaCl (Fig. 4B; representative plants are shown). The growth of WT plants was severely affected under these conditions as evidenced by their stunted growth and ultimate death. On the other hand, the transgenic plants grew, flowered, and produced normal viable seeds. Here, the growth and survival of double transgenics was much better as compared with the individual transgenics. In single-gene transformants, the older leaves showed signs of bleaching and the total foliage was also less as compared to the gly I+II transformants, whereas the double transgenics showed the least amount of symptoms of stress injury without any loss of chlorophyll and reduction in growth. These results demonstrate the ability of double transgenics to grow under saline soils.
Glyoxalase Transgenics Flowered and Produced Viable Seeds Under Salinity. Because the glyoxalase transgenic plants were found to sustain growth under salinity, it was crucial to analyze its ultimate effect on seed production and yield. For this purpose, critical growth parameters like plant height, fresh weight of leaves, time required for flowering, and seed weight were scored. The T1 generation transgenic plants behaved similarly to the WT counterparts in all aspects analyzed when grown under nonstress conditions, indicating that overexpression of glyoxalase pathway genes do not pose any growth or yield penalty on the transgenic plants (data not shown).
To further check how these transgenic plants behave when grown under saline conditions, various parameters were scored for T1 generation transgenic plants growing under 200 mM NaCl vis-a-vis WT plants grown under water (Table 1). It should be noted here that similar data for WT plants grown under salinity could not be obtained as these plants failed to sustain growth in the presence of salt after 20 days. Here, no major difference in the overall performance or total seed yield of the double transgenic plants grown in the presence of 200 mM NaCl was found as compared with the WT grown in water. Each type of transgenic plant was able to enter the reproductive phase and set normal viable seeds under stress conditions, strongly indicating the ameliorating effect of the glyoxalase transgene on seed productivity and yield of the transgenic plants. The double transgenics under salinity were able to produce 95% of the total seeds when compared with the WT plants grown in water, whereas the gly I and gly II transgenic lines yielded 80% and 83%, respectively (Table 1). These data document that seed production under salt stress is not affected in gly I and II overexpression.
Table 1. Comparison of various growth parameters and seed production of the WT and glyoxalase transgenic tobacco plants (GI, gly I; GII, gly II; and GIGII, double transgenics) grown in the presence of water and 200 mM NaCl, respectively for 4 months.
| H2O
|
200 mM NaCl
|
|||
|---|---|---|---|---|
| Parameter | WT | GI | GII | GIGII |
| Height, cm | 110.3 ± 6.88 | 120.1 ± 8.43 | 90.6 ± 4.66 | 100.9 ± 5.04 |
| Fresh weight of leaf, g* | 3.9 ± 0.60 | 3.9 ± 0.42 | 3.9 ± 0.31 | 4.0 ± 0.75 |
| No. of days to flower | 115.2 ± 3.15 | 90.8 ± 4.07 | 125.6 ± 5.19 | 117.1 ± 2.59 |
| Seed weight per pod, mg | 162.9 ± 4.58 | 129.8 ± 5.16 | 135.6 ± 3.38 | 154.9 ± 4.96 |
The average weight of the fourth leaf from top. Each value is the mean of ± SD (n = 15).
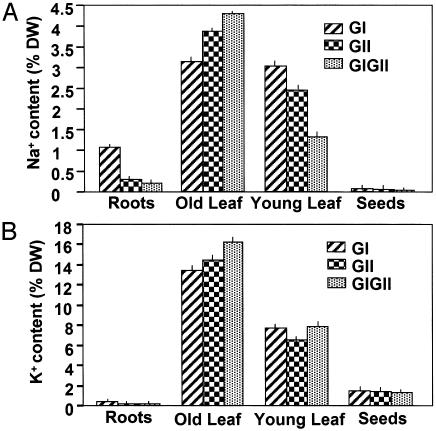
Glyoxalase Transgenic Plants Retain More Cations than WT. It has been reported that the sodium ion sequestration in transgenic plants can be altered to improve stress tolerance without causing a major effect on seed or fruit quality (6, 7). To determine whether glyoxalase transgenic plants accumulate Na+, we carried out analysis related to accumulation of cations. These studies showed that Na+, K+, and Ca2+ accumulate to different levels in various tissues, with old leaves accumulating greater amounts than young leaves, roots, and seeds. In transgenic plants, accumulation of sodium ions in young leaves and seeds was significantly lower than in the old leaves (Fig. 5A), which seem to function as ion sinks, thus keeping the seeds essentially free from the additional uptake of Na+ ions.
Fig. 5.
Na+ (A) and K+ (B) content [calculated as percent dry weight (DW) of the tissue] in various tissues of the glyoxalase transgenic plants (GI, gly I; GII, gly II; and GIGII, double transgenics) grown under the continued presence of 200 mM NaCl. In the histogram, each of the transgenic types is indicated by different patterned bars as shown. For each determination, roots, old leaf (fourth leaf from the bottom), young leaf (second leaf from the top), and seeds were collected from three different plants of each type. Values are the mean ± standard deviation (n = 3). Similar data for WT plants could not be obtained as these plants did not grow further in the presence of 200 mM NaCl. However, the relative values for Na+ in the WT plants grown in water were found to be 0.5% in roots, 0.1% in old leaf, 0.4% in young leaf, and 0.05% in seeds, and those for K+ were 1.5% in roots, 1.8% in old leaf, 2.0% in young leaf, and 1.0% in seeds (data not shown).
