Abstract
With recent interest in seeking new biologically inspired device-fabrication methods in nanotechnology, a new biological approach was examined to fabricate Cu nanotubes by using sequenced histidine-rich peptide nanotubes as templates. The sequenced histidine-rich peptide molecules were assembled as nanotubes, and the biological recognition of the specific sequence toward Cu lead to efficient Cu coating on the nanotubes. Cu nanocrystals were uniformly coated on the histidine-incorporated nanotubes with high packing density. In addition, the diameter of Cu nanocrystal was controlled between 10 and 30 nm on the nanotube by controlling the conformation of histidine-rich peptide by means of pH changes. Those nanotubes showed significant change in electronic structure by varying the nanocrystal diameter; therefore, this system may be developed to a conductivity-tunable building block for microelectronics and biological sensors. This simple biomineralization method can be applied to fabricate various metallic and semiconductor nanotubes with peptides whose sequences are known to mineralize specific ions.
The size and shape of nanocrystals have significant effects on catalytic, optical, and electronic properties (1–10). To apply nanocrystals as building blocks for practical electronic, magnetic, and optical devices, nanocrystals must be assembled. Although various nanocrystals have been assembled on flat substrates (11–17), the self-assembly of nanocrystals on cylindrical nanotube surfaces has also been reported recently (18–25). When the coupling strength of overlapped wave functions between neighboring nanocrystals is tuned by compressing the lattice of nanocrystals, the range of tuning the electronic structures is considerable because the overlap depends exponentially on the interdot distance and the size (26, 27). If nanocrystals can be formed on nanotube geometry in controlled diameters and packing densities, one may be able to produce nanotubes with tunable electronic properties from one type of nanocrystal. Although this type of material is expected to serve as a smart building block to interconnect nanometer-sized electronic components in microelectronics and biological/chemical sensors, the controls of nanocrystal size and packing density are not straightforward on nanotubes.
Biological systems control mineralizations and nanocrystal synthesis of various metals in exact shapes and sizes with high reproducibility and accuracy (28–33). Therefore, it is logical to use biological nanotubes as templates on which to grow monodisperse nanocrystals by biomineralization (20, 23, 34–38). Complexation of metals and histidine-containing peptides has been studied extensively because their high affinities to metal ions damage central nervous systems by altering peptide conformations into abnormal forms, and this protein deformation may cause Parkinson's disease and Alzheimer's disease (39). Therefore, a nanotube form of the sequenced histidine-rich peptides has potential to serve as an efficient template for metallic nanotube synthesis, because the specific sequences of peptides mineralize specific metals/semiconductors to produce highly crystalline nanocrystals (32, 40–43). In addition, the peptide conformations and charges on nanotubes, which can be controlled by experimental conditions such as pH, ion concentration, and temperature, determine the size and packing density of nanocrystals (23, 40, 44–46). Therefore, the size and packing density of nanocrystals on histidine-rich peptide nanotube surfaces are potentially controlled by simply tuning those experimental conditions. Because metallic nanocrystals in diameters <10 nm are in the size domain to observe a significant conductivity change by tweaking the nanocrystal size (11, 47, 48), this system may be developed to a conductivity-tunable nanotube-building block.
By applying this principle, we have mineralized Au nanocrystals on the sequenced peptide [Ala-His-His-Ala-His-His-Ala-Ala-Asp (HRE)]-functionalized nanotubes, and the packing density of Au nanocrystals was controlled by pH of the growth solution (23). Although the charge-distribution change of the sequenced HRE peptides by pH change controlled the Au nanocrystal-packing density, the Au nanocrystal size was constant over various pH values, because the conformation of HRE peptide could not be altered by pH change because of the stiffness of HRE peptide backbone (49).
