Abstract
Aim
To determine the cellular location and expression of MUC17 mucin in specimens of normal, inflamed and neoplastic colon.
Methods
Immunohistochemical analysis of human surgical resection specimens (n=106) was performed with a specific antibody to the MUC17 apomucin protein. A semi-quantitative scoring system was used to measure MUC17 expression. In various colon cancer cell lines, the MUC17 expression was examined by immunoblot analysis and normal RT-PCR.
Results
MUC17 was highly expressed on the surface epithelium and crypts of colonic mucosa. In contrast, the expression of MUC17 was significantly decreased in colonic mucosa of chronic ulcerative colitis (p<0.0001) and ischaemic colitis (p=0.003). Similarly, MUC17 expression was decreased in hyperplastic polyps (p=0.0003), tubular and tubulovillous adenomas (p<0.0001) and colon cancers (p<0.0001). Furthermore, of eight different colon cancer cell lines, MUC17 expression was only detected in LS174T and LS180 cells.
Conclusion
Results indicate that the potential protective effects of this membrane-bound mucin are primarily or secondarily diminished in inflammatory and neoplastic conditions. Further research is needed to determine the specific role of MUC17 in the pathogenesis of these conditions.
INTRODUCTION
Mucins comprise a family of large O-linked glycoproteins expressed by epithelial cells of tubular organs in the human body. Thus far, a total of 21 mucin genes have been identified including MUC1, MUC2, MUC3A, MUC3B, MUC4, MUC5AC, MUC5B, MUC6–9, MUC11–13, MUC15–17 and MUC19–21.1–5 The mucins are classified into two subgroups: the membrane-bound mucins that are expressed at the apical cell surface of gastric pit cells, intestinal enterocytes; and colonic columnar cells and secreted mucins that are found in secretory cells such as gastric mucous cells and intestinal goblet cells.6 Due to their specialised structure, mucins play various roles in normal and pathological conditions.7 The protective mucous barrier of the intestine is composed of membrane bound mucins anchored at the surface of the epithelial cells and by a viscous gel composed of secreted mucins. Deregulated mucin production has been associated with various types of cancer and inflammatory disorders.
Inflammatory bowel disease (IBD) results from a complex and abnormal mucosal immune response to commensal microorganisms primed by infection with a specific pathogen or an impaired mucosal barrier.8, 9 An intact intestinal mucus layer is thought to be essential for protection, and an altered mucus composition has been identified in patients with IBD. Numerous studies have demonstrated alterations in mucin expression, reduced O-glycosylation and sulphation, and increased mucin sialylation in IBD, likely resulting in altered viscoelastic properties of mucus, hence reducing the protective function of the mucus layer.10, 11 MUC2 synthesis, secretion, and sulphation are all reduced in active ulcerative colitis, which would make the colonic mucosa more accessible to toxic agents and pathogens. Mice lacking MUC2 spontaneously develop colitis, and over time develop adenocarcinoma.12 In humans, the region of the membrane-bound mucin gene cluster (MUC3A/B, MUC12 and MUC17) has been implicated in genetic susceptibility to IBD.13, 14 The rare variable number of tandem repeat (VNTR) allele of the MUC3A mucin gene was more common in patients with ulcerative colitis compared with controls. Therefore, the mutated MUC3 gene may result in a defective protein that would increase susceptibility to IBD.15
Clinical evidence has shown that patients with long lasting IBD are at an increased risk for developing colon cancer. Inflammation of the colon is hypothesised to predispose to abnormal cell growth which over time can give rise to adenoma (dysplasia) and adenocarcinoma. However, colon cancer is a heterogeneous and multifactorial disease. Adenomatous polyps (tubular adenoma and tubulovillous adenoma) are recognised as precursor lesions to colon cancer. Additionally, through a different molecular mechanism, non-adenomatous polyps (eg, hyperplastic polyps) may develop into serrated adenomas (an aggressive type of adenoma). Alterations in expression and post-translational modifications of several membrane bound mucins including MUC1, MUC2, MUC3 and MUC4 have been reported previously in colorectal polyps and other colonic neoplasms.16