Under salt stress, the glyoxalase transgenic lines also accumulated higher amounts of K+ ions in the tissues (Fig. 5B), whereas no major change in the level of total Ca+ was noted (data not shown). It is generally known that the maintenance of K+/Na+ homeostasis is an important aspect of salinity tolerance and that transgenic plants showing higher K+/Na+ levels are able to tolerate salinity stress (8, 22, 23).
Conclusions
Significant progress has been made toward developing salinity-tolerant plants via biotechnology. Reports have suggested that although abiotic stress is a multigenic trait, salinity stress-tolerant plants could be produced by transgenic approaches by the transfer of a single gene (6, 7, 24, 25). In this article, we provide experimental evidence to indicate that manipulation of two genes of the glyoxalase pathway enhances tolerance to salinity and that transgenic plants are able to complete their life cycle and set normal viable seeds under stress conditions without yield penalty.
Three different mechanisms for stress tolerance have been suggested: maintaining ion and osmotic homeostasis, regulating cell division and growth, and detoxification and cellular repair (26). It appears that the glyoxalase pathway might be operating through detoxification and cellular repair. Glutathione is one of the major redox potential-regulating components of the cell, whose level is altered by MG to form S-d lactoyl glutathione. The overexpression of glyoxalases could enhance the level of reduced glutathione that presumably helps to detoxify reactive oxygen species. However, we have yet to determine how glyoxalase metabolism is linked with the salinity-tolerance mechanism in plants. This pathway probably interacts with other physiological processes in the cell for selective uptake and sequestration of ions during salinity stress.
Finally, this study presents an additional role of the glyoxalase pathway under stress conditions in plants and also provides an example of the exploitation of this biochemical pathway for engineering salinity tolerance. The improved performance of double transformants vis-a-vis single transformed lines with respect to stress tolerance and productivity demonstrates that engineering of the entire pathway is more effective than overexpressing either of the components alone. These findings suggest that engineering the glyoxalase pathway in crop plants can result in improved salinity-stress tolerance.
Acknowledgments
We thank Dr. N. B. Sarin and Mr. Mukesh, School of Life Sciences, and Drs. V. Rajamani and J. K. Tripathi, School of Environmental Sciences, JawaharLal Nehru University, New Delhi for extending help in the work related to antibody production and ionic content measurements, respectively. This work was supported by grants from the International Centre for Genetic Engineering and Biotechnology.
Abbreviations: MG, methylglyoxal; gly I, glyoxalase I; gly II, glyoxalase II.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY054407).
References
- 1.Altman, A. (1999) Electronic J. Biotechnol. 2, 51–55. [Google Scholar]
- 2.Cushman, J. C. & Bohnert, H. J. (2000) Curr. Opin. Plant Biol. 3, 117–124. [DOI] [PubMed] [Google Scholar]
- 3.Hasegawa, P. M., Bressan, R. A., Zhu, J. K. & Bohnert, H. J. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. [DOI] [PubMed] [Google Scholar]
- 4.Singla-Pareek, S. L., Reddy, M. K. & Sopory, S. K. (2001) Proc. Indian Natl. Sci. Acad. 67, 265–284. [Google Scholar]
- 5.Datta, S. K. (2002) in Genetic Engineering of Crop Plants for Abiotic Stress, JIRCAS Working Report, ed. Iwanaga, M. (Japan International Research Center for Agricultural Sciences, Ibaraki), pp. 43–53.
- 6.Zhang, H. K. & Blumwald, E. (2001) Nat. Biotechnol. 19, 765–768. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, H. K., Hodson, J. N., Williams, J. P. & Blumwald, E. (2001) Proc. Natl. Acad. Sci. USA 98, 12832–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg, A. K., Kim, J. K., Owens, T. G., Ranwala, A. P., Choi, Y. D., Kochian, L. V. & Wu, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornalley, P. J. (1990) Biochem. J. 269, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veena, Reddy, V. S. & Sopory, S. K. (1999) Plant J. 17, 385–395. [DOI] [PubMed] [Google Scholar]
- 11.Paulus, C., Knollner, B. & Jacobson, H. (1993) Planta 189, 561–566. [DOI] [PubMed] [Google Scholar]
- 12.Espartero, J., Sanchez-Aguayo, I. & Pardo, J. M. (1995) Plant Mol. Biol. 29, 1223–1233. [DOI] [PubMed] [Google Scholar]
- 13.Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y. & Shinozaki, K. (2001) Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229–1231. [Google Scholar]
- 15.Pareek, A., Singla, S. L. & Grover, A. (1995) Plant Mol. Biol. 29, 293–301. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, E. & Lane, D. (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab Press, Plainview, NY).
- 17.Zivy, M., Thiellement, H., de Vienne, D. & Hofmann, J. P. (1983) Theor. Appl. Genet. 66, 1–7. [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 19.Ramaswamy, O., Guha-Mukherjee, S. & Sopory, S. K. (1983) Biochem. Int. 7, 307–318. [Google Scholar]
- 20.Maiti, M. K., Krishnasamy, S., Owen, H. A. & Makaroff, C. A. (1997) Plant Mol. Biol. 35, 471–481. [DOI] [PubMed] [Google Scholar]
- 21.Arnon, D. I. (1949) Plant Physiol. 24, 1–15. Ridderstrom, M. & Mannervik, B. (1997) Biochem. J. 322, 449–454. [Google Scholar]
- 22.Epstein, E. (1998) Science 280, 1906–1907. [DOI] [PubMed] [Google Scholar]
- 23.Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., Koiwa, H., Zhu, J. K., Bressan, R. A. & Hasegawa, P. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1999) Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- 25.Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K. & Izui, K. (2000) Plant J. 23, 319–327. [DOI] [PubMed] [Google Scholar]
- 26.Zhu, J. K. (2002) Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]