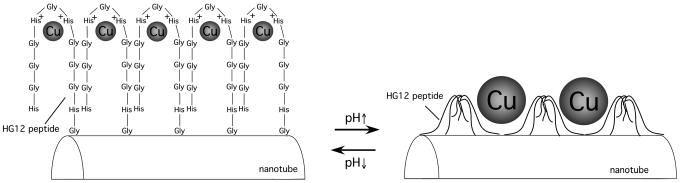
Here we report Cu nanocrystal growth on the nanotubes functionalized by a histidine-rich peptide, whose sequence is His-Gly-Gly-Gly-His-Gly-His-Gly-Gly-Gly-His-Gly (HG12). The fabrication process is illustrated in Fig. 1. In brief, the sequenced HG12 peptides were immobilized onto amide groups of the template nanotubes, self-assembled from bolaamphiphile peptide monomers (50), by hydrogen bonding (Fig. 1a) (51). Then, the HG12 peptides coordinate Cu(II) as the nucleation site (Fig. 1b) for further Cu nanocrystal growth by reduction of trapped ions (Fig. 1c). This peptide sequence has been reported to fold into multiple conformations by coordinating Cu ions at various positions, dependent on pH values (39). Therefore, the HG 12 peptide, immobilized on the nanotube surfaces, has potential to control the size of Cu nanocrystals due to the flexible conformations by way of pH change. Indeed, we observed that the pH change altered the HG12 peptide conformation and the grown Cu nanocrystal size on the nanotube, shown by Fourier transform IR spectroscopy and transmission electron microscopy (TEM). Although the self-assembly of Cu nanocrystals on flat substrates has been studied extensively for microelectronic, sensor, and catalytic applications (7, 52, 53), this study is a previously uninvestigated example of growing monodisperse and isotropic Cu nanocrystals on the nanotube surfaces by biomineralization.
Fig. 1.
Scheme of the Cu nanotube fabrication. (a) Immobilization of the sequenced HG12 peptide at the amide-binding sites of the template nanotubes. (b) The Cu ion–HG12 peptide complexation on the nanotube surfaces. (c) Cu nanocrystal growth on the nanotubes nucleated at Cu ion-binding sites after reducing trapped Cu ions with NaBH4.
Materials and Methods
To prepare templates for the Cu nanocrystal coating, bis(N-α-amido-glycylglycine)-1,7-heptane dicarboxylate molecules (10 mM) were self-assembled into nanotubes in a citric acid/ NaOH (pH 5.5) solution. Details of this bolaamphiphile peptide monomer synthesis and the nanotube self-assembly have been described (50, 54, 55). The HG12 peptide was sequenced by Applied Biosystems Peptide Synthesizer 432A and purified by using Beckman 110 HPLC equipped with a C-18 reverse-phase chromatographic column at the Center for Study of Gene Structure and Function, City University of New York. To immobilize the HG12 peptide on the template nanotubes, 50 μl of the HG12 peptide solution in buffer was incubated to 100 μl of the nanotube solution, and the HG12 peptides were bound on amide sites of the nanotube surfaces by means of hydrogen bonding (Fig. 1a). The reaction mixture was then stirred slowly for 24 h, the coated nanotubes were washed with deionized water to remove any unbound HG12 peptides, and then centrifuged for 30 min (14,500 rpm) to collect the HG12 peptide-coated nanotubes. The HG12 peptide coating on the nanotubes was confirmed by using a Raman microscope and atomic force microscope (23). To grow Cu nanocrystals on the HG12 peptide nanotubes, 50 μl of 50 mM CuCl2 solution was added to the peptide nanotube solution to form Cu(II)–HG12 complexes on the nanotubes to create Cu nanocrystal nucleation sites (Fig. 1b). The concentration of Cu(II) was maintained at a metal-to-ligand ratio of 1:1 [0.0012 mM Cu(II)], and the pH was varied between 4 and 10 to study the pH effect on Cu nanocrystal growth. In all cases, the reaction mixtures were allowed to sit undisturbed overnight under nitrogen to complete Cu-ion immobilization on the nanotubes. The Cu(II) on the peptide nanotubes was then reduced by 50 μmol of NaBH4 to produce Cu nanocrystals (Fig. 1c). This reduction step was carried out under nitrogen, and the resulting solutions were aged for 24 h after reduction with sodium boronhydride. After 24 h, the nanotubes were washed with nanopure water and then centrifuged twice to remove the excess reducing agent and nanocrystals that were not coated on the nanotubes. The nanotube solutions (3–5 μl) were then dropped on TEM grids for further analysis by TEM (model JEOL 1200 EX). Two sets of control experiments were carried out as follows. In the first set, Cu nanocrystals were grown directly on the neat nanotubes at pH 6 with no HG12 peptides coating on the nanotubes. The experimental condition was the same as above except that no HG12 was coated on the nanotubes before incubating Cu ions. In the second set, 50 μl of 50 mM CuCl2 solution was incubated with 50 μl of the peptides in buffer solutions of pH 6 and pH 8 under nitrogen in the absence of the nanotubes. These solutions were then reduced by sodium boronhydride and washed in the same procedure as above. These samples were analyzed by TEM.