The primary structure of the MUC17 protein harbours a signal peptide, a large tandemly repeated central domain (TR), two epidermal growth factor (EGF)-like domains, a SEA domain, a transmembrane domain (TM) and an 80 amino acid cytoplasmic tail. The long N-terminal extracellular domain of MUC17 can potentially affect cell-to-cell adhesion by altering the interaction of cell adhesion molecules and is a site of extensive glycosylation. The cytoplasmic domain has many predicted phosphorylation sites that may mediate signal transduction. The role(s) of the two EGF-like domains is not completely understood. The similarity of human MUC17 to rodent Muc3 (mouse and rat) was first reported by Gum et al.17 Further, the analysis suggests that MUC17 diverged from human MUC3 earlier in evolution than the divergence of primates and rodents.17 Mouse Muc3 (mMuc3) is therefore currently believed to be the orthologue of human MUC17 (separated by a speciation event). A higher degree of sequence similarity exists between Muc3 and MUC17 than between Muc3 and MUC3.18 The N-terminal domain for MUC17 is coded by two exons, whereas for MUC3, it is coded by a single exon. Therefore, the degree of sequence homology between the carboxy-extremity of MUC17 and Muc3 is higher than that between MUC3 and Muc3.17
Little is known concerning the specific functions of the intestinal membrane-bound mucins. A recombinant protein corresponding to the cysteine-rich EGF-like domains of the mouse Muc3 mucin and the human MUC17 mucin have been shown to inhibit apoptosis and stimulate cell migration in intestinal cell models; and rectal treatment with exogenous recombinant Muc3 or MUC17 cysteine-rich domain (CRD) proteins ameliorates experimental models of colitis, suggesting that these mucins play an active role in mucosal restitution.19, 20 To date, very little data exists regarding the expression of the MUC17 membrane-bound mucin in inflammatory diseases of the intestine. We hypothesised that the down-regulation of the MUC17 mucin may occur in intestinal inflammation, thus potentially contributing to a reduced mucosal barrier.
Therefore, our aim is to determine the specific cell type expression and subcellular localisation of the MUC17 intestinal mucin in normal human colon specimens, and compare this with ulcerative colitis, acute ischaemic colitis, hyperplastic and neoplastic colon. The results of our study indicate that MUC17 is highly expressed in normal colonocytes both in the proximal and distal colon, and that the expression of this protein is markedly reduced in inflammatory and neoplastic conditions. These results suggest that MUC17 plays a pathogenetic role in inflammatory and neoplastic conditions of the colon. Further detail studies are warranted to determine the specific functional significance of MUC17 in these conditions.
MATERIALS AND METHODS
Antibody
A polyclonal antibody against a synthetic peptide corresponding to a portion of the MUC17 tandem repeat sequence (PTTAEGTSMPTSTPSE) was developed, as reported previously.18
Collection of tissue specimens
Human tissue specimens were obtained from surgical resections and biopsies in accordance with human studies guidelines and approval by the ethics committee of the University of California, San Diego. Clinical data was recorded from medical records and then the identifying information was removed. Tissues for immunohistochemical use were fixed in buffered formalin and embedded in paraffin using standard methods. Normal colon specimens were obtained from patients undergoing colonoscopy and who had normal colonoscopic exams and normal histology.
Immunohistochemistry
The slides were baked at 58°C overnight and then deparaffinised by washing in xylene (4×10 min). Sections were hydrated through graded alcohol and endogenous peroxidase activity was quenched by incubating the sections in 3% H2O2 in methanol for 30 min. After further washing with tap water for 15 min, antigen retrieval was done by heating the slides in citrate buffer (0.01M, pH 6.0) at 80°C for 15 min. After heating, the samples were allowed to cool for 15–20 min at room temperature. This was followed by washing with phosphate-buffered saline (PBS) (2×5 min). Non-specific binding was blocked by incubating the sections with 2.5% normal horse serum for 30 min (ImmPress Reagent Kit, Vector, California, USA). Sections were then incubated with anti-MUC17 polyclonal antibody (1:200) at 4°C overnight. Then slides were washed with phosphate buffered saline with 0.05% tween 20 (PBST) (4×10 min) and incubated with secondary antibody (peroxidase labelled universal anti-mouse/anti-rabbit IgG, ImmPress reagent Kit, Vector Laboratories, Burlingame, California, USA) for 30 min. Then sections were washed with PBST (4×10 min) followed by treatment with DAB reagent (Vector Laboratories, Burlingame, California, USA) as per the manufacturer’s instructions. After washing with PBS, counterstaining was done using haematoxylin (Vector Laboratories, Burlingame, California, USA). With further washing in tap water, sections were dehydrated in graded alcohol and after air drying; the slides were mounted in permount mounting media (Fisher Scientific, Fair Lawn, New Jersey, USA).
Assessment of antigen staining
All the slides were observed under a Nikon Eclipse E400 light microscope (Nikon Corporation, Tokyo, Japan) and representative photos were taken. The intensity of MUC17 immunoreactivity was scored by a gastrointestinal pathologist (PS). Staining intensity was graded and a composite score was obtained as detailed elsewhere.21 Negative controls consisted of slides stained without primary antibody, which resulted in negative staining.
RNA isolation and reverse transcription-PCR analysis
Total RNA from different colon cancer cell lines was isolated by RNeasy mini kit (Qiagen, Maryland, USA) according to the manufacturer’s instructions. The normal PCR was carried out as reported previously.22 Actin was used as an internal control. The PCR products (of 517 bp) were electrophoretically resolved on 1% agarose gels stained with ethidium bromide. Photographs were taken under UV light, using the gel documentation system (Bio-Rad, Hercules, California, USA). The sequence of the forward primer for MUC17 is: 5′-GCC AGT TCA TCT CCT CCC A-3′ and that of the reverse primer is: 5′-AGG AAG TGT TGA AGC TGT G-3′.
Immunoblot assay