Results and Discussion
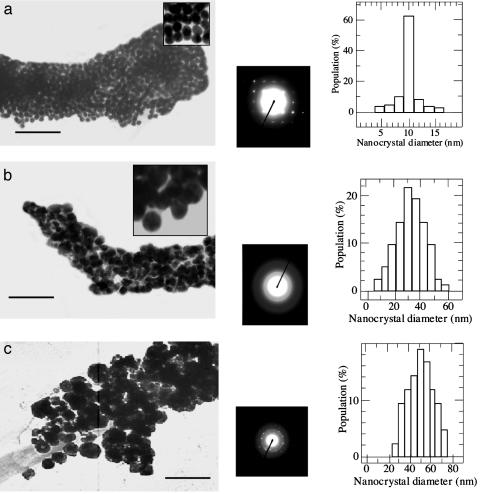
Because the charge distribution of histidine changes dramatically at about pH 6 (56), the HG12 is expected to undergo a significant conformation change below and above pH 6. When Cu nanocrystals were grown on the HG12 peptide nanotubes between pH 4 and pH 6, the Cu nanocrystals were monodisperse and packed in high density as shown in the TEM image in Fig. 2a Left. Inset, a magnified TEM image, shows the isotropic Cu nanocrystal shape at pH 6. In this pH range, the Cu nanocrystals were grown in an average diameter of 10 nm (Fig. 2a Right), determined by the TEM images. The electron-diffraction pattern of Cu nanocrystals on the nanotubes shows (111) and (220) planes for a face-centered-cubic crystal (Fig. 2a Center). The diffraction pattern of copper oxide was not observed in Fig. 2a, and the HG12 peptide seems to protect the Cu nanocrystals on the nanotubes from oxidation (52, 53).
Fig. 2.
(a) Cu nanocrystals grown on the nanotube at pH 6. (Left) TEM image. (Center) Electron-diffraction pattern. (Right) Size distribution. (Inset) The TEM image in higher magnification. (b) Cu nanocrystals grown on the nanotube at pH 8. (Left) TEM image. (Center) Electron-diffraction pattern. (Right) Size distribution. (Inset) The TEM image in higher magnification. (c) Cu nanocrystals grown on the nanotube without the HG12 peptide at pH 6. (Left) TEM image. (Center) Electron-diffraction pattern. (Right) Size distribution. (Scale bar = 100 nm.)