All the cell lines (HT-29, LS174T, SW480, HCT-15, LoVo, LS-180, COLO-205 and WiDr) were processed for extraction of whole cell protein; using a standard protocol western blotting was carried out. Due to the large size of MUC17, the protein samples were resolved by electrophoresis on a 2% sodium dodecylsulphate–agarose gel under reducing conditions and processed as reported previously.18
RESULTS
MUC17 immunoreactivity in normal colon tissue
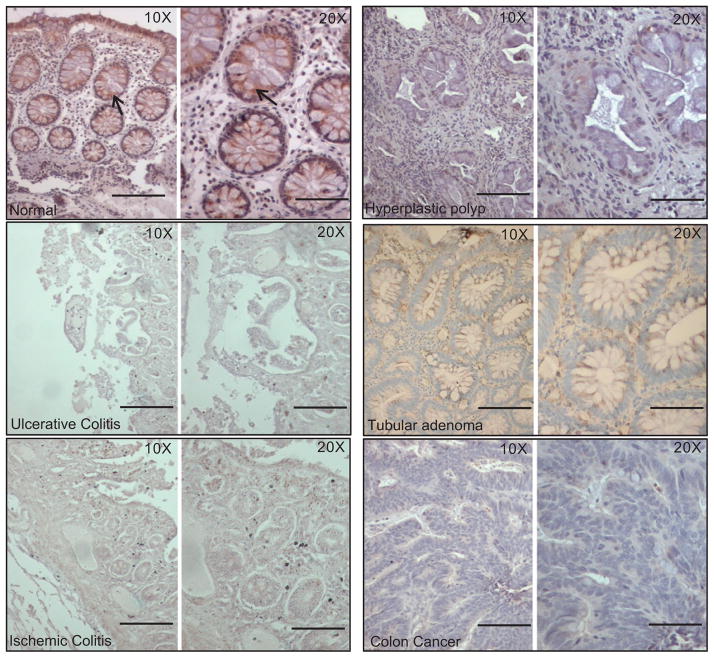
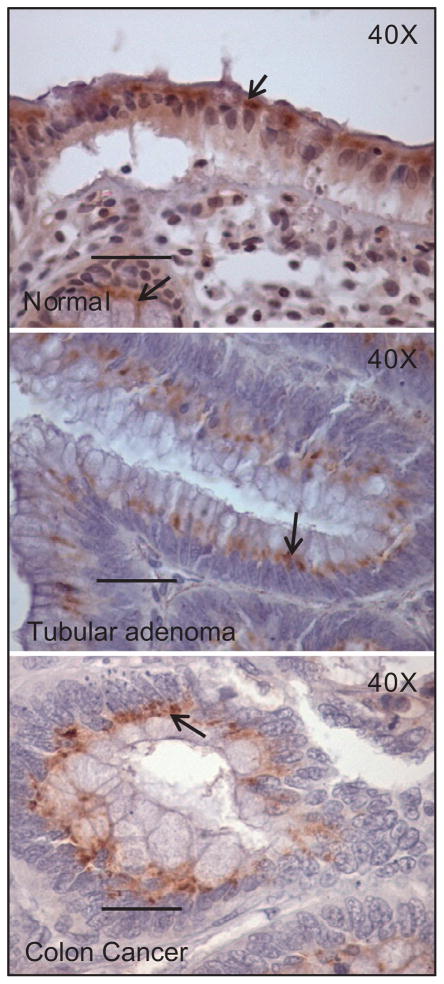
MUC17 was highly expressed on surface and crypt columnar epithelial cells (figure 1, n=12). As shown in figure 2, MUC17 expression was primarily localised to the supranuclear area, with some specimens exhibiting a diffuse cytoplasmic staining in columnar cells. The apical membrane and glycocalyx of most columnar cells did not contain MUC17 immunoreactivity. With respect to inter- and intra-specimen variation, specimens derived from healthy individuals confirmed that there was no significant difference in intensity of MUC17 expression.
Figure 1.
The expression of MUC17 in normal and pathological colon tissues (ulcerative colitis, ischaemic colitis, hyperplastic polyp, tubular adenoma and colon cancer) by immunohistochemistry. The colon tissue sections were cut from the paraffin-embedded blocks and stained with anti-MUC17 polyclonal antibody. All sections were examined under a microscope and the immunoreactivity was judged by dark brown staining. Anti-MUC17 antibody showed very faint or no staining in different pathological slides. However, the normal colonic epithelial cells showed a strong staining (arrows). Bar=0.5 inches.
Figure 2.
Subcellular localisation of MUC17 in normal and pathological colon tissues (tubular adenoma and colon cancer). Immunohistochemistry showed that in both normal and pathological colons, such as tubular adenoma and colon cancer (in which sporadic staining was found), MUC17 was mostly detected in apical columnar cells, and with a supranuclear pattern of staining (arrow).
MUC17 immunoreactivity in ulcerative and ischaemic colitis
As indicated in figure 1 and table 1, the expression of MUC17 was significantly decreased in tissues with colonic inflammation (both in ulcerative (n=25, p<0.0001) and ischaemic colitis (n=7, p=0.003)). There was no significant difference of MUC17 expression between ulcerative and ischaemic colitis patients (p=0.1). In contrast with normal colon specimens, the expression of MUC17 in ulcerative and ischaemic colitis specimens was markedly diminished, with only sparse staining in the regenerating columnar cells on the surface colon, and loss of expression in cells within the crypts (data not shown). The loss of MUC17 staining was similar in specimens with dysplasia and specimens without dysplasia.
Table 1.
Comparison of the mean composite scores in normal, ulcerative colitis and ischaemic colitis tissue samples
| Type of tissue | n | Staining score (mean±SE) | t test |
|---|---|---|---|
| Normal (N) | 12 | 6.67±0.86 | N/UC, p<0.0001 |
| Ulcerative colitis (UC) | 25 | 1.56±0.55 | N/IC, p=0.003 |
| Ischaemic colitis (IC) | 7 | 1.57±1.13 | N/UC+IC, p<0.0001 |
| UC/IC, p=0.1 |
MUC17 immunoreactivity in hyperplastic and neoplastic colon specimens
In adenoma tissues, MUC17 expression was significantly decreased compared to normal tissues (figure 1 and table 2). The loss of expression was uniform in adenomas with tubular and tubulovillous histology, in both small (≤1 cm) and large (>1 cm) adenomas, and in adenomas with and without foci of high grade dysplasia (data not shown). When present, focal supranuclear MUC17 staining occurred at the base of the polyp near the transition from normal to adenoma tissue (figure 2). MUC17 expression was rare in colon adenocarcinomas compared with normal colon (n=10, p<0.0001), and expression did not differ between adenoma and adenocarcinoma tissues (p=0.2), and between colitis and adenoma or adenocarcinoma tissues (table 2). We also observed that in transitional mucosa adjacent to adenocarcinomas, MUC17 expression was variable, equal to or less than that of normal mucosa, but occasionally completely absent. MUC17 expression in adenoma and adenocarcinoma tissues was focal and localised to a supranuclear pattern of staining, as observed in normal colon tissues (figure 2). Interestingly, MUC17 expression was also largely absent in the surface and crypts of hyperplastic polyps (figure 1 and table 2).
Table 2.
Comparison of the mean composite scores in normal, hyperplastic and neoplastic colon tissue samples
| Type of tissue | n | Staining score (mean±SE) | t test |
|---|---|---|---|
| Normal (N) | 12 | 6.7±0.86 | HP/C, p=0.89 |
| Colon cancer (CC) | 10 | 1.4±0.52 | T/C, p=0.2 |
| Hyperplastic polyp (HP) | 8 | 1.25±0.85 | TV/C, p=0.42 |
| Tubular adenoma (T) | 10 | 2.9±1 | CC/N, p<0.0001 |
| Tubulovillous adenomas (TV) | 34 | 0.9±0.4 | HP/N, p=0.0003 |
| T/N, p=0.01 | |||
| TV/N, p<0.0001 |
MUC17 expression in colon cancer cell lines
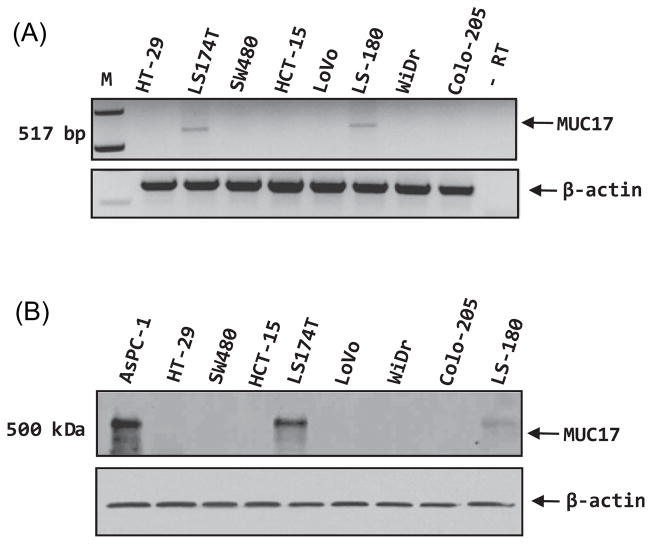
The expression of MUC17 was examined at the transcript level in eight different colon cancer cell lines using RT-PCR. None of the cell lines, except LS174Tand LS180, have a detectable level of MUC17 expression (figure 3A). A similar finding was also found in the western blot analysis (figure 3B).
Figure 3.
(A) RT-PCR analysis of MUC17 expression in various colon cancer cell lines. Actin was used as an internal control. Except for LS174T and LS-180 cells, none of the colon cancer cells have a detectable expression of MUC17. (B) Protein lysates from all the colon cancer cell lines and the AsPC-1 pancreatic cancer cell line (as a positive control), were resolved on 2% agarose gel. The MUC17 protein was only detected in LS174T and LS-180 cells. β-actin was used as a loading control.