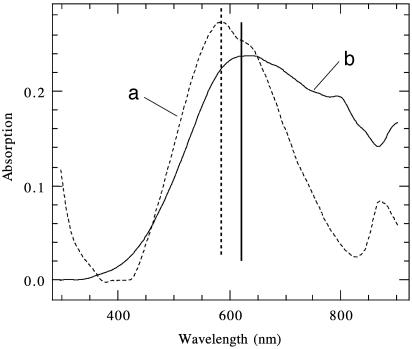
A striking difference in Cu nanocrystal growth was observed when the HG12 peptide nanotubes with Cu(II) were reduced between pH 7 and pH 10. In this pH range, the average diameter of Cu nanocrystals was increased to 30 nm (Fig. 2b Left). Although the statistical size distribution of Cu nanocrystals in the basic condition is fairly monodisperse in general (Fig. 2b Right), those Cu nanocrystals were less monodisperse than the Cu nanocrystals grown in the pH range between 4 and 6. The electron-diffraction pattern of Cu nanocrystals grown on the HG 12 peptide nanotubes at pH 8 (Fig. 2b Center) shows that the crystallinity of those Cu nanocrystals is equivalent to the Cu nanocrystals in the acidic condition except for the orientation of nanocrystals, which is more aligned in the acidic condition. The significant difference of electronic structures between the 10-nm Cu nanocrystal-coated nanotube and the 30-nm Cu nanocrystal-coated nanotube was also observed in UV-visible absorption spectra (Fig. 3). The absorption maximum of the 30-nm Cu nanocrystal-coated nanotube, 620 nm (Fig. 3b), was shifted to 585 nm for the 10-nm Cu nanocrystal-coated nanotube (Fig. 3a). This comparison indicates that the Cu nanocrystal size on the nanotubes, controlled by the pH change, alters the electronic property of nanotube.
Fig. 3.
UV-visible spectra of the nanotubes coated with Cu nanocrystals in a diameter of 10 nm, grown in pH 6 solution (dotted line) (a) and Cu nanocrystals in a diameter of 30 nm, grown in pH 8 solution (solid line) (b).
To confirm that the HG12 peptide plays a role in controlling the size of Cu nanocrystals on the nanotube, we examined a control experiment to grow Cu nanocrystals on the template nanotubes without the HG12 peptide at pH 6 in the same procedure described above. The Cu nanocrystals were grown in much larger sizes, 50 nm in an average diameter, and they were polydisperse, as shown in Fig. 2c Left and Right. The Cu nanocrystals grown without the HG12 peptide are less oriented, as shown in the diffraction pattern (Fig. 2c Center). The comparison between Fig. 2 a and c indicates that the HG12 peptide on the nanotube certainly regulates the size and the monodispersity of Cu nanocrystals.
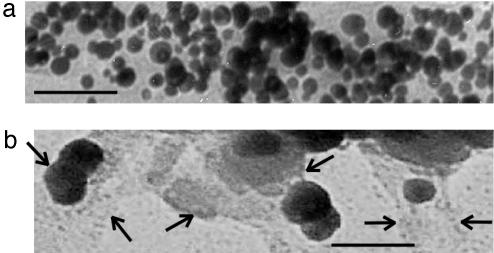
Another control experiment was examined to confirm the role of template nanotubes in the Cu nanocrystal growth. This time, we produced Cu nanocrystals in the HG12 peptide solution without the template nanotubes. The TEM image in Fig. 4a shows Cu nanocrystals grown in the HG12 peptide solution at pH 6. The average diameter of Cu nanocrystals was 14 nm grown in the pH 6 growth solution, which is similar to the size of Cu nanocrystals formed on the HG12 peptide nanotubes. But the particle-size distribution of Fig. 4a is more polydisperse than the one observed on the HG12 peptide nanotubes. This comparison indicates that the monodispersity of Cu nanocrystals is regulated by the template nanotubes. When we reported Au nanocrystal growth on the sequenced HRE peptide-coated nanotubes, the regular spacing of the HRE peptide binding on the nanotube was observed to be crucial to control the monodispersity and size of Au nanocrystals (49). Therefore, it is reasonable that the monodispersity of Cu nanocrystals on the HG12 peptide nanotubes in the lower pH range is also regulated with the same growth mechanism that Cu nanocrystals are grown between the regularly spaced HG12 peptide chains on the nanotubes. When the same nonnanotube experiment was examined at pH = 8, Cu nanocrystals were grown in an average diameter of 50 nm (Fig. 4b), which shows the trend similar to the Cu nanocrystal size observed on the HG12 peptide nanotubes. In this basic condition, the HG12 peptides were found to aggregate, shown between arrows in Fig. 4b. This observation suggests that the HG12 peptides also aggregate on the nanotubes to induce the larger Cu nanocrystal growth in the higher pH range.