DISCUSSION
The present study examined the pattern of expression of MUC17 in the normal colon and compared it with that in inflamed and neoplastic colonic tissues. In the normal colon, a prominent MUC17 expression was found on the surface and crypt columnar epithelial cells. In contrast, the expression of MUC17 was significantly decreased in colonic epithelial samples with associated inflammation. Similarly, MUC17 expression was also decreased in hyperplastic, tubular and tubulovillous adenomas, and in colonic adenocarcinoma. These results indicate that MUC17 was highly expressed on the luminal side of the colonic epithelial cells under normal conditions, but was markedly reduced with the development of colonic inflammation and early in the transition to hyperplastic and neoplastic states (figure 4). The decrease of MUC17 expression in both ulcerative and ischaemic colitis indicates that irrespective of the cause of inflammation, one or more inflammation-related pathways common to both conditions that negatively regulate the expression of this mucin are activated. Involvement of inflammatory signalling events in the regulation of MUC17 expression has been described,23 however further studies are warranted. Furthermore, the absence of a difference in MUC17 expression between adenomatous polyps (both tubular and tubulovillous adenomas) and colon cancer suggests that the loss of MUC17 may be an early event in colon cancer development. Additionally, the absence of a detectable MUC17 expression in the majority of colon cancer cell lines corroborates the immunohistochemical analysis data that MUC17 expression is strongly down-regulated in colon cancer tissues.
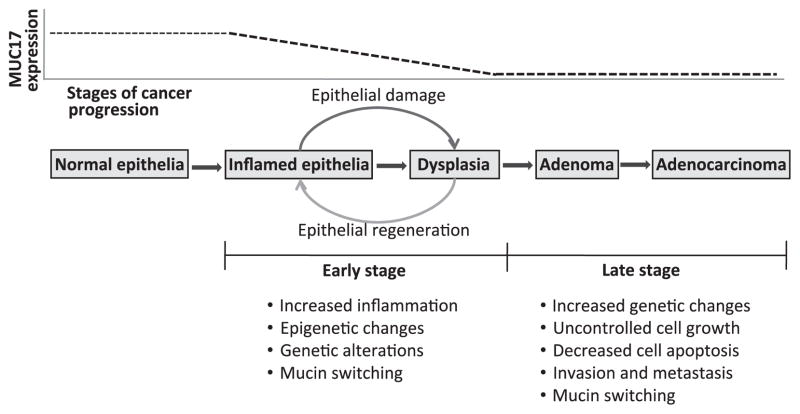
Figure 4.
Schematic representations of different stages of colitis-associated colon cancer progression, and the status of MUC17 expression in those stages. Colon carcinogenesis mediated trough different colonic inflammatory conditions is a consequence of sequential molecular changes. Inflamed colonic mucosa harbours many altered cancer associated genetic/molecular changes even before any histological evidence of dysplasia or cancer is detected. Among those changes, alteration in the normal expression of mucin (mucin switching) may have functional significance in colitis-associated colon cancer progression. The present study showed that normal colonic MUC17 expression steadily goes down in inflammatory conditions. However, in further advanced stages of cancer progression, MUC17 expression does not differ significantly from inflammatory conditions.
Previous studies have shown that MUC17 is primarily expressed in intestinal tissues, with the highest levels found in the duodenum and the transverse colon (85% of the level detected in duodenum) and lower expression in the stomach and fetal kidney.17 In-situ hybridisation showed that MUC17 is expressed predominantly in the apical region of the absorptive villi, but it was barely detectable in the immature cells of the crypts, while no expression was detected in the goblet cells.17 Therefore, MUC17 is localised predominantly to the mature, absorptive cells of intestinal epithelium.17 Other studies have shown that the MUC17 protein is expressed in the stomach, small intestine, colon and pancreas ductal cells.18, 23 The specific function of MUC17 is unknown, but recombinant protein corresponding to the cysteine-rich EGF-like domains of mouse Muc3 and human MUC17 have been shown to inhibit apoptosis and stimulate cell migration in cell culture models.19, 20 In addition, stable transfection of small hairpin RNA targeting MUC17 in colonic cell lines resulted in reduced cell aggregation, reduced cell–cell adherence and migration, and increased apoptosis.19 These data suggest that Muc3/MUC17 plays a role in mucosal protection related to cellular restitution.