Fig. 4.
TEM images of the Cu nanocrystals grown in the HG12 peptide solution without the template nanotubes at pH 6 (a) and at pH 8 (b). Arrows show the edges of aggregated HG12 peptides. (Scale bar = 100 nm.)
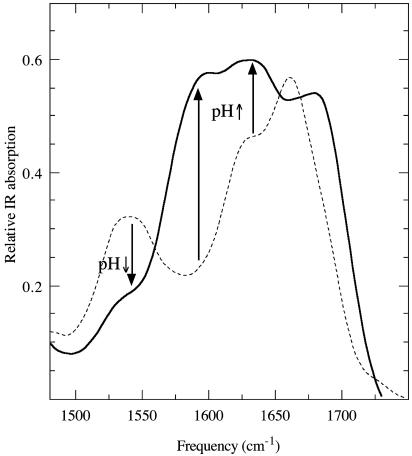
When Au nanocrystals were grown on the sequenced HRE peptide-coated nanotubes, the Au nanocrystal size was unchangeable with pH change because of the rigidity of HRE peptide backbone (49). Because the size of Cu nanocrystals was changed on the HG12 peptide nanotubes by pH change, it is likely that the HG12 peptide–Cu(II) complex undergoes the conformation change on the nanotubes between the lower pH range and the higher pH range. This conformation change is supported by the report that the free HG12 peptide molecules in aqueous solution formed complexes with Cu(II) and folded into the compact form by means of ligation between histidines and Cu(II) in the lower pH range, whereas the HG12 peptide–Cu(II) complexes were aggregated in the higher pH range (39, 57). To confirm that this peptide conformation change by pH change also occurs on the nanotubes, Fourier transform IR spectra of the HG12 peptide–Cu(II) complexes on the nanotubes at pH 6 and pH 8 were compared in Fig. 5. The glycine amide I mode of the N(Gly)–Cu(II)–N(His) complex at 1,657 cm–1 in pH 6 (Fig. 5, dotted line) is blue-shifted to 1,674 cm–1 in pH 8 (Fig. 5, solid line), indicating that fewer Cu ions bind the glycine amides in the basic condition (58). The 1,625 cm–1 peak is the amide vibration of β-sheet conformation of peptide backbone aggregation by hydrogen bonding (57, 60), and the increase of this IR intensity by increasing pH indicates that the HG12 peptide chains on the nanotubes were aggregated in the higher pH range. The increase of IR intensity at a 1,604 cm–1 peak by pH increase is due to the formation of intermolecular N(His)–Cu(II)–N(His) bonds between the peptide chains by means of the aggregation (57, 58). This observation is consistent with the loss of IR intensity at a 1,550 cm–1 peak, the imidazole side chain vibrational mode of histidine without the Cu(II) ligation, by way of pH increase (61). Although this vibrational analysis does not reveal the complete Cu growth mechanism vs. pH change on the nanotubes, the combination of this analysis and the control experiments in Fig. 4 strongly supports the hypothesis that the peptide aggregation is one of the major factors controlling the size of Cu nanocrystal. This hypothesis is summarized in Fig. 6. In the lower pH range, Cu(II) is coordinated with histidines and glycines to fold the HG12 peptides on the nanotubes, and Cu nanocrystals nucleate on the folded peptide chains (Fig. 6 Left). This folded conformation of the HG12 peptide–Cu(II) complex is adapted from the complex conformation observed in the free suspension (39, 57). The compacted HG12 peptide chain by folding may also contribute reduction of the aggregation between the regularly spaced peptide chains on the nanotubes. In the higher pH range, intermolecular His–Cu(II)–His bonds are formed between the neighboring aggregated HG12 peptides on the nanotube to nucleate Cu nanocrystals between the HG12 peptide aggregates (Fig. 6 Right). The peptide configuration in Fig. 6 Right may increase the space to grow Cu nanocrystals, which induces larger Cu nanocrystal growth in the basic condition. The degree of peptide chain aggregation seems not to be uniform on the entire nanotube surface, and the variation in the peptide chain aggregations likely induces the uneven HG12 peptide chain spacing for the Cu nanocrystal nucleation on the nanotubes, which makes Cu nanocrystals less monodisperse in the higher pH range.