The primary structure of MUC17 suggests that it is a type 1 membrane bound protein. However, in the present study, MUC17 expression was observed in the supranuclear region of colonic epithelial cells, suggesting one or more of the following possibilities: (a) the anti-MUC17 antibody that is used in the present study was generated by using a synthetic peptide against the TR region, therefore this antibody may be only detecting the un/under-glycosylated precursor forms of the MUC17 present in the early compartment of Golgi; or (b) a possible heavy glycosylation of the mature form of MUC17 (membrane bound), particularly in colonic cells may block the epitope for the antibody. To address these issues, our ongoing efforts to generate more antibodies to different unglycosylated regions of MUC17 will be very instrumental. Confirming a reduction of MUC17 gene expression in cancer was the finding that MUC17 protein and RNA were only detected in two of eight colon cancer cell lines, LS174T and LS-180. Further studies of the mechanism of reduced MUC17 expression in human colonic diseases are needed. In this regard, a computational study by Moehle et al showed that certain cis elements are over-represented and can effectively distinguish the promoter regions of mucin genes from the promoters of other genes whose expression is not changed in inflammatory bowel disease.23 Most strikingly, NFκB binding sites were identified in all mucin promoters.23 Recently, our database analysis of the MUC12-MUC17 intergenic region and upstream sequence (encompassing MUC12 intron 11 and exon 12) showed the presence of potential NFκB, SMAD3 and SMAD4 binding sites (data not shown). Jono et al have shown a cooperation of NFκB and TGFβ signalling in the regulation of MUC2 in non-typeable Haemophilus influenzae infection.24 Therefore, in future we will be confirming the MUC17 expression pattern in the presence of different inflammatory cytokines and bacterial components in vitro.
In summary, our findings indicate that the expression of MUC17, a membrane-bound mucin, is lost in both inflammatory and early and late neoplastic conditions of the colon. Due to the importance of MUC17 in processes related to cellular restitution, further research is needed to determine the mechanisms of loss of this glycoprotein, and to elucidate if a reduction of MUC17 expression plays a specific role in the initiation or prolongation of inflammation and neoplastic transformation in the colon.
Take-home messages.
MUC17 expression is primarily or secondarily diminished in inflammatory and neoplastic conditions of the colon.
MUC17 may play a role in colon-mucosal protection.
Inflammatory signals may regulate MUC17 expression in the colon.
Acknowledgments
Funding This work was supported by NIH grant CA78590, CA111294, CA133774, CA131944, Department of Defense PC081409 (SKB), a Veterans Affairs Merit Review Award (SBH), NIH Center grant DK080506 (SBH), and the Veterans Affairs Research Service.
Footnotes
Competing interests None.