Fig. 5.
IR spectra of the HG12 peptide–Cu(II) complexes on the nanotubes at pH 6 (dotted line) and pH 8 (solid line).
Fig. 6.
A proposed structure of the Cu nanocrystal–HG12 peptide complex on the template nanotube.
In conclusion, Cu nanocrystals can be mineralized in different sizes via the HG12 peptide conformation changes on the nanotubes, controlled by pH values. This system may be developed as smart nanotubes that can tune their electronic properties by simple environmental controls and applied as smart building blocks for microelectronic, sensor, and catalytic applications.
Acknowledgments
We thank Prof. K. Fath at Core Facilities for Imaging Cellular and Molecular Biology at Queens College, City University of New York, for the use of the transmission electron microscope and Mr. C. Matthaeus and Dr. M. Diem at Hunter College, City University of New York, for the assistance with Fourier transform IR studies. This work was supported by U.S. Department of Energy Grant DE-FG-02-01ER45935.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HRE, Ala-His-His-Ala-His-His-Ala-Ala-Asp; HG12, His-Gly-Gly-Gly-His-Gly-His-Gly-Gly-Gly-His-Gly; TEM, transmission electron microscope.
References
- 1.Puntes, V. F., Krishnan, K. M. & Alivisatos, A. P. (2001) Science 291, 2115–2117. [DOI] [PubMed] [Google Scholar]
- 2.Puntes, V. F., Zanchet, D., Erdonmez, C. K. & Alivisatos, A. P. (2002) J. Am. Chem. Soc. 124, 12874–12880. [DOI] [PubMed] [Google Scholar]
- 3.El-Sayed, M. A. (2001) Acc. Chem. Res. 34, 257–264. [DOI] [PubMed] [Google Scholar]
- 4.Sun, Y. & Xia, Y. (2002) Science 298, 2176–2179. [DOI] [PubMed] [Google Scholar]
- 5.Ertl, G. & Knozinger, H. (1997) Handbook of Heterogeneous Catalysis (Wiley, New York).
- 6.Wang, Z. L., Dai, Z. R. & Sun, S. H. (2000) Adv. Mater. 12, 1944–1946. [Google Scholar]
- 7.Hansen, P. L., Wagner, J. B., Helveg, S., Rostrup-Nielsen, J. R., Clausen, B. S. & Topsoe, H. (2002) Science 295, 2053–2055. [DOI] [PubMed] [Google Scholar]
- 8.Jin, R. C., Cao, Y. W., Mirkin, C. A., Kelly, K. L., Schatz, G. C. & Zheng, J. G. (2001) Science 294, 1901–1903. [DOI] [PubMed] [Google Scholar]
- 9.Gou, L. F. & Murphy, C. J. (2003) Nano Lett. 3, 231–234. [Google Scholar]
- 10.Filankembo, A. & Pileni, M. P. (2000) Appl. Surf. Sci. 164, 260–267. [Google Scholar]
- 11.Murray, C. B., Kagan, C. R. & Bawendi, M. G. (2000) Annu. Rev. Mater. 30, 545–610. [Google Scholar]
- 12.Whetten, R. L., Shafigullin, M. N., Khoury, J. T., Schaaff, T. G., Vezmar, I., Alvarez, M. M. & Wilkinson, A. (1999) Acc. Chem. Res. 32, 397–406. [Google Scholar]
- 13.Markovich, G., Collier, C. P., Henrichs, S. E., Remacle, F., Levine, R. D. & Heath, J. R. (1999) Acc. Chem. Res. 32, 415–423. [Google Scholar]