Ethics approval This study was conducted with the approval of the Department of Medicine, University of California, San Diego and VA San Diego Healthcare System, San Diego, California, USA.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Dekker J, Rossen JW, Buller HA, et al. The MUC family: an obituary. Trends Biochem Sci. 2002;27:126–31. doi: 10.1016/s0968-0004(01)02052-7. [DOI] [PubMed] [Google Scholar]
- 2.Gum JR, Byrd JC, Hicks JW, et al. Molecular cloning of human intestinal mucin cDNAs. Sequence analysis and evidence for genetic polymorphism. J Biol Chem. 1989;264:6480–7. [PubMed] [Google Scholar]
- 3.Gum JR, Hicks JW, Swallow DM, et al. Molecular cloning of cDNAs derived from a novel human intestinal mucin gene. Biochem Biophys Res Commun. 1990;171:407–15. doi: 10.1016/0006-291x(90)91408-k. [DOI] [PubMed] [Google Scholar]
- 4.Lan MS, Batra SK, Qi WN, et al. Cloning and sequencing of a human pancreatic tumor mucin cDNA. J Biol Chem. 1990;265:15294–9. [PubMed] [Google Scholar]
- 5.Porchet N, Dufosse J, Audie JP, et al. Structural features of the core proteins of human airway mucins ascertained by cDNA cloning. Am Rev Respir Dis. 1991;144:S15–18. doi: 10.1164/ajrccm/144.3_pt_2.S15. [DOI] [PubMed] [Google Scholar]
- 6.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 7.Senapati S, Das S, Batra SK. Mucin-interacting proteins: from function to therapeutics. Trends Biochem Sci. 2010;35:236–45. doi: 10.1016/j.tibs.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol. 1998;161:5733–44. [PubMed] [Google Scholar]
- 9.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 10.Raouf AH, Tsai HH, Parker N, et al. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83:623–6. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- 11.Tytgat KM, van der Wal JW, Einerhand AW, et al. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem Biophys Res Commun. 1996;224:397–405. doi: 10.1006/bbrc.1996.1039. [DOI] [PubMed] [Google Scholar]
- 12.Van der SM, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Satsangi J, Parkes M, Louis E, et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 14.Satsangi J, Jewell DP, Bell JI. The genetics of inflammatory bowel disease. Gut. 1997;40:572–4. doi: 10.1136/gut.40.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyo K, Muto T, Nagawa H, et al. Associations of distinct variants of the intestinal mucin gene MUC3A with ulcerative colitis and Crohn’s disease. J Hum Genet. 2001;46:5–20. doi: 10.1007/s100380170118. [DOI] [PubMed] [Google Scholar]
- 16.Biemer-Huttmann AE, Walsh MD, McGuckin MA, et al. Immunohistochemical staining patterns of MUC1, MUC2, MUC4, and MUC5AC mucins in hyperplastic polyps, serrated adenomas, and traditional adenomas of the colorectum. J Histochem Cytochem. 1999;47:1039–48. doi: 10.1177/002215549904700808. [DOI] [PubMed] [Google Scholar]
- 17.Gum JR, Jr, Crawley SC, Hicks JW, et al. MUC17, a novel membrane-tethered mucin. Biochem Biophys Res Commun. 2002;291:466–75. doi: 10.1006/bbrc.2002.6475. [DOI] [PubMed] [Google Scholar]
- 18.Moniaux N, Junker WM, Singh AP, et al. Characterization of human mucin MUC17. Complete coding sequence and organization. J Biol Chem. 2006;281:23676–85. doi: 10.1074/jbc.M600302200. [DOI] [PubMed] [Google Scholar]
- 19.Luu Y, Junker W, Rachagani S, et al. Human intestinal MUC17 mucin augments intestinal cell restitution and enhances healing of experimental colitis. Int J Biochem Cell Biol. 2010;42:996–1006. doi: 10.1016/j.biocel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho SB, Dvorak LA, Moor RE, et al. Cysteine-rich domains of muc3 intestinal mucin promote cell migration, inhibit apoptosis, and accelerate wound healing. Gastroenterology. 2006;131:1501–17. doi: 10.1053/j.gastro.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Singh AP, Chauhan SC, Bafna S, et al. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate. 2006;66:421–9. doi: 10.1002/pros.20372. [DOI] [PubMed] [Google Scholar]
- 22.Moniaux N, Chakraborty S, Yalniz M, et al. Early diagnosis of pancreatic cancer: neutrophil gelatinase-associated lipocalin as a marker of pancreatic intraepithelial neoplasia. Br J Cancer. 2008;98:1540–7. doi: 10.1038/sj.bjc.6604329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moehle C, Ackermann N, Langmann T, et al. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med. 2006;84:1055–66. doi: 10.1007/s00109-006-0100-2. [DOI] [PubMed] [Google Scholar]
- 24.Jono H, Shuto T, Xu H, et al. Transforming growth factor-beta -Smad signaling pathway cooperates with NF-kappa B to mediate nontypeable Haemophilus influenzae-induced MUC2 mucin transcription. J Biol Chem. 2002;277:45547–57. doi: 10.1074/jbc.M206883200. [DOI] [PubMed] [Google Scholar]