- 14.Korgel, B. A., Fullam, S., Connolly, S. & Fitzmaurice, D. (1998) J. Phys. Chem. B 102, 8379–8388. [Google Scholar]
- 15.Forster, S. & Antonietti, M. (1998) Adv. Mater. 10, 195–217. [Google Scholar]
- 16.Talapin, D. V., Schevchenko, E. V., Kornowski, A., Gaponik, N., Haase, M., Rogach, A. L. & Weller, H. (2001) Adv. Mater. 13, 1868–1871. [DOI] [PubMed] [Google Scholar]
- 17.Gittins, D. I., Susha, A. S., Schoeler, B. & Caruso, F. (2002) Adv. Mater. 14, 508–512. [Google Scholar]
- 18.Dujardin, E., Peet, C., Stubbs, G., Culver, J. N. & Mann, S. (2003) Nano Lett. 3, 413–417. [Google Scholar]
- 19.Demir, M. & Stowell, M. H. B. (2002) Nanotechnology 13, 541–544. [Google Scholar]
- 20.Behrens, S., Rahn, K., Habicht, W., Bohm, K.-J., Rosner, H., Dinjus, E. & Unger, E. (2002) Adv. Mater. 14, 1621–1625. [Google Scholar]
- 21.Jiang, K., Eitan, A., Schadler, L. S., Ajayan, P. M., Siegel, R. W., Grobert, N., Mayne, M., Reyes-Reyes, M., Terrones, H. & Terrones, M. (2003) Nano Lett. 3, 275–277. [Google Scholar]
- 22.Banerjee, S. & Wong, S. S. (2002) Nano Lett. 2, 195–200. [Google Scholar]
- 23.Djalali, R., Chen, Y.-f. & Matsui, H. (2002) J. Am. Chem. Soc. 124, 13660–13661. [DOI] [PubMed] [Google Scholar]
- 24.Matsui, H., Pan, S. & Douberly, G. E. J. (2001) J. Phys. Chem. B 105, 1683–1686. [Google Scholar]
- 25.Banerjee, I. A., Yu, L. & Matsui, H. (2003) Nano Lett. 3, 283–287. [Google Scholar]
- 26.Remacle, F. & Levine, R. D. (2000) J. Am. Chem. Soc. 122, 4084–4091. [Google Scholar]
- 27.Remacle, F., Beverly, K. C., Heath, J. R. & Levine, R. D. (2002) J. Phys. Chem. B 106, 4116–4126. [Google Scholar]
- 28.Baeuerlein, E. (2000) Biomineralization (Wiley, New York).
- 29.Mann, S. (2001) Biomineralization (Oxford Univ. Press, New York).
- 30.Niemeyer, C. M. (2001) Angew. Chem. Int. Ed. Engl. 40, 4128–4158. [DOI] [PubMed] [Google Scholar]
- 31.Dujardin, E. & Mann, S. (2002) Adv. Mater. 14, 775–788. [Google Scholar]
- 32.Slocik, J. M., Moore, J. T. & Wright, D. W. (2002) Nano Lett. 2, 169–173. [Google Scholar]
- 33.Gardea-Torresdey, J. L., Parsons, J. G., Gomez, E., Peralta-Videa, J., Troiani, H. E., Santiago, P. & Yacaman, M. J. (2002) Nano Lett. 2, 397–401. [Google Scholar]
- 34.Douglas, T. & Young, M. (1999) Adv. Mater. 11, 679–681. [Google Scholar]
- 35.Shenton, W., Douglas, T., Young, M., Stubbs, G. & Mann, S. (1999) Adv. Mater. 11, 253–256. [Google Scholar]
- 36.Lee, S. W., Mao, C. B., Flynn, C. E. & Belcher, A. M. (2002) Science 296, 892–895. [DOI] [PubMed] [Google Scholar]
- 37.Hartgerink, J. D., Beniash, E. & Stupp, S. I. (2001) Science 294, 1684–1688. [DOI] [PubMed] [Google Scholar]
- 38.Field, M., Smith, C. J., Awshalom, D. D., Mayes, E. L., Davis, S. A. & Mann, S. (1998) Appl. Phys. Lett. 73, 1739–1741. [Google Scholar]
- 39.Pappalardo, G., Impellizzeri, G., Bonomo, R. P., Campagna, T., Grasso, G. & Saita, M. G. (2002) New J. Chem. 26, 593–600. [Google Scholar]
- 40.Whitling, J. M., Spreitzer, G. & Wright, D. W. (2000) Adv. Mater. 12, 1377–1380. [Google Scholar]
- 41.Seeman, N. C. & Belcher, A. M. (2002) Proc. Natl. Acad. Sci. USA 99, 6451–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naik, R. R., Stringer, S. J., Agarwal, G., Jones, S. E. & Stone, M. O. (2002) Nat. Mater. 1, 169–172. [DOI] [PubMed] [Google Scholar]
- 43.Brown, S., Sarikaya, M. & Johnson, E. (2000) J. Mol. Biol. 299, 725–735. [DOI] [PubMed] [Google Scholar]
- 44.Whaley, S. R., English, D. S., Hu, E. L., Barbara, P. F. & Belcher, A. M. (2000) Nature 405, 665–668. [DOI] [PubMed] [Google Scholar]
- 45.Ziegler, J., Chang, R. T. & Wright, D. W. (1999) J. Am. Chem. Soc. 121, 2395–2400. [Google Scholar]
- 46.Spreitzer, G., Whitling, J. M., Madura, J. D. & Wright, D. W. (2000) Chem. Commun. 209–210.
- 47.Beverly, K. C., Sample, J. L., Sampaio, J. F., Remacle, F., Heath, J. R. & Levine, R. D. (2002) Proc. Natl. Acad. Sci. USA 99, 6456–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sample, J. L., Beverly, K. C., Chaudhari, P. R., Remacle, F., Heath, J. R. & Levine, R. D. (2002) Adv. Mater. 14, 124–128. [Google Scholar]
- 49.Djalali, R., Chen, Y.-f. & Matsui, H. (2003) J. Am. Chem. Soc. 125, 5873–5879. [DOI] [PubMed] [Google Scholar]
- 50.Matsui, H. & Gologan, B. (2000) J. Phys. Chem. B 104, 3383–3386. [Google Scholar]
- 51.Douberly, G. E., Jr., Pan, S., Walters, D. & Matsui, H. (2001) J. Phys. Chem. B 105, 7612–7618. [Google Scholar]
- 52.Pileni, M. P. (2001) Langmuir 17, 7476–7486. [Google Scholar]
- 53.Ziegler, K. J., Doty, R. C., Johnston, K. P. & Korgel, B. A. (2001) J. Am. Chem. Soc. 123, 7797–7803. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu, T., Kogiso, M. & Masuda, M. (1996) Nature 383, 487–488. [Google Scholar]
- 55.Kogiso, M., Ohnishi, S., Yase, K., Masuda, M. & Shimizu, T. (1998) Langmuir 14, 4978–4986. [Google Scholar]
- 56.Berg, J. M., Tymoczko, J. T. & Stryer, L. (2002) Biochemistry (Freeman, New York).
- 57.Casolaro, M., Chelli, M., Ginanneschi, M., Laschi, F., Muniz-Miranda, M., Papini, A. M. & Sbrana, G. (1999) Spectrochim. Acta A 55, 1675–1689. [Google Scholar]
- 58.Miura, T., Hori-i, A., Mototani, H. & Takeuchi, H. (1999) Biochemistry 38, 11560–11569. [DOI] [PubMed] [Google Scholar]
- 59.Arrondo, J. L. R., Blanco, F. J., Serrano, L. & Goni, F. M. (1996) FEBS Lett. 384, 35–37. [DOI] [PubMed] [Google Scholar]
- 60.Halverson, K. J., Sucholeiki, I., Ashburn, T. T. & Lansbury, P. T. (1991) J. Am. Chem. Soc. 113, 6701–6703. [Google Scholar]
- 61.Miura, T., Suzuki, K., Kohata, N. & Takeuchi, H. (2000) Biochemistry 39, 7024–7031. [DOI] [PubMed] [Google Scholar]